Cover Picture: A Suite of Solid-State NMR Experiments for RNA Intranucleotide Resonance Assignment in a 21 kDa Protein–RNA Complex (Angew. Chem. Int. Ed. 38/2013)
Graphical Abstract
Resonance assignment is the first step of RNA structure determination by NMR spectroscopy. In their Communication on page 9996 ff., T. Carlomagno and co-workers present a suite of solid-state NMR experiments for RNA intranucleotide resonance assignment. These experiments open the way to structural studies of RNA by solid-state NMR spectroscopy.
Resonance assignment is the first step of RNA structure determination by NMR spectroscopy. In their Communication on page 9996 ff., T. Carlomagno and co-workers present a suite of solid-state NMR experiments for RNA intranucleotide resonance assignment. These experiments open the way to structural studies of RNA by solid-state NMR spectroscopy.
Nanoscale Assemblies
In their Communication on page 9932 ff., R. E. P. Winpenny et al. describe assemblies of polymetallic components that are linked together, such as six octametallic rings surround a dodecametallic central ring.1
Anion–π Interactions
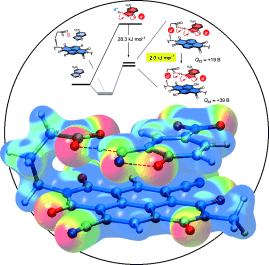
S. Matile et al. provide experimental evidence in their Communication on page 9940 ff. that anion–π interactions can contribute to catalysis. They also use simulations to show how the negative charge slides over the π-acidic surface of the catalyst.1
Actuators
When subjected to a light stimulus, certain crystals jump to release the strain that accumulates in their interior as a result of a photochemical reaction, as P. Naumov, E. V. Boldyreva, et al. describe in their Communication on page 9990 ff.1





