The Role of Rotational Motion in Diffusion NMR Experiments on Supramolecular Assemblies: Application to Sup35NM Fibrils
Dr. Boris B. Kharkov
Laboratory of Biomolecular NMR, St. Petersburg State University, 199034 St. Petersburg, Russia
These authors contributed equally to this work.
Search for more papers by this authorIvan S. Podkorytov
Laboratory of Biomolecular NMR, St. Petersburg State University, 199034 St. Petersburg, Russia
These authors contributed equally to this work.
Search for more papers by this authorDr. Stanislav A. Bondarev
Department of Genetics and Biotechnology, St. Petersburg State University, 199034 St. Petersburg, Russia
Search for more papers by this authorDr. Mikhail V. Belousov
Department of Genetics and Biotechnology, St. Petersburg State University, 199034 St. Petersburg, Russia
Laboratory for Proteomics of Supra-Organismal Systems, All-Russia Research Institute for Agricultural Microbiology (ARRIAM), 196608 St. Petersburg, Russia
Search for more papers by this authorVladislav A. Salikov
Laboratory of Biomolecular NMR, St. Petersburg State University, 199034 St. Petersburg, Russia
Search for more papers by this authorProf. Dr. Galina A. Zhouravleva
Department of Genetics and Biotechnology, St. Petersburg State University, 199034 St. Petersburg, Russia
Search for more papers by this authorCorresponding Author
Prof. Dr. Nikolai R. Skrynnikov
Laboratory of Biomolecular NMR, St. Petersburg State University, 199034 St. Petersburg, Russia
Department of Chemistry, Purdue University, West Lafayette, IN, 47907 USA
Search for more papers by this authorDr. Boris B. Kharkov
Laboratory of Biomolecular NMR, St. Petersburg State University, 199034 St. Petersburg, Russia
These authors contributed equally to this work.
Search for more papers by this authorIvan S. Podkorytov
Laboratory of Biomolecular NMR, St. Petersburg State University, 199034 St. Petersburg, Russia
These authors contributed equally to this work.
Search for more papers by this authorDr. Stanislav A. Bondarev
Department of Genetics and Biotechnology, St. Petersburg State University, 199034 St. Petersburg, Russia
Search for more papers by this authorDr. Mikhail V. Belousov
Department of Genetics and Biotechnology, St. Petersburg State University, 199034 St. Petersburg, Russia
Laboratory for Proteomics of Supra-Organismal Systems, All-Russia Research Institute for Agricultural Microbiology (ARRIAM), 196608 St. Petersburg, Russia
Search for more papers by this authorVladislav A. Salikov
Laboratory of Biomolecular NMR, St. Petersburg State University, 199034 St. Petersburg, Russia
Search for more papers by this authorProf. Dr. Galina A. Zhouravleva
Department of Genetics and Biotechnology, St. Petersburg State University, 199034 St. Petersburg, Russia
Search for more papers by this authorCorresponding Author
Prof. Dr. Nikolai R. Skrynnikov
Laboratory of Biomolecular NMR, St. Petersburg State University, 199034 St. Petersburg, Russia
Department of Chemistry, Purdue University, West Lafayette, IN, 47907 USA
Search for more papers by this authorGraphical Abstract
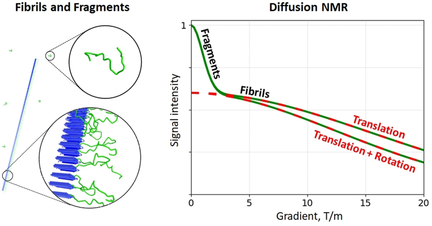
Diffusion of amyloid fibrils that contain disordered domains can be captured by pulsed-field-gradient NMR experiments. A new theory is presented that interprets the results of these measurements in terms of fibrils’ translational and rotational motion. Application to Sup35NM fibrils demonstrates the feasibility of diffusion-based sorting in complex amyloidogenic samples.
Abstract
Pulsed-field gradient (PFG) NMR is an important tool for characterization of biomolecules and supramolecular assemblies. However, for micrometer-sized objects, such as amyloid fibrils, these experiments become difficult to interpret because in addition to translational diffusion they are also sensitive to rotational diffusion. We have constructed a mathematical theory describing the outcome of PFG NMR experiments on rod-like fibrils. To test its validity, we have studied the fibrils formed by Sup35NM segment of the prion protein Sup35. The interpretation of the PFG NMR data in this system is fully consistent with the evidence from electron microscopy. Contrary to some previously expressed views, the signals originating from disordered regions in the fibrils can be readily differentiated from the similar signals representing small soluble species (e.g. proteolytic fragments). This paves the way for diffusion-sorted NMR experiments on complex amyloidogenic samples.
Conflict of interest
The authors declare no conflict of interest.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie202102408-sup-0001-misc_information.pdf2.1 MB | Supplementary |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1E. O. Stejskal, J. E. Tanner, J. Chem. Phys. 1965, 42, 288–292.
- 2J. Christodoulou, G. Larsson, P. Fucini, S. R. Connell, T. A. Pertinhez, C. L. Hanson, C. Redfield, K. H. Nierhaus, C. V. Robinson, J. Schleucher, et al., Proc. Natl. Acad. Sci. USA 2004, 101, 10949–10954.
- 3L. Shen, R. Soong, M. F. Wang, A. Lee, C. Wu, G. D. Scholes, P. M. Macdonald, M. A. Winnik, J. Phys. Chem. B 2008, 112, 1626–1633.
- 4B. R. Szymczyna, L. Gan, J. E. Johnson, J. R. Williamson, J. Am. Chem. Soc. 2007, 129, 7867–7876.
- 5A. J. Baldwin, S. J. Anthony-Cahill, T. P. J. Knowles, G. Lippens, J. Christodoulou, P. D. Barker, C. M. Dobson, Angew. Chem. Int. Ed. 2008, 47, 3385–3387; Angew. Chem. 2008, 120, 3433–3435.
- 6G. W. Platt, W. F. Xue, S. W. Homans, S. E. Radford, Angew. Chem. Int. Ed. 2009, 48, 5705–5707; Angew. Chem. 2009, 121, 5815–5817.
- 7A. J. Baldwin, J. Christodoulou, P. D. Barker, C. M. Dobson, G. Lippens, J. Chem. Phys. 2007, 127, 114505.
- 8
- 8aH. C. Torrey, Phys. Rev. 1956, 104, 563–565;
- 8bW. S. Price, NMR studies of translational motion: principles and applications, Cambridge University Press, Cambridge, 2009.
10.1017/CBO9780511770487 Google Scholar
- 9D. Sinnaeve, Concepts Magn. Reson. Part A 2012, 40, 39–65.
- 10
- 10aS. Broersma, J. Chem. Phys. 1981, 74, 6989–6990;
- 10bM. M. Tirado, C. L. Martinez, J. G. de la Torre, J. Chem. Phys. 1984, 81, 2047–2052;
- 10cJ. Newman, H. L. Swinney, L. A. Day, J. Mol. Biol. 1977, 116, 593–603.
- 11
- 11aG. Zhouravleva, L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, M. Philippe, EMBO J. 1995, 14, 4065–4072;
- 11bI. Stansfield, K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, M. F. Tuite, EMBO J. 1995, 14, 4365–4373.
- 12
- 12aM. D. Ter-Avanesyan, A. R. Dagkesamanskaya, V. V. Kushnirov, V. N. Smirnov, Genetics 1994, 137, 671–676;
- 12bJ. R. Glover, A. S. Kowal, E. C. Schirmer, M. M. Patino, J. J. Liu, S. Lindquist, Cell 1997, 89, 811–819.
- 13B. H. Toyama, M. J. S. Kelly, J. D. Gross, J. S. Weissman, Nature 2007, 449, 233–238.
- 14A. I. Sulatskaya, I. M. Kuznetsova, M. V. Belousov, S. A. Bondarev, G. A. Zhouravleva, K. K. Turoverov, PLoS One 2016, 11, e0156314.
- 15A. Kishimoto, K. Hasegawa, H. Suzuki, H. Taguchi, K. Namba, M. Yoshida, Biochem. Biophys. Res. Commun. 2004, 315, 739–745.
- 16B. Hoffmann, C. Eichmuller, O. Steinhauser, R. Konrat in Nuclear Magnetic Resonance of Biological Macromolecules Part C, Vol. 394 (Ed.: T. L. James), Academic Press, New York, 2005, pp. 142–175.
- 17
- 17aR. J. Morris, K. Eden, R. Yarwood, L. Jourdain, R. J. Allen, C. E. MacPhee, Nat. Commun. 2013, 4, 1891;
- 17bT. Doussineau, C. Mathevon, L. Altamura, C. Vendrely, P. Dugourd, V. Forge, R. Antoine, Angew. Chem. Int. Ed. 2016, 55, 2340–2344; Angew. Chem. 2016, 128, 2386–2390.
- 18C. A. Schneider, W. S. Rasband, K. W. Eliceiri, Nat. Methods 2012, 9, 671–675.
- 19
- 19aD. K. Wilkins, S. B. Grimshaw, V. Receveur, C. M. Dobson, J. A. Jones, L. J. Smith, Biochemistry 1999, 38, 16424–16431;
- 19bJ. A. Marsh, J. D. Forman-Kay, Biophys. J. 2010, 98, 2383–2390.
- 20
- 20aM. Nick, Y. B. Wu, N. W. Schmidt, S. B. Prusiner, J. Stohr, W. F. DeGrado, Biopolymers 2018, 109, e23096;
- 20bH. Konno, T. Watanabe-Nakayama, T. Uchihashi, M. Okuda, L. W. Zhu, N. Kodera, Y. Kikuchi, T. Ando, H. Taguchi, Proc. Natl. Acad. Sci. USA 2020, 117, 7831–7836.
- 21P. Alam, L. Bousset, R. Melki, D. E. Otzen, J. Neurochem. 2019, 150, 522–534.
- 22V. V. Kushnirov, I. M. Alexandrov, O. V. Mitkevich, I. S. Shkundina, M. D. Ter-Avanesyan, Methods 2006, 39, 50–55.
- 23J. E. Tanner, J. Chem. Phys. 1970, 52, 2523–2526.
- 24
- 24aI. Teraoka, N. Ookubo, R. Hayakawa, Phys. Rev. Lett. 1985, 55, 2712–2715;
- 24bI. Teraoka, R. Hayakawa, J. Chem. Phys. 1988, 89, 6989–6995;
- 24cI. Teraoka, R. Hayakawa, J. Chem. Phys. 1989, 91, 2643–2648;
- 24dJ. Käs, H. Strey, J. X. Tang, D. Finger, R. Ezzell, E. Sackmann, P. A. Janmey, Biophys. J. 1996, 70, 609–625.
- 25S. Leitmann, F. Hofling, T. Franosch, Phys. Rev. E 2017, 96, 012118.
- 26J. H. Lee, Y. Okuno, S. Cavagnero, J. Magn. Reson. 2014, 241, 18–31.
- 27
- 27aF. Hasecke, T. Miti, C. Perez, J. Barton, D. Scholzel, L. Gremer, C. S. R. Gruning, G. Matthews, G. Meisl, T. P. J. Knowles, et al., Chem. Sci. 2018, 9, 5937–5948;
- 27bR. Mishra, K. Sorgjerd, S. Nystrom, A. Nordigarden, Y. C. Yu, P. Hammarstrom, J. Mol. Biol. 2007, 366, 1029–1044;
- 27cK. Ono, M. M. Condron, D. B. Teplow, Proc. Natl. Acad. Sci. USA 2009, 106, 14745–14750;
- 27dS. Shimonaka, T. Nonaka, G. Suzuki, S. Hisanaga, M. Hasegawa, J. Biol. Chem. 2016, 291, 8896–8907;
- 27eY. P. Wang, J. Biernat, M. Pickhardt, E. Mandelkow, E. M. Mandelkow, Proc. Natl. Acad. Sci. USA 2007, 104, 10252–10257.
- 28C. S. Johnson, Prog. Nucl. Magn. Reson. Spectrosc. 1999, 34, 203–256.