Molecular Basis for Two Stereoselective Diels–Alderases that Produce Decalin Skeletons**
A previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.1101/2021.02.01.429105).
Abstract
Enzymes catalyzing [4+2] cycloaddition have attracted increasing attention because of their key roles in natural product biosynthesis. Here, we solved the X-ray crystal structures of a pair of decalin synthases, Fsa2 and Phm7, that catalyze intramolecular [4+2] cycloadditions to form enantiomeric decalin scaffolds during biosynthesis of the HIV-1 integrase inhibitor equisetin and its stereochemical opposite, phomasetin. Computational modeling, using molecular dynamics simulations as well as quantum chemical calculations, demonstrates that the reactions proceed through synergetic conformational constraints assuring transition state-like substrates folds and their stabilization by specific protein-substrate interactions. Site-directed mutagenesis experiments verified the binding models. Intriguingly, the flexibility of bound substrates is largely different in two enzymes, suggesting the distinctive mechanism of dynamics regulation behind these stereoselective reactions. The proposed reaction mechanism herein deepens the basic understanding how these enzymes work but also provides a guiding principle to create artificial enzymes.
Introduction
Molecular chirality, discovered by Louis Pasteur in the middle of the 19th century,1 is found in most primary and secondary metabolites. Particularly, the so-called natural products are rich in chiral centers.2 The stereochemistry of natural products is strictly recognized in living organisms, and is thus closely related to their biological functions. Enzymes that create complex carbon frameworks with multiple chiral centers, such as polyketide synthase (PKS) and terpene cyclase, are gaining increasing attention not only in natural product chemistry but also in the chemical industry.3 Enzymes catalyzing [4+2] cycloadditions, or the Diels–Alder reactions, referred to as Diels–Alderases in some cases, form two C−C bonds and up to four chiral centers to generate a cyclohexene from a conjugated diene and substituted alkene; these enzymes play key roles in controlling stereochemistry during the formation of polycyclic structures (Figure S1).4 Since the discovery of SpnF, the first monofunctional enzyme reported to catalyze a [4+2] cycloaddition,5 many enzymes have been identified in the biosynthetic pathways of bacterial, fungal, and plant origins. Unlike PKSs, nonribosomal peptide synthetases (NRPSs), and terpene cyclases, which share active site-containing domain structures involved in their specific functions,3b, 6 DAases have no common structural features and are derived from distinct progenitor enzymes or proteins. SpnF, which catalyzes DA reaction in the spinosyn A biosynthesis,5 contains S-adenosylmethionine (SAM), and its overall structure belongs to the SAM-dependent methyltransferase family,7 and the overall structures of PyrI4 and PyrE3, which are involved in the pyrroindomycin biosynthesis,8 are very similar to those of lipocalin family proteins and FAD-dependent monooxygenases, respectively.9 Although such enzymes evolved independently from their own progenitors, they all exhibit high stereoselectivity and catalytic efficiency. The molecular basis of this emerging group of enzymes is gradually being revealed by genetic, biochemical and structural analyses, in combination with computational investigations.10 However, the mechanisms underlying the remarkable features of naturally occurring Diels–Alderases, such as the origin of stereoselectivity and their performance as catalysts, have remained elusive.
Fsa2-family decalin synthases (DSs) found in filamentous fungi11 catalyze stereoselective [4+2] cycloaddition during the biosynthesis of decalin-containing pyrrolidin-2-ones (DPs), which exhibit various biological activities.12 This class of molecules includes the HIV-1 integrase inhibitors equisetin (1) and phomasetin (2)13 and the telomerase inhibitor UCS1025A14 (Figure S2). It is noteworthy that all the six stereocenters of 1 and 2 are opposite. The first one at C6 is introduced by the action of a highly reducing, iterative type I PKS module of PKS/NRPS hybrid enzyme in collaboration with a trans-acting enoyl reductase. The second one at C5′ is derived from an amino acid precursor, l/d-serine, likely determined by a NRPS module of the hybrid enzyme. The remaining four chiral centers at C2, C3, C8, and C11, are installed by Fsa2 and its homolog Phm7 via intramolecular [4+2] cycloaddition to yield enantiomeric decalin scaffolds from similar linear polyenoyl tetramic acids (e.g., 3 and 4)11, 15 (Figure 1 a). We have shown that replacement of phm7 in a 2-producing fungus with fsa2 resulted in the production of a 1-type decalin scaffold (2S,3R,8S,11R),15 indicating that these enzymes determine the stereochemistry of the decalin scaffold during DP biosynthesis. Another homologous enzyme, MycB,16 also produces a 1-type decalin scaffold, and the 2-type decalin scaffold is produced by CghA17 and UscH,18 which are involved in the biosynthesis of Sch 210 972 and UCS1025A, respectively. Interestingly, but not surprisingly, another decalin scaffold (2R,3S,8S,11R) is produced by PvhB, which is involved in the varicidin A biosynthesis19 (Figure S2). Thus, structural comparisons of the DSs that differ in function, i.e., stereochemical output, provide insight into the mechanisms of the stereoselective [4+2] cycloaddition.
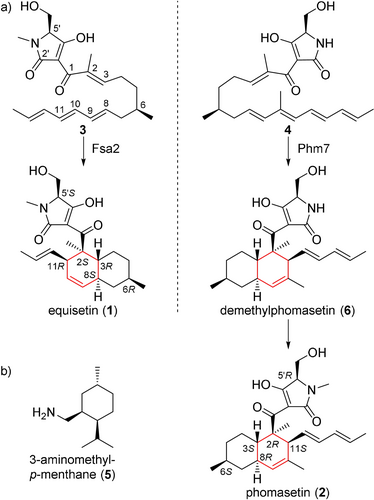
a) The [4+2] cycloadditions catalyzed by Fsa2 and Phm7 to form enantiomeric decalin scaffolds. Linear tetraenoyl tetramic acid 3 was chemically synthesized and confirmed to be a Fsa2 substrate to form 1,20 whereas pentaenoyl tetramic acid 4 is a likely Phm7 substrate to yield N-demethylphomasetin (6), which is further converted to 2. b) A decalin synthase inhibitor, 3-aminomethyl-p-menthane (5).
In this study, we revealed the molecular basis of two stereoselective enzymes, Fsa2 and Phm7, that catalyze [4+2] cycloaddition to form enantiomeric decalin scaffolds. X-ray crystal structures of substrate-free Fsa2 and Phm7, and Phm7 bound to an inhibitor (hereafter referred to as inhibitor-bound Phm7), were determined at 2.17, 1.62, and 1.61-Å resolution, respectively. The substrate-bound poses were modelled using docking simulations followed by all-atom molecular dynamics (MD) simulations. We employed the generalized replica-exchange with solute tempering (gREST) method,21 which allows extensive sampling of possible binding poses of the substrates.22 Site-directed mutagenesis studies were performed to verify the binding models for stereoselective synthesis and examine the amino acid residues involved in substrate interactions. Density functional theory (DFT) calculations were performed to reveal the reaction mechanism in detail, particularly the stereoselectivity and rate acceleration by Phm7. This powerful combination of experimental methods and calculations, which we used to investigate two enzymes that produce enantiomeric decalin scaffolds, provides insight into the molecular mechanism of enzyme-mediated [4+2] cycloaddition and its stereoselectivity.
Results
Structure Analyses of Fsa2, Phm7, and Inhibitor-Bound Phm7
The crystal structure of Fsa2, which produces the 2S,3R,8S,11R decalin scaffold found in equisetin (1, Figure 1 a), has a β-sandwich and a β-barrel domain at the N- and C-termini, respectively (Figure 2 a). The structures of both domains exhibit structural similarity with those of lipocalin family proteins that bind heme, steroids, and other hydrophobic ligands in the pocket located in their β structures.23 Interestingly, however, Fsa2 does not have a pocket in either the N- or C-domains (Figure S3). Instead, a large pocket is present between the two domains. Considering the likely substrate structure and volume, we speculate that the large cavity created by the two domains is an active- and substrate-binding site. Phm7 catalyzes intramolecular [4+2] cycloaddition to produce the enantiomeric decalin scaffold (Figure 1 b). Despite their distinct stereoselectivity and low sequence similarity (36 % sequence identity), the crystal structure of Phm7 is similar to that of Fsa2 (RMSD=0.849 Å), and the shape and volume of their large pockets are also similar (Figure 2 b).
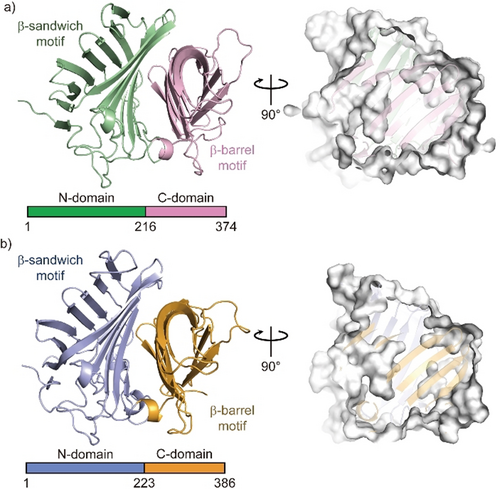
Crystal structures of Fsa2 (a) and Phm7 (b). Both DSs consist of two domains: N-domains for residues 1–215 of Fsa2 and 1–222 of Phm7, and C-domains for residues 216–374 and 223–386. Surface models (right panels) show shapes of the pocket between the two domains of both enzymes.
To probe the active site of the DSs, we performed Phm7 ligand screening using microscale thermophoresis (MST)24 (Figure S4). We found that 3-aminomethyl-p-menthane (5, Figure 1 b), which was similar to the phomasetin (2) substructure, dose-dependently inhibited Phm7 activity in vitro and production of 2 in the fungus Pyrenochaetopsis sp. RK10-F058 (Figure S5). Compound 5 also inhibited equisetin (1) production in the fungus Fusarium sp. FN080326 (Figure S6). We next performed co-crystallization of Phm7 and 5 and obtained the crystal structure of the inhibitor-bound Phm7 at 1.61-Å resolution (Figure S7). The inhibitor was located on the lower side of the pocket and formed hydrophobic interactions with Y178, W223, F226, L245, and L381. The amino group of 5 was surrounded by Y68, E51, and Y178. Because 5 can bind to both Phm7 and Fsa2 to inhibit their functions (Figures S4–S6), the lower side of the pocket between the two domains would form the active site of these enzymes. Indeed, CghA, exhibiting the same stereoselectivity as Phm7 (Figure S2),17 was recently reported to have two β domains, as in Phm7 and Fsa2.25 The product binding site is almost the same as the 5-binding site of Phm7.
Docking and MD Simulations for Binding Pose Determination
To determine whether the pocket would be large enough to accommodate the substrates, substrate 4 was docked into the crystal structure of the inhibitor-bound form of Phm7 using AutoDock Vina. Substrate 4 fits within the pocket in various binding poses, including a folded form in which a U-shaped folded alkyl chain including asymmetric C6 of 4 was located at the lower side of the pocket where 5 can bind, and the tetramic acid and polyene moieties were extended into the inner upper part of the pocket (Figure S8a). Likewise, folded substrate 3 fits within the pocket of Fsa2 (Figure S8b). These docking simulations using the crystal structures indicated that the pocket between the N- and C-domains of Phm7 and Fsa2 has enough room for binding of the folded substrates.
To explore the binding poses of the substrates and their dynamics in the pockets, we carried out all-atom MD simulations using the gREST method,21 which extensively samples possible binding poses otherwise elusive in conventional simulations. A variety of poses were obtained (see Movies S1 and S2 for collected binding poses of Phm7 and Fsa2, respectively), and clustering analysis of the simulation trajectories resulted in four major bound poses, including “folded” and “extended” conformations, for both 4 and 3 (Figure S9). In Figure 3, the major cluster of Phm7 (63 % of Phm7, pA in Figure S9a) shows a well-defined bound pose for 4. In this cluster, the tetramic acid moiety and polyene tail of the folded poses were located at the upper front and back of the pocket, respectively, whereas the U-shaped part was found at the lower side of the pocket. Similar tetramic acid-front and polyene-back poses were also found in the folded conformations in Fsa2 (33 % of Fsa2, fA in Figure S9b), although bound 3 fluctuated in the pocket to a greater extent than in Phm7 (Figure 3 b). In both Phm7 and Fsa2, the electrostatic potential inside the pocket had a large negative value as it moved deeper into the pocket (Figure S10). The bottom surface of the pocket was significantly hydrophobic, whereas the hydrophilic surface was found in the upper wall of the pocket (Figure S11). These inhomogeneous electrostatic, hydrophobic, and hydrophilic environments of the pocket coincide with the common tetramic acid-front and polyene-back orientation.
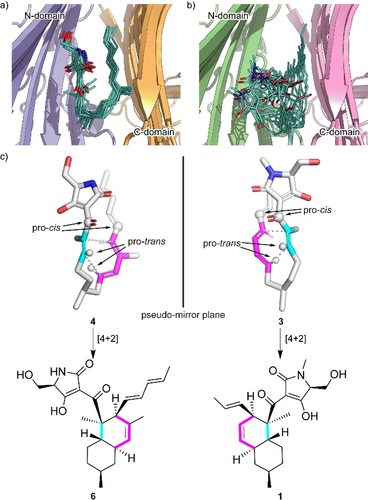
Predicted binding model. a,b) A collective view of 12 representative snapshots taken from the top 70 % of the main folded clusters (chosen based on the RMSD values from the cluster center in ascending order) for Phm7 (a) and Fsa2 (b), respectively. c) Conformations of the representative poses of 4 (left) and 3 (right) in the pockets. The diene and dienophile moieties of the substrate are indicated in pink and cyan, respectively. The corresponding cycloadducts are also shown.
The representative folded conformation of the major cluster of Phm7, which dominates the cluster (Figure S12a), explains the configuration of the stereoselective cycloadduct with the 2R,3S,8R,11S decalin scaffold (Figure 3 c). In contrast to Phm7, the pocket of Fsa2 allows various “folded” conformations of 3 (Figure S12b). Nevertheless, a certain number of poses are consistent with the configuration of the 1-type (2S,3R,8S,11R) decalin scaffold (Figure 3 c). Comparison of the carbon chain conformation (C-C-C-C dihedral angles) between substrates 3 and 4 revealed that the diene and dienophile moieties exhibit pseudo-enantiomeric conformations, consistent with the stereochemical relationship of the transition state structures corresponding to decalin scaffolds of 1 and 2 (Figure S13). The LigPlot analysis of the representative pose of 4 shows that the tetramic acid moiety forms hydrogen bonds with the side chains of E51, N84, and K356, and with the main chain carbonyl of G64. The U-shaped part and polyene tail of the substrate fit with the hydrophobic region of the pocket lined by L49, W223, Y232, L245, W342, F377, L381, and I383 (Figure S14a). Similarly, the hydrophobic U-shaped part and polyene tail of 3 are surrounded by many hydrophobic side chains of Fsa2 (W45, V169, Y171, W216, Y225, and M238), and the tetramic acid moiety is hydrogen-bonded with N346 (Figure S14b). Therefore, MD simulations based on the crystal structures of the two enzymes suggested distinct substrate-enzyme interactions and their resultant substrate poses corresponding to the enantiomeric decalin scaffolds.
Functional Analyses of Phm7 and Fsa2
To examine the substrate-enzyme interactions predicted from the MD simulations, we first established an in vitro enzyme assay system using cell lysates prepared from fungal mycelia lacking the phm7 gene.15 The cell lysates were directly incubated with Phm7 or Fsa2, and their reaction products were analyzed by liquid chromatography/electrospray ionization mass spectrometry (LC/ESI-MS). Linear polyenoyl tetramic acid 4 was less abundant in the presence of the enzymes, and the expected products of Phm7 and Fsa2, N-demethylphomasetin (6) and its derivative containing 1-type trans-decalin (7), respectively, were formed (Figure 4 a,b). The reaction selectivity of Phm7 and Fsa2 shown in the in vitro assay was consistent with that observed in the producer fungus and a mutant in which phm7 was replaced with fsa2.15 Furthermore, we observed no significant difference in the amount of cis-decalin containing derivative 8 between the presence and absence of enzyme. The time course of the in vitro reaction confirmed the linear formation of product 6 for the first several minutes, and no cycloaddition in the absence of enzyme under the conditions tested, suggesting that 8 detected in the reaction mixtures were present in the cell lysates from the beginning and likely to be formed via Phm7-independet manner in the fungal cells during 3-day culture (Figure S15). These results indicated that products 6 and 7 were exclusively formed by the action of Phm7 and Fsa2, respectively, from the linear polyene substrate found in the fungal cell lysate. Therefore, the in vitro assay using the fungal cell lysate allowed us to evaluate the enzyme activities by measuring the products formed.
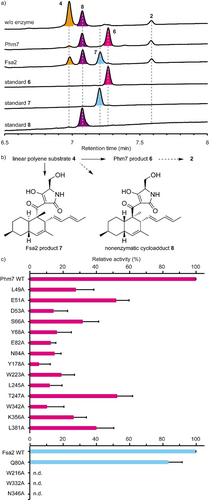
In vitro analysis of Ala-substituted Phm7 and Fsa2 mutants. a) UPLC traces of the in vitro Phm7 and Fsa2 reaction products. The cell lysates prepared from the Δphm7 mycelia were incubated with Phm7 and Fsa2 and analyzed by LC/ESI-MS. UV detection was carried out at 290 nm. b) Linear polyenoyl tetramic acid 4 was converted into 6 and 7 by Phm7 and Fsa2, respectively. Conversions indicated by broken arrows were not detected under the conditions tested: non-enzymatic formation of 8 from 4 was not observed in vitro, and sinefungin added in the reaction buffer inhibited N-methylation of 6. c) Comparison of the enzyme activities of the Ala-substituted mutants with those of the wild-type Phm7 and Fsa2. Data are the mean and error bars represent the standard deviation of four independent experiments. n.d., not detected.
Using the in vitro assay system, we carried out site-directed mutagenesis studies of Phm7 and Fsa2 to validate the substrate binding modes predicted by the MD simulations (Figures S14, S16). The key amino acid residues that interact with the tetramic acid moieties (K356 in Phm7 and N346 in Fsa2), the hydrophobic U-shaped parts (W223 in Phm7 and W216 in Fsa2), and the polyene tails (W342 in Phm7 and W332 in Fsa2) were substituted with Ala, and their enzyme activities were compared with those of the wild-type enzymes. Ala substitutions of these amino acid residues significantly decreased enzyme activity in vitro (Figure 4 c), indicating that the pockets between the N- and C-domains of Phm7 and Fsa2 were the active sites of both enzymes, and that the proposed binding modes were reliable.
Next, we focused on Phm7 to demonstrate the binding modes. We introduced additional single Ala substitutions into the amino acid residues close to substrate 4 (Figure S16a,b), and performed the enzyme assay. Ala substitutions of the residues in proximity to the tetramic acid moiety of 4 (Figure S17) significantly decreased the enzyme activity (Figure 4 c). In particular, we observed a marked decrease in the activity of mutants harboring substitutions in hydrophilic residues such as D53 and E82, suggesting that the hydrophilic upper wall of the pocket (Figure S10) was indeed responsible for trapping the tetramic acid moiety of 4, as predicted by the MD simulations. We also expected that the hydrogen bond donations from K356 to the carbonyl oxygen at C1, as well as those of E51 and N84 to the tetramic acid moiety, would activate the dienophile to accelerate the reaction. Ala substitution at Y178 and W223 also significantly decreased enzyme activity (Figure 4 c). A230, L245, and T247 are located near the substrate but do not interact directly with the substrate in the MD model. Substitution of these residues with smaller ones (L245V and T247A) yielded mutants that retained considerable activity, whereas substitution with longer and larger ones (A230F and T247F) significantly decreased activity, probably due to steric clash (Figure 4 c and Figure S17). These results confirmed that the 5-binding site formed by amino acid restudies such as Y178, W223, L245, and T247 is the reaction chamber in which the stereospecific [4+2] cycloaddition occurred.
To further validate the substrate-enzyme interactions in Phm7, we examined the effects of Ala substitutions of Phm7 on phomasetin (2) production in the producer fungus, in which the wild-type phm7 was replaced with the mutant genes. Phm7 Y68A and W223A mutants produced significantly lower levels of 2 and suppressed production of 8 in the fungus (Figure S18). In addition, a new peak 9 was detected in the culture extract of the W342A mutant. Structural analyses by NMR and MS revealed that 9 was a derivative of 2 with a hydroxy group at the terminus of polyene (Figure S18; see also the Supporting Information for structure determination of 9). Production of 9 in this gain-of-function mutant supported the role of W342 in the interaction with the polyene tail of 4. Taken together, our in-depth analyses of Phm7 mutants using an in vitro enzyme assay and in vivo production in fungus demonstrated that the MD simulation-based binding models were reliable, and that the predicted amino acid residues are involved in substrate binding.
DFT Calculations for the Molecular Mechanism in the Phm7 Pocket
Given that Phm7 promotes the intramolecular [4+2] cycloaddition of substrate 4 in a stereoselective manner, the detailed molecular mechanism in this event with Phm7 is of great importance. Hence, we focused on how Phm7 determined the stereoselectivity of the cycloaddition, as well as whether this enzyme can accelerate (i.e., catalyze) the reaction. MD simulations pointed out that the structure of 4 seemed to be convergent in the major folded conformations in the pocket of Phm7 (Figure S12a). The geometry between the dienophile and diene moieties in this “folded” structure is close to the conformation that would afford the decalin scaffold with the same configuration as Phm7 product 6 (Figure 3 c). We investigated the intrinsic stereoselectivity of the uncatalyzed [4+2] cycloaddition of 4 using DFT calculations at the M06-2X/6-311+G** (scrf=CPCM, water) level of theory (Figure 5 a). Among the four transition states to the corresponding decalin derivatives, TS1a, which affords 8, is located in the lowest energy state (ΔG≠ +16.1 kcal mol−1) relative to the linear conformation, whereas TS1b, which affords 6, requires slightly higher activation energy (ΔG≠ +16.5 kcal mol−1). The reactions that afford Fsa2-type product 7 and another cis-decalin derivative were found to be less feasible via TS1c and TS1d, respectively. These computational results suggested that the cycloaddition without any steric bias should give a mixture of 8 and 6, whereas 7 might also be present as a minor component. Thus, the combined experimental and theoretical results indicated that efficient folding of the substrate in the Phm7 pocket plays a pivotal role as a major determinant of stereoselectivity.
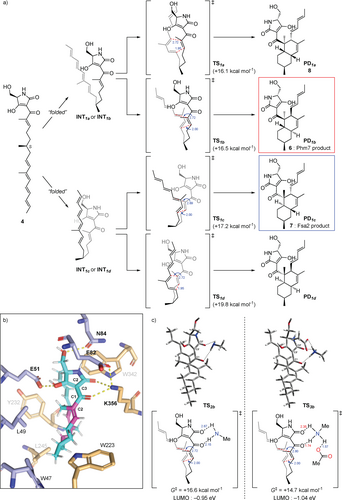
The DFT calculations for the reaction mechanism of the uncatalyzed and hydrogen bond-catalyzed [4+2] cycloaddition of 4. a) Reaction pathways for the uncatalyzed cycloaddition of substrate 4 to produce four types of decalin stereoisomers. The energy changes and bond lengths were calculated at the M06-2X/6-311+G** (scrf=CPCM, water) level of theory are shown in kcal mol−1 and Å, respectively. b) Illustration of the hydrogen bond network of 4 with amino acid residues in the Phm7 pocket obtained from the MD simulation (cf. Figure S14a). c) Transition state structures to give 6 with methylamine (TS2b) and both methylamine and acetic acid (TS3b) as models of the hydrogen bonding with K356 and E82 residues.
Next, to gain some insight into the rate acceleration mechanism, we investigated the amino acid residues that interact with 4 in the Phm7 pocket. In particular, we focused on the polar amino acid residues that trap the tetramic acid moiety in the upper part of the pocket. Similar to Lewis acid coordination, efficient hydrogen bond donation to the carbonyl group adjacent to the dienophile should lower the lowest unoccupied molecular orbital (LUMO) energy of the dienophile and facilitate the Diels–Alder reaction.26 From this point of view, the amino moiety of K356 is a hydrogen bond donor to the corresponding carbonyl group at the C1 of folded 4 (Figure 5 b). DFT calculations at the same level of theory were performed to track the course of the cycloaddition of 4 with methylamine as a model of the K356 residue. However, no significant acceleration was observed, and the activation barrier for TS2b was +16.6 kcal mol−1 relative to the folded conformation. In fact, no efficient decrease in LUMO energy was observed (−0.90 eV for TS1b vs. −0.95 eV for TS2b, Figure 5 a,c). Taking a closer look at the binding model, we discerned that the amino moiety of K356 makes another hydrogen bond with the carboxy group of E82 to form an ammonium architecture (Figure 5 b). We carried out DFT calculations by incorporating methylamine and acetic acid as models to reproduce the hydrogen bond network (i.e., 4-K356-E82).10a The modelled network provides a tight hydrogen bond on the carbonyl oxygen at the C1 of 4 (1.74 Å) due to the acidified proton of the amino group. Furthermore, because the amino protons became more electron-deficient, an additional weak hydrogen bond26 (2.35 Å) formed with the carbonyl oxygen at C2′. Therefore, the dihedral angle of the C1 and C2′ carbonyl groups in TS3b (25.4°) became much smaller than that of TS2b (33.9°), making the conjugation between the enone dienophile and tetramic acid moiety more efficient. Thus, both electronic and structural perturbation by the hydrogen bond network decreased the LUMO energy to −1.04 eV in TS3b, and the reaction was greatly facilitated by lowering the activation barrier for TS3b (+14.7 kcal mol−1). These results explain the marked decrease in the enzymatic activity of the E82A mutant (Figure 4 c). In addition, the direct incorporation of the carboxy group of E82 in hydrogen bonding with 4 in the absence of K356, which was also reproduced by the DFT calculation model (Figure S19), could explain why the Ala substitution of K356 did not cause a complete loss of activity. Computation revealed that other hydrogen bonds between the tetramic acid moiety and the amino acids, such as E51 and N84, did not lower the LUMO energy level (Figure S20), and should be devoted to the appropriate positioning of substrate 4 in the Phm7 pocket.
In sum, the triad of experiments, MD simulations, and DFT calculations revealed the molecular mechanism of the Phm7 pocket as the catalyst for stereoselective [4+2] cycloaddition. Key findings are as follows: (i) although the intrinsic selectivity of the reaction of 4 prefers the formation of 8 in the absence of the enzyme, the folding structure in the pocket defines the selective formation of 6; (ii) the sophisticated hydrogen bond network derived from K356 and E82, namely, Brønsted acid-activated hydrogen bonding catalysis, promotes the cycloaddition event very efficiently; and (iii) some polar amino acid residues guide the substrate to the opportune folding structure.
Discussion
Since the first report of direct evidence in biological Diels–Alder reaction in solanapyrones biosynthesis and crystal structure of macrophomate synthase,27 many enzymes catalyzing [4+2] cycloaddition have been discovered, and their crystal structures have been determined.7, 9, 10b, 28 Their structures tell us that these enzymes were derived from ones with distinct functions other than catalyzing the cycloadditions, implying that their active sites were subsequently converted into those for the cycloaddition. By sharp contrast, DSs do not use the ligand-binding site of lipocalin family protein as an active site for the [4+2] cycloaddition. Fusion of two lipocalin-family proteins generated two-β-domain proteins with a pocket between the two domains. By burying their pockets in the β-domains, these proteins then acquired the ability to bind unstable linear polyenoyl tetramic acids using the relatively relaxed pocket, and eventually catalyze the cycloaddition. A protein from Nitrosomonas europaea, NE1406, has a tertiary structure very similar to those of DSs.29 This protein, which consists of two lipocalin-like domains and has several small pockets in and between the two β-domains (Figure S21), is likely to be involved in the carotenoid biosynthesis. From the standpoint of molecular evolution, nature has utilized the fusion (or duplication) of lipocalin proteins to generate new functions in a pocket between two domains.
Several amino acid residues are conserved among the DSs whose functions have been characterized (Figure S22). Many of them are located inside the molecule and likely play roles in maintaining enzyme structure rather than in catalysis per se. Among the amino acid residues lining the active site pocket, only Trp at the bottom of the pocket (W223 in Phm7 and W216 in Fsa2) is highly conserved, suggesting that it plays a key role in the cycloaddition. Indeed, MD simulations predicted that the W223 and W216 form hydrophobic interactions with substrates 4 and 3, respectively (Figure S14c). The substitutions of the conserved Trp with Ala substantially retarded the enzyme activity for both Phm7 and Fsa2. Although the sequence homology among the DSs is limited, they seem to share amino acid distributions in the pockets between the two domains (Figure S23). It is likely that substrates bind to the pocket in a similar way (tetramic acid-front and polyene-back orientation), allowing stereoselective [4+2] cycloadditions to proceed. Considering that the substrate conformation in the enzyme pocket is correlated to the product stereochemistry, we propose a model of how DSs produce decalin scaffolds with four possible configurations via the cycloaddition (Figure 6). The conformations of TS1b and TS1c are enantiomeric to each other, except for the C6 methyl group (Figure 5 a), and all the C-C-C-C dihedral angels of the carbon chains are opposite (Figure S13). This can be described as a difference in the manner of folding the linear substrate, i.e., the linear substrate 3 is folded clockwise and 4 counter-clockwise to yield (pseudo)enantiomeric conformations in the enzyme pocket (Figure 3 c). When the folded substrate 3 is flipped horizontally, it binds to the Fsa2 pocket in the tetramic acid-front and polyene-back orientation. A key difference between the transition state structures for trans and cis-decalin scaffolds is the rotation at the C7-C8 bond. Thus, the substrate conformation in the enzyme pocket can be predicted for cis-decalins, such as varicidin A (2R,3S,8S,11R, Figure S2) and even an unidentified metabolite (2S,3R,8R,11S). Recent phylogenetic analysis suggested that approximately 100 sequences are potentially involved in DP biosynthesis.30 Therefore, this group of enzymes, which is widespread in Ascomycota fungi, are attractive targets for structure-function relationship studies, providing key structural features for determining the stereoselectivity of these reactions.
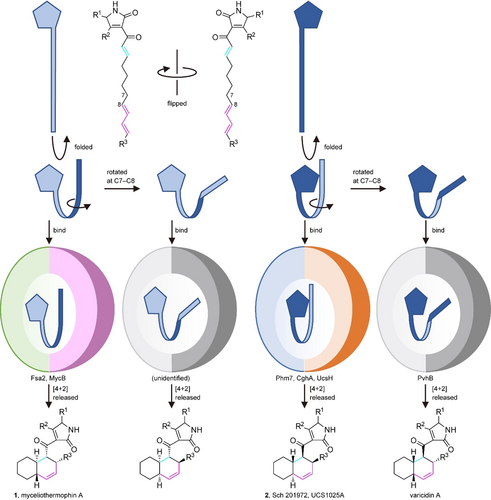
A model of the formation of four diastereomeric decalin scaffolds by decalin synthases. Linear polyenoyl tetramic acid substrates are shown schematically as a pentagon and a ribbon. Folded substrates bind to corresponding DSs in the tetramic acid-front and polyene-back manner, and are preorganized in the pocket. The substrates only with productive conformations are selected and undergo the reaction to produce respective decalin scaffolds.
The MD simulations showed that the pockets between the N- and C-domains of DSs were large enough to accommodate the substrates in various conformations. The “folded” cluster of substrate 4 contains a dominant pose (80 %) with an s-cis conformation at C9-C10, explaining the expected decalin configurations (Figure S24). The calculated C-C-C-C dihedral angles along the carbon chain of the dominant pose takes a single orientation at all the rotational bonds, and no pose corresponding to the other possible decalin configurations was observed. This indicates that the Phm7 pocket robustly regulates the conformation of the substrate to stabilize the folded pose with a specific conformation. On the contrary, substrate 3 in the Fsa2 pocket exhibits a rather large variation in folded poses (Figure S12b). This is likely due to the fact that Fsa2 binds a smaller substrate (the volumes of 3 and 4 are 379 Å3 and 402 Å3, respectively) in a larger pocket relative to Phm7 (the pocket size is approximately 150 Å3 larger than that of Phm7, Figure S25). In addition, unlike Phm7, which tightly holds the tetramic acid moiety by multiple hydrogen bonds, there is no hydrophilic amino acid residue in proximity to the C1 carbonyl, and only N346 is involved in hydrogen bonding with 3. Fsa2 likely retains the substrate affinity to the enzyme by accommodating various folded conformations and minimizing entropy loss upon binding. This enzyme still produces a reactive conformation for the cycloaddition with a desired stereochemistry. Our preliminary analysis suggests that both of the hydrogen bonding of substrate 3 with N346 and the indole ring orientation of W216 play key roles in stabilizing the reactive conformation (Figure S26). Taken together, both the regulation of substrate conformation and the chemical catalysis are likely important for the Phm7 enzyme, while neither of them seems to dominate the Fsa2 catalysis. Fsa2 possibly adopt a dynamic control of the reaction rate using fluctuations in the substrate structure. Further investigation on the enzymatic reaction mechanism, including activation free energies, using a hybrid quantum mechanics (QM)/ molecular mechanics (MM) method, should fill in the missing piece of the catalytic mechanisms. Given the structural similarity between Phm7 and Fsa2 and their opposite selectivity in the [4+2] cycloaddition, clarification of the detailed mechanism would be of great significance, and is the subject of ongoing research in our group.
Conclusion
Our investigations combining experimental and theoretical approaches highlighted the distinct molecular mechanisms underlying Phm7- and Fsa2-catalysed [4+2] cycloaddition. The folding of substrate 4 in the Phm7 pocket and 3 in the Fsa2 pocket, predicted by the gREST method based on X-ray crystal structures, exhibited stereoselectivity in the construction of decalin scaffolds. The results of site-directed mutagenesis studies and DFT calculations clarified how the hydrophilic amino acid residues in the Phm7 pocket regulate and catalyze the stereoselective cycloaddition. In addition, our results raise questions about the molecular tactics adopted by Fsa2, which should be addressed in future research: what determines the transition state from the flexible folded conformations in the pocket, and how dose Fsa2 accelerate the reaction without effective hydrogen bonding on the dienophile moiety?
Acknowledgements
We acknowledge the computational resources provided by the RIKEN Advanced Center for Computing and Communication (HOKUSAI GreatWave and BigWaterfall) and the HPCI system (Project ID: hp190181) for the MD simulations and the Research Center for Computational Science (Okazaki, Japan) for the DFT calculations. We also thank the beamline staff of BL26B1, BL41XU and BL32XU at SPring-8 (Hyogo, Japan) for assistance with the experiments. This work was supported by MEXT/KAKENHI (Grant Numbers 19H04665, 20K05872 (to NK), 19K12229 (to SR), 19K06992 (to RT), 20K15273 (to KW), 19H05645 (to YS), 20H00416 (ST), 19H04658, 19H05780 (to SN)), MEXT as “Priority Issue on Post-K computer” (Building Innovative Drug Discovery Infrastructure Through Functional Control of Biomolecular Systems), the Takeda Science Foundation (to RT), and the Naito Foundation (to RT), RIKEN Pioneering Research Projects (Dynamic Structural Biology/Glycolipidologue Initiative) (to YS), and the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) from the Agency for Medical Research and Development (AMED) under grant JP21am0101001 (to SN).
Conflict of interest
The authors declare no conflict of interest.




