Solid-state NMR and EPR Spectroscopy of Mn2+-Substituted ATP-Fueled Protein Engines
Dr. Thomas Wiegand
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
These authors contributed equally to this work.
Search for more papers by this authorDenis Lacabanne
Molecular Microbiology and Structural Biochemistry, Labex Ecofect, UMR 5086 CNRS/Université de Lyon, 69367 Lyon, France
These authors contributed equally to this work.
Search for more papers by this authorKatharina Keller
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
These authors contributed equally to this work.
Search for more papers by this authorRiccardo Cadalbert
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
Search for more papers by this authorDr. Lauriane Lecoq
Molecular Microbiology and Structural Biochemistry, Labex Ecofect, UMR 5086 CNRS/Université de Lyon, 69367 Lyon, France
Search for more papers by this authorDr. Maxim Yulikov
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
Search for more papers by this authorCorresponding Author
Dr. Laurent Terradot
Molecular Microbiology and Structural Biochemistry, Labex Ecofect, UMR 5086 CNRS/Université de Lyon, 69367 Lyon, France
Search for more papers by this authorCorresponding Author
Prof. Gunnar Jeschke
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
Search for more papers by this authorCorresponding Author
Prof. Beat H. Meier
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
Search for more papers by this authorCorresponding Author
Dr. Anja Böckmann
Molecular Microbiology and Structural Biochemistry, Labex Ecofect, UMR 5086 CNRS/Université de Lyon, 69367 Lyon, France
Search for more papers by this authorDr. Thomas Wiegand
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
These authors contributed equally to this work.
Search for more papers by this authorDenis Lacabanne
Molecular Microbiology and Structural Biochemistry, Labex Ecofect, UMR 5086 CNRS/Université de Lyon, 69367 Lyon, France
These authors contributed equally to this work.
Search for more papers by this authorKatharina Keller
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
These authors contributed equally to this work.
Search for more papers by this authorRiccardo Cadalbert
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
Search for more papers by this authorDr. Lauriane Lecoq
Molecular Microbiology and Structural Biochemistry, Labex Ecofect, UMR 5086 CNRS/Université de Lyon, 69367 Lyon, France
Search for more papers by this authorDr. Maxim Yulikov
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
Search for more papers by this authorCorresponding Author
Dr. Laurent Terradot
Molecular Microbiology and Structural Biochemistry, Labex Ecofect, UMR 5086 CNRS/Université de Lyon, 69367 Lyon, France
Search for more papers by this authorCorresponding Author
Prof. Gunnar Jeschke
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
Search for more papers by this authorCorresponding Author
Prof. Beat H. Meier
Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland
Search for more papers by this authorCorresponding Author
Dr. Anja Böckmann
Molecular Microbiology and Structural Biochemistry, Labex Ecofect, UMR 5086 CNRS/Université de Lyon, 69367 Lyon, France
Search for more papers by this authorGraphical Abstract
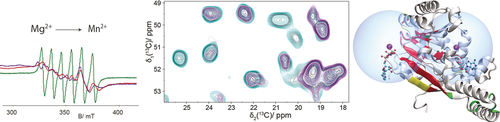
Metal location: Both EPR and solid-state NMR spectra change on the substitution of Mg2+ by Mn2+ in two different ATP:Mg2+ fueled protein engines. While EPR spectra report metal binding and metal-center geometry, NMR paramagnetic relaxation enhancements localize residues at the binding site. Both techniques indicate the location of paramagnetic ions in proteins.
Abstract
Paramagnetic metal ions deliver structural information both in EPR and solid-state NMR experiments, offering a profitable synergetic approach to study bio-macromolecules. We demonstrate the spectral consequences of Mg2+/ Mn2+ substitution and the resulting information contents for two different ATP:Mg2+-fueled protein engines, a DnaB helicase from Helicobacter pylori active in the bacterial replisome, and the ABC transporter BmrA, a bacterial efflux pump. We show that, while EPR spectra report on metal binding and provide information on the geometry of the metal centers in the proteins, paramagnetic relaxation enhancements identified in the NMR spectra can be used to localize residues at the binding site. Protein engines are ubiquitous and the methods described herein should be applicable in a broad context.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie201610551-sup-0001-misc_information.pdf17.7 MB | Supplementary |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1P. I. Hanson, S. W. Whiteheart, Nat. Rev. Mol. Cell Biol. 2005, 6, 519–529.
- 2D. C. Rees, E. Johnson, O. Lewinson, Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227.
- 3C. P. Jaroniec, J. Magn. Reson. 2015, 253, 50–59.
- 4S. J. Ullrich, S. Hölper, C. Glaubitz, J. Biomol. NMR 2014, 58, 27–35.
- 5I. Bertini, L. Emsley, M. Lelli, C. Luchinat, J. Mao, G. Pintacuda, J. Am. Chem. Soc. 2010, 132, 5558–5559.
- 6G. Pintacuda, N. Giraud, R. Pierattelli, A. Böckmann, I. Bertini, L. Emsley, Angew. Chem. Int. Ed. 2007, 46, 1079–1082; Angew. Chem. 2007, 119, 1097–1100.
- 7A. Bazin, M. V. Cherrier, I. Gutsche, J. Timmins, L. Terradot, Nucleic Acids Res. 2015, gkv 792-13.
- 8E. Steinfels, C. Orelle, J.-R. Fantino, O. Dalmas, J.-L. Rigaud, F. Denizot, A. Di Pietro, J.-M. Jault, J. Biol. Chem. 2004, 43, 7491–7502.
- 9C. Geourjon, C. Orelle, E. Steinfels, C. Blanchet, G. Deléage, A. Di Pietro, J. M. Jault, Trends Biochem. Sci. 2001, 26, 539–544.
- 10W. Bujalowski, M. M. Klonowska, J. Biol. Chem. 1993, 32, 5888–5900.
- 11M. J. Jezewska, U. S. Kim, W. Bujalowski, Biophys. J. 1996, 71, 2075–2086.
- 12A. Siarheyeva, R. Liu, F. J. Sharom, J. Biol. Chem. 2010, 285, 7575–7586.
- 13T. Wiegand, R. Cadalbert, C. Gardiennet, J. Timmins, L. Terradot, A. Böckmann, B. H. Meier, Angew. Chem. Int. Ed. 2016, 55, 14164–14168; Angew. Chem. 2016, 128, 14370–14375.
- 14C. Orelle, F. Gubellini, A. Durand, S. Marco, D. Lévy, P. Gros, A. Di Pietro, J.-M. Jault, J. Biol. Chem. 2008, 47, 2404–2412.
- 15H. Witt, A. Wittershagen, E. Bill, B. O. Kolbesen, B. Ludwig, FEBS Lett. 1997, 409, 128–130.
- 16M. Bennati, M. M. Hertel, J. Fritscher, T. F. Prisner, N. Weiden, R. Hofweber, M. Spörner, G. Horn, H. R. Kalbitzer, J. Biol. Chem. 2006, 45, 42–50.
- 17E. Bonneau, P. Legault, J. Biol. Chem. 2014, 53, 579–590.
- 18I. Bertini, C. Luchinat, G. Parigi, R. Pierattelli, ChemBioChem 2005, 6, 1536–1549.
- 19H. Kaur, A. Lakatos, R. Spadaccini, R. Vogel, C. Hoffmann, J. Becker-Baldus, O. Ouari, P. Tordo, H. Mchaourab, C. Glaubitz, Biol. Chem. 2015, 396, 1135–1149.
- 20G. Otting, Annu. Rev. Biophys. 2010, 39, 387–405.
- 21H. Tamaki, A. Egawa, K. Kido, T. Kameda, M. Kamiya, T. Kikukawa, T. Aizawa, T. Fujiwara, M. Demura, J. Biomol. NMR 2016, 64, 87–101.
- 22R. K. Soni, P. Mehra, N. R. Choudhury, G. Mukhopadhyay, S. K. Dhar, Nucleic Acids Res. 2003, 31, 6828–6840.
- 23H. Y. V. Ching, F. C. Mascali, H. C. Bertrand, E. M. Bruch, P. Demay-Drouhard, R. M. Rasia, C. Policar, L. C. Tabares, S. Un, J. Phys. Chem. Lett. 2016, 7, 1072–1076.
- 24R. J. P. Dawson, K. P. Locher, FEBS Lett. 2007, 581, 935–938.
- 25P. Lueders, S. Razzaghi, H. Jäger, R. Tschaggelar, M. A. Hemminga, M. Yulikov, G. Jeschke, Mol. Phys. 2013, 111, 2824–2833.
- 26S. Razzaghi, E. K. Brooks, E. Bordignon, W. L. Hubbell, M. Yulikov, G. Jeschke, ChemBioChem 2013, 14, 1883–1890.
- 27H. Yaginuma, S. Kawai, K. V. Tabata, K. Tomiyama, A. Kakizuka, T. Komatsuzaki, H. Noji, H. Imamura, Sci. Rep. 2014, 4, 6522.
- 28B. D. Bennett, E. H. Kimball, M. Gao, R. Osterhout, S. J. Van Dien, J. D. Rabinowitz, Nat. Chem. Biol. 2009, 5, 593–599.
- 29M. Pannier, S. Veit, A. Godt, G. Jeschke, H. W. Spiess, J. Magn. Reson. 2011, 213, 316–325.
- 30A. Doll, M. Qi, N. Wili, S. Pribitzer, A. Godt, G. Jeschke, J. Magn. Reson. 2015, 259, 153–162.
- 31D. Banerjee, H. Yagi, T. Huber, G. Otting, D. Goldfarb, J. Phys. Chem. Lett. 2012, 3, 157–160.
- 32H. Y. Vincent Ching, P. Demay-Drouhard, H. C. Bertrand, C. Policar, L. C. Tabares, S. Un, Phys. Chem. Chem. Phys. 2015, 17, 23368–23377.
- 33G. Jeschke, V. Chechik, P. Ionita, A. Godt, H. Zimmermann, J. Banham, C. R. Timmel, D. Hilger, H. Jung, Appl. Magn. Reson. 2006, 30, 473–498.
- 34M. S. Strycharska, E. Arias-Palomo, A. Y. Lyubimov, J. P. Erzberger, V. L. O'Shea, C. J. Bustamante, J. M. Berger, Mol. Cell 2013, 52, 844–854.
- 35C. Pliotas, R. Ward, E. Branigan, A. Rasmussen, G. Hagelueken, H. Huang, S. S. Black, I. R. Booth, O. Schiemann, J. H. Naismith, Proc. Natl. Acad. Sci. USA 2012, 109, E 2675-82.
- 36S. Valera, K. Ackermann, C. Pliotas, H. Huang, J. H. Naismith, B. E. Bode, Chem. Eur. J. 2016, 22, 4700–4703.
- 37K. Takegoshi, S. Nakamura, T. Terao, J. Chem. Phys. 2003, 118, 2325.
- 38T. Wiegand, C. Gardiennet, F. Ravotti, A. Bazin, B. Kunert, D. Lacabanne, R. Cadalbert, P. Güntert, L. Terradot, A. Böckmann, et al., Biomol. NMR Assign. 2015, 1–11.
- 39M. P. Williamson, Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16.
- 40T. A. Khrustaleva, Adv. Bioinf. 2014, 2014, 1–14.