Quantitative Mapping of Endosomal DNA Processing by Single Molecule Counting
Graphical Abstract
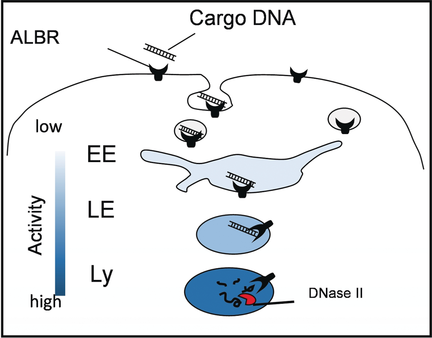
Elementary, my dear Watson: Organellar single-molecule, high-resolution localization and counting (oSHiRLoC), a fluorescence imaging method for the quantitative mapping of the endosomal processing of cargo DNA in innate immune cells with organelle-specific resolution, is reported. By using this method, it is shown that endosomal DNA degradation occurs mainly in lysosomes and is negligible in late endosomes.
Abstract
Extracellular DNA is engulfed by innate immune cells and digested by endosomal DNase II to generate an immune response. Quantitative information on endosomal stage-specific cargo processing is a critical parameter to predict and model the innate immune response. Biochemical assays quantify endosomal processing but lack organelle-specific information, while fluorescence microscopy has provided the latter without the former. Herein, we report a single molecule counting method based on fluorescence imaging that quantitatively maps endosomal processing of cargo DNA in innate immune cells with organelle-specific resolution. Our studies reveal that endosomal DNA degradation occurs mainly in lysosomes and is negligible in late endosomes. This method can be used to study cargo processing in diverse endocytic pathways and measure stage-specific activity of processing factors in endosomes.