A TriPPPro-Nucleotide Reporter with Optimized Cell-Permeable Dyes for Metabolic Labeling of Cellular and Viral DNA in Living Cells
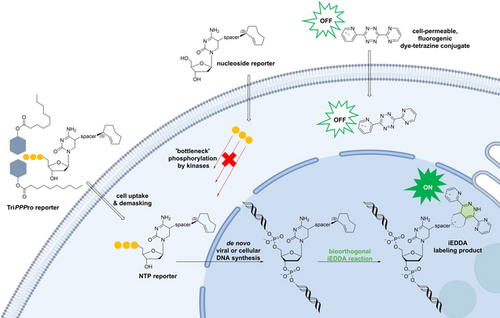
Graphical Abstract
The TriPPPro approach facilitates live-cell imaging of metabolically labeled cellular and viral DNA. The pronucleotide of a modified 2′-deoxycytidine triphosphate allows the incorporation of the fast bioorthogonal 2TCOa label belonging to the iEDDA repertoire. Subsequent staining of the tagged DNA with highly reactive dye-tetrazine conjugates within minutes to hours after dye addition enables the visualization of nucleic acids in real time.
Abstract
The metabolic labeling of nucleic acids in living cells is highly desirable to track the dynamics of nucleic acid metabolism in real-time and has the potential to provide novel insights into cellular biology as well as pathogen-host interactions. Catalyst-free inverse electron demand Diels–Alder reactions (iEDDA) with nucleosides carrying highly reactive moieties such as axial 2-trans-cyclooctene (2TCOa) would be an ideal tool to allow intracellular labeling of DNA. However, cellular kinase phosphorylation of the modified nucleosides is needed after cellular uptake as triphosphates are not membrane permeable. Unfortunately, the narrow substrate window of most endogenous kinases limits the use of highly reactive moieties. Here, we apply our TriPPPro (triphosphate pronucleotide) approach to directly deliver a highly reactive 2TCOa-modified 2′-deoxycytidine triphosphate reporter into living cells. We show that this nucleoside triphosphate is metabolically incorporated into de novo synthesized cellular and viral DNA and can be labeled with highly reactive and cell-permeable fluorescent dye-tetrazine conjugates via iEDDA to visualize DNA in living cells directly. Thus, we present the first comprehensive method for live-cell imaging of cellular and viral nucleic acids using a two-step labeling approach.
Introduction
Metabolic labeling of nucleic acids has been used since 1976, starting with 5-bromo-2′-deoxyuridine (BrdU).1 BrdU incorporates into newly synthesized DNA of dividing cells after being phosphorylated to its nucleoside triphosphate form (NTP) by endogenous kinases. The reporter can subsequently be detected with an anti-BrdU-antibody after fixation and denaturation of the cells to make the DNA accessible to the large antibodies.2 Three decades later, 5-ethynyl-2′-deoxyuridine (EdU) was developed to overcome the dependence on antibodies for detection.3 The small alkyne-functionality can react in a CuI-catalyzed [3+2] cycloaddition reaction with a small azide-sensor conjugate (copper-catalyzed azide-alkyne cycloaddition, CuAAC, k′=101–102 M−1s−1).4 Here, denaturation of the samples is not necessary anymore, but the fixation of the cells is still required due to the cytotoxicity of the catalyst. Moreover, the copper-containing buffer can quench fluorescent proteins, which makes it not ideal for many imaging approaches. A live-cell compatible click reaction would be a significant improvement and is the next critical step in furthering this technology. In 2014, Luedtke et al. described 5-vinyl-2′-deoxyuridine (VdU) as the first bioorthogonal reporter for cellular DNA, which made use of the catalyst-free inverse electron demand Diels–Alder reaction (iEDDA) with 1,2,4,5-tetrazines.5 The vinyl group is the smallest possible functional group used in iEDDA reactions, and VdU showed lower cytotoxicity than EdU. However, the vinyl group provides only slow reaction kinetics (k′ ≈10−2 M−1s−1), which initially required the denaturation of the samples and extensive washing steps to visualize VdU-containing DNA successfully. Later reports described a dual enhancement strategy utilizing a fluorogenic intercalating agent capable of undergoing iEDDA reactions with DNA containing VdU. Reversible high-affinity DNA-intercalation of a novel fluorogenic acridine-tetrazine agent increased the reaction rate of tetrazine-alkene iEDDA on duplex DNA by 60 000-fold (590 M−1s−1) and allowed for live-cell imaging of cellular nucleic acids.6
Recent studies have demonstrated that cellular polymerases can accept bulky modifications once the triphosphate is delivered into the cell. One study described a fluorescent TAMRA-dATP modified triphosphate, which spontaneously forms nanoparticles in Mg2+-containing buffers which are taken up via endocytosis into cells. Endosomal escape of the probe was then induced by photochemical activation using yellow laser light, which enabled subsequent incorporation into cellular DNA.7 Moreover, a recent study used SNTT1 to bring 2′-deoxycytidine based nucleoside triphosphates with reactive 2- or 4-ene trans-cyclooctene (TCOs) and bicyclononyne (BCN) groups into living cells.8
Here, we set out to develop tools to track de novo synthesized nucleic acids in real-time to study fundamental biological processes such as genome replication and pathogen-host cell interactions at the single particle level. To achieve this goal, multiple requirements need to be met. Ideal bioorthogonal reactions in vivo should be at least as fast as cellular processes (103–106 M−1s−1) to enable efficient labeling at physiological concentrations (μM to nM) and time scales.9 Hence, modified nucleotides should be used that carry a highly reactive moiety. Moreover, a live-cell compatible, cell-permeable, fast-reacting fluorescent dye is essential for an efficient bioorthogonal reaction in living cells that matches the reactivity of the incorporated reporter. Finally, the dye should produce low cellular background when not conjugated to the incorporated nucleotide reporter, ideally minimizing wash protocols that stress living systems. Strained trans-cyclooctenes (TCO), belonging to the iEDDA repertoire, provide sufficiently high reaction rates (k′=103–105 M−1s−1).10 Still, the motifs are sterically demanding and significantly alter the properties of the carrier nucleoside. Due to the narrow substrate window of most endogenous kinases, such modifications to the nucleobase are likely not accepted by these enzymes, resulting in inefficient phosphorylation to yield the active NTP.11 Consequently, incorporation of the reporter nucleotide by cellular or viral polymerases would be impeded. This obstacle is well-known in nucleoside drug discovery and fueled the development of pronucleotide concepts in the past decades.12 The main emphasis was on delivering nucleoside monophosphates (NMPs) into cells by masking the polar phosphate group to allow for diffusion across the cell membrane. In contrast, we recently reported the TriPPPro approach as the first nucleoside triphosphate delivery system, which is membrane permeable and independent of the action of cellular kinases.13 This approach is based on masking the terminal γ-phosphate group with two enzymatically cleavable lipophilic acyloxybenzyl (AB) groups. We applied this concept to directly deliver nucleoside triphosphates carrying a bulky reactive bioorthogonal axial 2-ene TCO (2TCOa) group into living cells (Figure 1). We show that intracellularly released 2TCOa-dCTP is incorporated into de novo synthesized cellular DNA and viral genomes by staining it with different fast-reacting fluorescent dye-tetrazine conjugates. The efficient turn-off properties of these dye-conjugates allowed us to visualize genomes in living cells within minutes to hours with low background and without any subsequent washing steps.
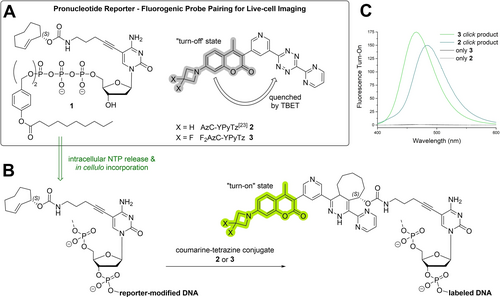
A) Overview of the pronucleotide reporter and fluorogenic probe pairing utilized for live-cell imaging of cellular and viral DNA. B) The TriPPPro delivery system enables the intracellular release of the active nucleoside triphosphates and facilitates the in vivo incorporation of the reporter nucleoside into de novo synthesized DNA. Subsequent fluorescent labeling with fluorogenic coumarin-tetrazine conjugates results in der fluorescent “turn-on” upon bioorthogonal reaction due to the transformation of the quenching tetrazine moiety into the dihydropyridazine reaction product. C) The fluorescence emission spectra of unreacted coumarin-YPyTz-conjugates 2 and 3 (gray lines) and their respective click products with the reporter nucleoside S2 (green lines) showing fluorescence turn-on upon reaction (for more detail regarding measurements, see Figure S1).
Results and Discussion
Synthesis of 2TCOa-TriPPPro and low-background dyes
Since modifications at the C5-position of pyrimidine nucleosides are known to be well accepted by polymerases, we decided to anchor our reactive labeling site to this position.14 To synthesize a TriPPPro reporter for labeling, 5-iodo-2′-deoxycytidine (4, IdC) was used as the starting material. The reason for choosing a cytidine derivative was two-fold. First, both prokaryotes and eukaryotes, as well as some DNA viruses (e.g., herpes simplex virus 1, HSV-1) and some retroviruses (e.g., HIV-1), encode dUTPases. This enzyme lowers the intracellular bioavailability of dUTP by hydrolyzing the triphosphate to yield dUMP and pyrophosphate.15 Since most DNA polymerases misincorporate dUTP instead of TTP into DNA, which causes mutations, it can be essential for the viability of these organisms or pathogens to reduce the intracellular dUTP/TTP ratio.16 Thus, choosing a cytidine-based reporter over a 2′-deoxyuridine derivative might enhance labeling efficiency. Second, the genome of HSV-1, which serves as a model virus, is GC-rich,17 potentially resulting in a better incorporation of a cytidine-based reporter.
In the first step, IdC 4 was transferred to its 5′-O-phosphate 5 by applying the Sowa & Ouchi protocol with P(O)Cl3, pyridine, and H2O in acetonitrile.18 After neutralization with aqueous NH4HCO3 and RP18 column chromatography, the counter ions were exchanged to triethylammonium (TEA) by Et3N addition and subsequent evaporation of all volatiles. Next, the linker motif was introduced using Sonogashira cross-coupling conditions. 1-Amino-4-pentyne was selected as a building block since it provides some distance between the nucleobase and a terminal amino group for later functionalization. Hence, the C−C-bond formation between the TEA salt of IdCMP 5 and 1-amino-4-pentyne was achieved by a catalyst system formed from Pd2(dba)3 and tri(2-furyl)phosphine in the presence of copper(I)-iodide and Et3N in DMSO. The precipitation of the formed nucleoside phosphate from cold acetone and subsequent RP18 column chromatography of the collected precipitate with 0.05 M tetraethylammonium bicarbonate (TEAB) in acetonitrile provided the TEA salt of 5-(5-aminopent-1-yn-1-yl)-dCMP 6 in a yield of 89 %. The amino group of the linker motif allowed the introduction of the 2TCOa via N-hydroxysuccinimide (NHS) chemistry. The carbamate formation was performed in DMSO in the presence of Et3N within 30 min at room temperature. Volatiles were removed in an oil pump vacuum at 50 °C. Subsequent RP18 column chromatography with 0.05 M TEAB in acetonitrile provided the TEA salt of 2TCOa-dCMP 7.
The axial 2-ene TCO provides significantly improved stability compared to the commonly used 4-ene TCO, which rapidly isomerizes to the less reactive cis-cyclooctene in the presence of thiols under physiological conditions.19 While the bicyclic reaction product of 2TCOa and 1,2,4,5-tetrazines undergo a β-elimination reaction, resulting in the undesired dissociation of the fluorophore after successful staining, this reaction can be minimized by choosing appropriate 3,6-substituents on the tetrazine.
For example, the reaction product of 3,6-dimethyl-1,2,4,5-tetrazine (Me2Tz) led to a much faster elimination reaction than 3,6-di(pyridin-2-yl)-1,2,4,5-tetrazine (Py2Tz), which was, as well as the 3-(pyridin-3-yl)-6-(pyrimidin-2-yl)-1,2,4,5-tetrazine (YPyTz) motif, used in this work.20 Hence, a compromise was accepted between the stability of the TCO motif and the stability of the bioorthogonal reaction product.
The TriPPPro-compound γ-(bis-C9AB)-2TCOa-dCTP 1 was synthesized via the H-phosphonate route previously described.21 4-(Hydroxymethyl)phenyldecanoate 8 was synthesized from 4-hydroxybenzyl alcohol and decanoyl chloride. Bis(4-decanoyloxybenzyl)-H-phosphonate 9 was prepared by displacing the phenoxy groups of diphenyl H-phosphonate (DPP) by 4-(hydroxymethyl)phenyldecanoate 8 in pyridine. Bis(4-decanoyloxybenzyl)-H-phosphonate 10 was obtained in 54 % yield after recrystallization from i-PrOH. This compound was oxidatively chlorinated with N-chlorosuccinimide (NCS) and treated with an excess of tetrabutylammonium phosphate in CH3CN. The bis(4-decanoyloxybenzyl) pyrophosphate 7 was isolated by extraction with CH2Cl2 from ammonium acetate and centrifugation. The TriPPPro-synthesis was initiated by a stepwise activation of the pyrophosphate with trifluoroacetic acid anhydride (TFAA) and 1-methylimidazole in acetonitrile. After the activation, the TEA salt 2TCOa-dCMP 7 was added, dissolved in DMF, RP18 column chromatography, Dowex ion-exchange chromatography (NH4+ form), and a second, RP18 column chromatography gave the pronucleotide reporter γ-(bis-C9AB)-2TCOa-dCTP 1 in a yield of 21 % (Scheme 1). In the following, the compound will be referred to as 2TCOa-TriPPPro 1. Moreover, the parent nucleoside 2TCOa-dC S2 was synthesized from IdC 4 over two steps as a control compound to validate the TriPPPro-approach. Furthermore, various novel and literature-known membrane-permeable fluorophore-tetrazine conjugates were synthesized (Figure 2). To lower the background fluorescence caused by non-reacted dye, a methyl-substituted SiR was attached to a methyl tetrazine (MeTz) by a trans-ethylene linkage resulting in SiR-(E)-MeTz 11. The connected π-system allows the efficient quenching of the fluorophore by the tetrazine via a through-bond energy transfer mechanism (TBET). Upon the reaction of the tetrazine with a TCO, the dye's fluorescence can “turn on”.22 To increase reactivity, a benzyl tetrazine (BzTz) was coupled to silicon rhodamine (SiR) by NHS chemistry via an amide bond (SiR-BzTz 12), as well as an azetidine-substituted SiR was coupled to the Py2Tz motif shown above via a short diamide linker resulting in SiR-Py2Tz 13. We also coupled the polar and membrane-impermeable AlexaFluor488 dye to the Py2Tz motif (AF488-Py2Tz 14) to generate a cell-impermeable, hydrophilic derivative which we used for labeling of fixed and permeabilized samples. Furthermore, we synthesized a 7-azetidinylcoumarin that we coupled to a pyrimidine(Y)-pyridine(Py) substituted tetrazine (AzC-YPyTz 2) according to the literature.23 Finally, we improved the fluorescence properties of the coumarin by introducing fluorine atoms to the 7-azetidinyl moiety resulting in the novel derivative F2AzC-YPyTz 3. Detailed synthesis protocols for these novel fluorescent probes are described in the Supporting Information.
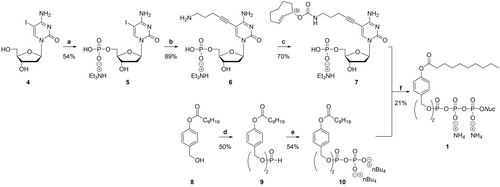
Synthesis of the TriPPPro compound γ-(bis-C9AB)-2TCOa-dCTP 1. Reagents and conditions: a) i) P(O)Cl3 (4.4 equiv), pyridine (4.8 equiv), H2O (2.8 equiv) in CH3CN (18.9 equiv), 0 °C, 10 min; ii) 5-iodo-2′-deoxycytidine, 0 °C; iii) ice water, NH4HCO3 (pH=8); iv) RP18 column chromatography v) ion-exchange to TEA; b) Pd2(dba)3 (5.4 mol%), tri(2-furyl) phosphine (12 mol%), CuI (15 mol%), 1-amino-4-pentyne, Et3N (7 equiv), DMSO, 55 °C, 1 h; c) 2TCOa-NHS, Et3N, DMSO, rt, 30 min; d) diphenyl phosphonate, pyridine, 40 °C; e) i) NCS, CH3CN, 50–60 °C; ii) 0.4 M mono-tetra-n-butylammonium phosphate in CH3CN, rt; f) i) TFAA, Et3N, CH3CN, 0 °C, 10 min; ii) 1-methylimidazole, Et3N, DMF, 0 °C to rt; iii) NMP (TEA salt) in DMF, CH3CN, rt, 3 h; iv) RP18 column chromatography, ion-exchange to ammonia, RP18 column chromatography.

Utilized fluorophore-tetrazine conjugates listed with increasing reactivity from left to right. The SiR-(E-MeTz and coumarin-YPyTz conjugates are quenched by a TBET mechanism. The laser wavelength used during fluorescence image acquisition is indicated.
Cellular and viral DNA can be marked with the 2TCOa-TriPPPro in living cells and labeled in fixed cells
First, we tested if the TriPPPro-technique enabled the incorporation of 2TCOa-modified 2′-deoxycytidine reporters into newly synthesized cellular and viral DNA in living cells. To this end, we decided to uncouple the development of cell-permeable pronucleotide reporters from the development of compatible cell-permeable dyes and use non-permeable dyes first that can be used after cell fixation and permeabilization. We chose Vero cells as a cellular model and HSV-1 as a viral model. HSV-1 is an important and well-characterized human pathogen and can be grown in Vero cells. To test 2TCOa-dCTP incorporation into the genomes of living cells as well as viral genomes, Vero cells were either left uninfected (mock) or infected with an HSV-1 mutant which codes for a fusion of the red fluorescent protein mCherry to its small capsid protein VP26 (mCherry-VP2624).
We used a multiplicity of infection (MOI) of 3 plaque-forming units (PFU) per cell in 10 % FCS-containing DMEM. After 1 h, the medium was exchanged to VP-serum-free medium (VP-SFM). This media does not contain serum and thus no carboxyesterases or lipases that can cleave the TriPPPro's AB-masks prior to internalization.
At 4 hours post-infection (hpi), the cells were incubated with 5 μM of 2TCOa-TriPPPro 1 for 3–4 hours. After incorporation, the samples were washed briefly, fixed, and permeabilized. The incorporated 2TCOa motif was conjugated with the polar AF488-Py2Tz dye 14. After the “click”-labeling, the samples were washed again, counterstained with Hoechst 33342 to label DNA, and imaged using confocal spinning disk microscopy.
As shown in Figure 3A, we found specific AF488 signal in both infected and uninfected TriPPPro-treated Vero cells. In uninfected cells, the AF488 signal colocalized with the Hoechst signal, as shown by a line plot across a representative nucleus in Figure 3C. This indicates that 2TCOa-dCTP was incorporated into the cellular genomes of dividing cells. Moreover, we compared our labeling approach with the well-established CuAAC approach utilizing a commercially available EdC click-kit with an AF488-azide conjugate and observed identical labeling patterns (Figure S4). To provide additional evidence for successful reporter incorporation into genomic DNA, we synthesized the corresponding nucleoside triphosphate 2TCOa-dCTP S5 and performed primer extension assays (Figure S5). We observed successful primer elongation for human pol β and γ, whereas pol α showed no elongation. Additionally, we tested HIV-1 RT polymerase, which also accepted 2TCOa-dCTP S5 as a substrate suggesting that the released 2TCOa-modified NTP shows good substrate properties towards a wide variety of DNA polymerases. Moreover, a cell viability assay also confirmed that up to 10 μM of 2TCOa-TriPPPro 1 did not significantly affect cell growth when incubated for 3 hours (Figure S6).
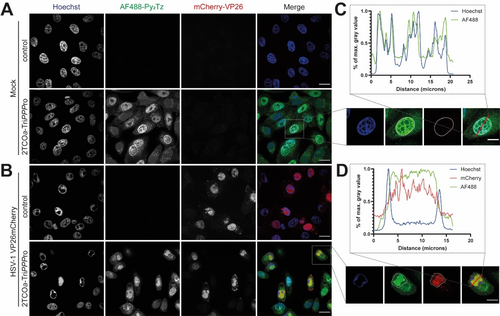
Labeling of cellular and viral DNA with 2TCOa-TriPPPro. Vero cells were either A) mock- or B) HSV-1 mCherry-VP26 infected at an MOI of 3. The capsids of HSV-1 are labeled with mCherry-VP26 (red). 1 hpi, medium was changed to VP-SFM, and 4 hpi nascent DNA was tagged with 5 μM 2TCOa-TriPPPro 1 for 4 hours. The TriPPPro medium was renewed once at 6 hpi. 8 hpi, cells were fixed and processed: Genomes containing incorporated 2TCOa-dC were stained overnight with AF488-Py2Tz 14 (green). Nuclei were stained with Hoechst 33342 (blue), and samples were imaged. C) insert one representative cell with a line plot illustrating the colocalization of AF488 and Hoechst signals. D) insert of one representative cell with a line plot illustrating the colocalization of AF488 and mCherry-VP26. Nuclear outlines are circled. Individual channels are shown in greyscale, and the merged image is shown in color. Scalebar=20 μm. Enlarged section: Scalebar=10 μm.
In infected cells, HSV-1 creates replication compartments (RCs) that increase the nuclear volume and displace the host chromatin to the nuclear periphery. RCs can be identified by areas without Hoechst signal as alphaherpesvirus RCs are not labeled by Hoechst25). The rearrangement of the nucleus facilitates the synthesis of viral genomic DNA, transcription of viral mRNA, and assembly of new capsids.26 The mCherry-VP26 fusion accumulates in nuclear RCs and can be used as a marker for cells in the late stage of virus replication. We found specific AF488 signal colocalizing with viral RCs labeled by mCherry-VP26 (Figure 3B/D), indicating that our new reporter was incorporated into viral genomes. To ensure that the viral DNA was indeed localized to viral RCs, we also stained for the infected cell protein 8 (ICP8) by immunofluorescence. ICP8 is the single-stranded DNA binding protein of HSV-1 and is routinely used as a viral RC marker.27 We found strong colocalization between the signals of the metabolic label and ICP8, further supporting our conclusion that viral genomes can be labeled by our TriPPPro reporter 1 (Figure S7).
To confirm that the delivery of the triphosphate reporter into living cells is needed, we tested the parent nucleoside, which cellular kinases must still phosphorylate to form the active triphosphate. We treated uninfected and HSV-1-infected Vero cells with up to 30 μM of 2TCOa-dC S2, as described in Figure 4. As shown, we could not detect any DNA signal in the AF488 channel, stressing the importance of the TriPPPro-approach to deliver nucleoside triphosphates with bulky modification into cells. Off note, cells incubated with the lipophilic TriPPPro-nucleotides showed increased cytoplasmatic fluorescence, which might be in part due to the unspecific attachment of the lipophilic pronucleotides onto intracellular membranes as well as the incorporation of the reporter into mitochondrial DNA. Based on our experience, limited optimization of the labeling protocol is needed to fine-tune the labeling protocol to the experimental system.
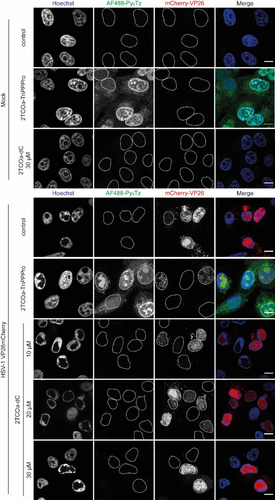
The TriPPPro-approach is necessary to bypass the “bottleneck” phosphorylation to the active nucleoside triphosphate reporter. Confluent Vero cells were either mock-infected or infected with HSV-1 mCherry-VP26 at an MOI of 3. The capsids of HSV-1 are labeled with mCherry (red). 1 hpi, the medium was changed to VP-SFM and 4 hpi nascent DNA was tagged with 5 μM of the pronucleotide 2TCOa-TriPPPro 1. As controls, mock-infected cells were treated with 30 μM of the parent nucleoside 2TCOa-dC S2 in VP-SFM, and HSV-1 infected cells were treated with 10, 20, or 30 μM of the parent nucleoside in VP-SFM. The medium was renewed once at 6 hpi. 8 hpi, cells were fixed and processed: Genomes containing incorporated 2TCOa-dC were stained overnight with AF488-Py2Tz 14 (green). Nuclei were stained with Hoechst (blue) and samples were imaged. Individual channels are shown in greyscale and the merged image is shown in color. Nuclear outlines are circled. Scalebar=10 μm. The representative images originate from the same assay presented in Figure 3 and are comparable.
Cellular and viral DNA can be visualized with 2TCOa TriPPPro-compounds in living cells using highly reactive, membrane-permeable dyes
Next, we wanted to establish highly reactive, permeable dyes which show minimal fluorescence in the unconjugated state to label and image DNA in living cells. Based on these premises, we initially developed three SiR-conjugates carrying tetrazines with different reactivities (compounds 11–13). To test them, we used uninfected and HSV-1 mCherry-VP26 infected Vero cells and incubated them with 5 μM of 2TCOa-TriPPPro 1 at 4 hpi in VP-SFM. After 6 hpi, the TriPPPro-compound-containing medium was replenished to ensure the integrity of the AB masks throughout the incorporation phase. At 8 hpi, cells were washed with VP-SFM, stained with 5 μM of the respective SiR-tetrazine conjugate, and directly imaged without further washing. As shown in Figure S8, the most reactive conjugate, SiR-Py2Tz 13, stained both cellular and viral DNA within one hour after dye addition. In contrast, we could not detect any signal for the less reactive SiR-(E)-MeTz 11 or SiR-BzTz 12 conjugates even after prolonged incubation. Unfortunately, all SiR dyes showed unspecific staining of cytoplasmic compartments, which was even more prominent in uninfected cells (Figure S8).
We concluded that fast tetrazine probes are the key to the successful visualization of DNA in living cells and searched for potential dye-tetrazine conjugates with increased reactivity and improved background. Galeta et al. recently described a HELIOS probe consisting of a 7-azetidinylcoumarin dye directly coupled to a highly reactive tetrazine motif (YPyTz), which facilitated the staining of their cytoplasmic targets within tens of seconds.23
The direct conjugation of the coumarin dye to the aromatic tetrazine π-system allows effective fluorescence quenching by TBET, resulting in a “turn-on” of fluorescence upon reaction with an axial 4-ene TCO.23 In addition to the AzC-YPyTz 2, we synthesized its derivative F2AzC−YPyTz 3, which contained a 3,3-difluoroazetidine moiety, as done previously to optimize xanthene dyes.28 We determined both coumarin-tetrazine conjugates’ “turn-on” values upon reaction with the 2TCOa-dC reporter nucleoside S2. Here, the AzC−YPyTz 2 and F2AzC−YPyTz 3 probes showed similar “turn-on” values of 150 and 175, respectively (see Figure 1 and S1).
The in vitro reaction rates of the YPyTz-based conjugates with 2TCOa-dC S2 were not improving compared to the less reactive Py2Tz-functionalized silicon-rhodamine (although the reactivity trend is true for BCNendo alcohol S21, Table 1 and Figure S2–S3).
|
SiR−Py2Tz 13 |
AzC−YPyTz 2 |
F2AzC−YPyTz 3 |
|---|---|---|---|
BCNendo S21 |
20±3 M−1s−1 |
901±38 M−1s−1 |
721±51 M−1s−1 |
2TCOa-dC S2 |
8.7±0.3 M−1s−1 |
7.3±0.8 M−1s−1 |
17.1±0.4 M−1s−1 |
However, both coumarin-based probes resulted in clear staining of cellular and viral DNA within minutes post-dye addition compared to within one hour for SiR−Py2Tz 13, underscoring their utility in the more complex cellular context. The coumarin-YPyTz dyes were efficiently excitable at their second excitation shoulder (Figure S9) with 405 nm lasers, commonly found on contemporary confocal microscopes. The fluorinated coumarin derivative 3 was dimmer than the parent fluorogenic probe 2, but the F2AzC−YPyTz 3 appeared to show less cytoplasmic background (Figure 5), and we proceeded with this derivate.
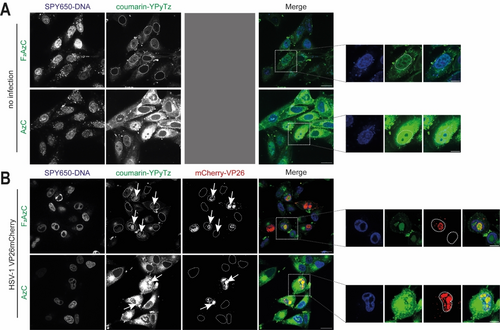
The fluorinated F2AzC−YPyTz conjugate shows less cytoplasmic background in living cells. A) Confluent Vero cells were washed in VP-SFM, and nascent DNA was tagged with 5 μM 2TCOa-TriPPPro 1 for 4 hours. The TriPPPro medium was renewed 2 h after the first addition. After 4 h, the samples were washed twice with VP-SFM and stained with 5 μM of AzC−YPyTz 2 or F2AzC−YPyTz 3 (green). SPY650-DNA (blue, 1 : 1000) was added simultaneously. The samples were imaged live 15–30 min after dye addition. B) Confluent Vero cells were infected with HSV-1 mCherry-VP26 at an MOI of 3. The capsids of HSV-1 are labeled with mCherry (red). 1 hpi, the medium was changed to VP-SFM, and 4 hpi nascent DNA was tagged with 5 μM 2TCOa-TriPPPro 1 for 4 hours. The TriPPPro medium was renewed once at 5.5 hpi. 7 hpi, the samples were washed in VP-SFM (2x). 7.5 hpi, the samples were washed once again with VP-SFM and stained with 5 μM of AzC−YPyTz 2 or F2AzC−YPyTz 3 (green) under simultaneous addition of SPY650-DNA (blue, 1 : 1000). 8.5 hpi, samples were imaged live. Individual channels are shown in greyscale, and the merged image is shown in color. Scalebar=20 μm. Enlarged section: Scalebar=10 μm. Nuclear outlines are circled.
To optimize staining conditions, we quantified the fluorescent signal after treatment with increasing concentrations of 2TCOa-TriPPPro 1 (0, 0.5, 1.5, 5, and 10 μM) and labeling with F2AzC−YPyTz 3 in living cells. As shown in Figures S10, S11, and Figure S12 for cellular and viral DNA, respectively, we found the best staining results at a concentration of 5 μM. The highest TriPPPro dose (10 μM) caused cell dissociation from the growth support, although cell viability was not significantly affected on this time scale (compare Figure S6).
To ensure that labeling was due to the incorporation of the reporter into cellular and viral DNA, we used aphidicolin29 and phosphonoacetic acid (PAA)30 as inhibitors of cellular and viral replication, respectively (Figure 6). We did not detect any DNA labeling when cellular DNA synthesis was blocked, demonstrating that the F2AzC signals are specific and depend on cellular polymerase activity to incorporate 2TCOa-dCTP. The same was true for viral DNA labeling: PAA, an inhibitor of viral replication, abrogated 2TCOa-dCTP incorporation into viral genomes and RCs, as we did not detect any specific F2AzC signals. Finally, we also tested DNA labeling in MDCK cells, and the results suggest that the presented approach can be generalized (Figure S13).
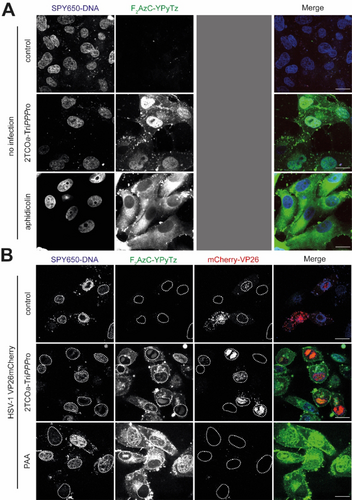
Inhibitor controls with aphidicolin and PAA demonstrate that 2TCOa-dCTP incorporation is dependent on polymerase activity. A) For the inhibitor control, aphidicolin was added 1 h after cell seeding in a concentration of 10 μg/mL. Confluent Vero cells were washed in VP-SFM (2x) and nascent DNA was tagged with 5 μM 2TCOa-TriPPPro 1. The TriPPPro medium was renewed once at 1.5 h after the first addition. After 3 h, the samples were washed in VP-SFM (2x). After 3.5 h, the samples were washed once again and stained with 5 μM of F2AzC−YPyTz 3 (green) under simultaneous addition of SPY650-DNA (blue, 1 : 1000). After 4 h, the media were changed to VP-SFM and samples were imaged live. B) Confluent Vero cells were infected with HSV-1 mCherry-VP26 at an MOI of 3. For the inhibitor control, PAA (400 μg/ml) was added at the point of infection, and the concentration was held until staining. The capsids of the HSV-1 are tagged with mCherry (red). 1 hpi, the medium was changed to VP-SFM and 4 hpi nascent DNA was tagged with 5 μM 2TCOa-TriPPPro 1. The TriPPPro medium was renewed once at 5.5 hpi. 7 hpi, the samples were washed in VP-SFM (2x). 7.5 hpi, the samples were washed once again with VP-SFM and stained with 5 μM of F2AzC−YPyTz 3 (green) under simultaneous addition of SPY650-DNA (blue, 1 : 1000). 8.5 hpi, samples were imaged live. Individual channels are shown in greyscale and the merged image is shown in color. Nuclear outlines are circled. Scalebar=20 μm.
Future work will be targeted expand the concept to label RNA, to improve bioorthogonal partner pairing for faster reaction rates and to optimize fluorophores to minimize cytotoxicity and expand the “photon budget” available for live-cell imaging.
Conclusion
Our data illustrates that de novo synthesized cellular and viral DNA can be imaged in living cells using iEDDA-mediated bioorthogonal labeling. We demonstrated three crucial advances: First, our TriPPPro pronucleotide technique allowed the delivery of highly reactive nucleotide reporters, which do not need to be phosphorylated but can be directly utilized by polymerases. Second, we find it vital to use labeling partners which react fast in the complex cellular environment since dye-tetrazine conjugates with less reactive dienes did not result in detectable DNA in living cells. Third, using fluorogenic probes with “turn-off” properties seems essential for improved cellular background—such as our F2AzC−YPyTz dye. We believe that our bioorthogonal DNA live-cell labeling approach should be of considerable utility for use in cell biology and virology to study processes such as genome replication and trafficking in live cells.
Acknowledgments
We thank Roland Thünauer (CSSB and LIV microscopy facilities) and Prof. Joachim Hauber (LIV) for support, as well as Maike Voges, Enrico Caragliano, and Heidi Meisner for testing of initial compounds. This project was partly funded by the Leibniz Science Campus InterACt (grant agreement no. W6/2018). C.M. is funded by the University of Hamburg and by the Deutsche Forschungsgemeinschaft (DFG) (Me1161/17-1); J.B.B. is funded by DFG under Germany's Excellence Strategy EXC 2155—project no. 390874280, by the Wellcome Trust through a Collaborative Award (209250/Z/17/Z), and also in the framework of the Research Unit FOR5200 DEEP-DV (443644894) project BO 4158/5-1. Open Access funding enabled and organized by Projekt DEAL.
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.