A Liquid Chromatography with Tandem Mass Spectrometry-Based Proteomic Analysis of Primary Cultured Cells and Subcultured Cells Using Mouse Adipose-Derived Mesenchymal Stem Cells
Abstract
Adipose-derived mesenchymal stem cells (MSC-ATs) are representative cell sources for cell therapy. However, how cell stress resulting from passage influences the MSC-AT protein expression has been unclear. In this study, a protein expression analysis was performed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) using mouse primary cultured cells (P0) and cells passaged three times (P3) as samples. A total of 256 proteins were classified as cellular process-related proteins, while 179 were classified as metabolic process-related proteins in P0. These were considered to be adaptive responses of the cells to an in vitro environment. However, seven proteins of growth were identified (Csf1, App, Adam15, Alcam, Tbl1xr1, Ninj1, and Sbds) in P0. In addition, four proteins of antioxidant activity were also identified (Srxn1, Txndc17, Fam213b, and Apoe) in P0. We identified 1139 proteins expressed in both P0 and P3 cells that had their expression decreased to 69.4% in P3 cells compared with P0 cells, but 1139 proteins are very likely proteins that are derived from MSC-AT. The function of MSC-ATs was maintained after three passages. However, the LC-MS/MS analysis data showed that the protein expression was degraded after three passages. MSC-ATs retained about 70% of their protein expression ability in P3 cells.
1. Introduction
Mesenchymal stromal stem cells (MSCs) are considered to have the ability to differentiate into mesenchymal cells, such as osteoblasts, adipocytes, muscle cells, and chondrocytes [1, 2]. These cells are also expected to have an immunosuppressive effect and are regarded as promising cellular therapeutic agents for immunological diseases resistant to treatment. MSCs have been established from various tissues (umbilical cord blood, placenta, adipose tissue, etc.), among which adipose tissue contains a particularly large amount of cells.
Clinical research and treatment using adipose-derived mesenchymal stem cells (MSC-ATs) [3] is already underway in many medical institutions around the world [4]. The clinical practical application of islet cell transplantation therapy was reported in 2000 in the Edmonton protocol [5] and many subsequent papers [6-10]. The technology of islet transplantation is thought to be useful for the processing of therapeutic cells using MSC-ATs. We recently reported that the University of Wisconsin (UW) [11] organ preservation solution has a better cell survival/proliferation ability than Hank’s balanced salt solution (HBSS) [12]. There is a possibility that the adipose tissue collected through the patient’s skin may be infected with skin bacteria. One method of sterilizing tissues collected from a living body involves immersing and storing such tissue for 16 h using HBSS [13], which also contains antibiotics. After such storage, MSC-ATs can be isolated from adipose tissue. In addition, adipose tissue collected from a patient can be transported to a remote location.
It was recently reported that the stress of long-term culture of cells in vitro also occurs in stem cells, such as induced pluripotent stem (iPS) cells, causing DNA damage and cellular carcinogenesis [14]. Some researchers recommend reducing the number of passages of MSC-ATs to maintain the quality of primary cultured cells. However, MSC-ATs of primary cultured cells are reportedly contaminated with various types of cells, such as blood cells, through the process of cell isolation. This is because the stromal vascular fraction (SVF) [15] obtained when collecting MSC-ATs using centrifugation contains many kinds of cells (e.g., adipocytes, fibroblasts, smooth muscle cells, endothelial cells, blood cells, endothelial progenitor cells, preadipocytes, vascular progenitors, hematopoietic progenitors, and hematopoietic stem cells) [16, 17]. For MSC-ATs isolated from adipocytes collected from patients, the number of cells can be increased by increasing the number of passages. Because this processing can be done outside the body, the patient can thus obtain many of her/his own cells after undergoing only one procedure of fat collection surgery. However, it is also important to maintain the quality of the cells. Therefore, researchers and clinicians have fervently discussed how many passaging operations of clinical MSC-ATs should be performed.
With liquid chromatography (or high-performance liquid chromatography (HPLC)) with tandem mass spectrometry (LC-MS/MS), the components to be analyzed are separated by a liquid chromatograph (LC) and ionized via a dedicated interface (ion source) and the generated ions are then separated by MS. LC-MS/MS is an analytical technique that dissociates and fragments mass ions and detects them with MS [18]. Recently, an online LC-MS/MS system for quantitative proteomics based on data-dependent protein IDs and shotgun-based quantitative proteomics methods was developed [19-23] by connecting the measuring equipment for a protein analysis to a computer and linking to an online protein database. In this way, a comprehensive protein expression analysis can be performed by checking the peptide sequence data of the protein contained in the sample.
A comprehensive expression analysis of the protein expressed by the cell is important for accurately understanding the mechanism underlying the effect of cell therapy accompanying the administration of the culture supernatant of cells and the cells themselves. The present study was performed to identify the functional protein components in mouse MSC-ATs of primary cultured cells (P0) and mMSC-ATs passaged three times (P3) using LC-MS/MS. The proteins specifically contained in primary cultured cells were identified as those expressed only in the SVF derived from AT. By examining the proteins expressed in both P0 and P3 cells, we can identify the proteins expressed by MSC-ATs. Determining the protein component of MSC-ATs that exerts a therapeutic effect is expected to be useful for cell therapy in the future.
2. Materials and Methods
2.1. Reagents
Fetal bovine serum (FBS) was obtained from Biowest (Nuaille, France). DMEM (high glucose) with L-glutamine, phenol red, and sodium pyruvate was obtained from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan). Plastic dishes were obtained from TPP (Trasadingen, Switzerland). All other materials used were of the highest commercial grade.
2.2. Animal Care
All experimental protocols were in accordance with the guidelines for the care and use of laboratory animals set by Research Laboratory Center, Faculty of Medicine, and the Institute for Animal Experiments, Faculty of Medicine, University of the Ryukyus (Okinawa, Japan). The experimental protocol was approved by the Committee on Animal Experiments of University of the Ryukyus (permit number: A2017101). C57BL/6 male mice (8 weeks of age; Japan SLC, Shizuoka, Japan) were maintained under controlled temperature (23 ± 2°C) and light conditions (lights on from 08:30 to 20:30). Animals were fed standard rodent chow pellets with ad libitum access to water. All efforts were made to minimize the suffering of the animals.
2.3. Isolation of MSC-ATs from Mouse via the Inguinal Pad Fat
AT was obtained from the inguinal pad fat of three 8-week-old mice. The method of isolating MSC-ATs from AT was in accordance with the AT-derived stem cell product standard document (RMRC-A 01: 2015) of Ryukyus Regenerative Medicine Research Center. In brief, these ATs were stored in cold HBSS and washed vigorously using HBSS three times before starting digestion. Next, the tissues were cut into small fragments with a scalpel for enzymatic digestion (2 mg collagenase type IV/ml; HBSS) in 50 ml tubes (rotation speed: 20 × 37°C × 60 min) using the shaker (BioShaker BR-42FM; TAITEC, Saitama, Japan). These tubes were then centrifuged (800g) for 5 minutes. The SVF [24] containing various kinds of cells, including MSC-ATs, was confirmed at the bottom of the tube after centrifugation. The MSC-ATS were collected as a cell pellet and then washed with fresh DMEM containing 10% FBS to remove the enzyme after the digestion. The digested tissue was then incubated in a T25 flask.
All of the mouse studies were approved by the Institutional Animal Care and Use Committee of University of the Ryukyus.
2.4. Preparation of mMSC-ATs
Mouse MSC-ATs were cultured (37°C, 5% CO2) in an uncoated T25 flask (TPP 90026). The passage of cells was performed every 3 to 4 days after reaching 80% confluence following sowing. The cells were then washed with PBS (calcium, magnesium-free), and mouse MSC-ATs were dissociated using a dissociation solution (Trypsin/EDTA (Lonza CC-3232)). Subculturing was carried out by plating on an uncoated T25 flask. DMEM containing 10% FBS was used for the culture medium.
2.5. Flow Cytometry
Cell flow cytometry was performed as described previously [25], using specific antibodies for CD34, CD 44, CD45, and CD90.2.
2.6. Cell Differentiation
Adipogenic and osteogenic differentiations were performed as described previously [25].
2.7. Protein Identification by a Nano-LC-MS/MS Analysis
We used an EzRIPA Lysis kit (ATTO Corporation, Tokyo, Japan) for cell lysis according to the manufacturer’s instructions. A protein solution of 4493 μg/ml (P0) and 3105 μg/ml (P3) was obtained from mADSCs, and 6.0 μg protein was used for sample preparation. Finally, 0.4 μg of protein was used for nano-LC-MS/MS. The comprehensive expression analysis of proteins using LC-MS/MS and data analyses were performed according to the method reported previously [25].
3. Results and Discussion
The application of cell therapy in regenerative medicine is expected to be useful for the treatment of many kinds of diseases. For example, MSC-ATs, which can be collected from AT, have been applied to the treatment of a wide range of diseases in light of the low invasiveness compared with surgery. It is generally recognized that MSC-ATs are stable in quality from P0 cells to P5 cells [26]. As such, many manuals of commercially available MSC-ATs state that the quality is guaranteed for five passages. It was reported that multiple passaging processes reduce both the cell proliferative activity and the cell surface marker expression [27] and induce chromosome abnormalities [15]. In addition, increasing the number of passages also increases the risk of microbial infection of cells. Therefore, the general perception among clinical researchers dealing with therapeutic cells is that MSC-ATs are only useful as therapeutic cells within the first five passages [28].
However, it is easy to imagine that preparing cells collected from a living organism without subculturing may enable the production of therapeutic cells with new and special functions. Therefore, clinicians may try using MSC-ATs isolated from AT without any culturing or P0 MSC-ATs. Close attention must be paid in such cases, as immune rejection can be caused when using therapeutic cells not only for autologous transplantation but also other transplantations as well.
3.1. The Characteristics and Cell Quality of mMSC-ATs (P0)
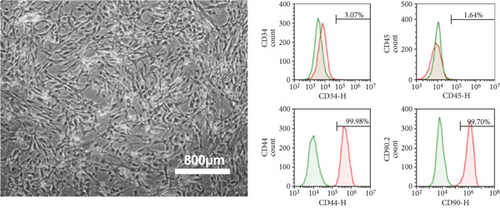
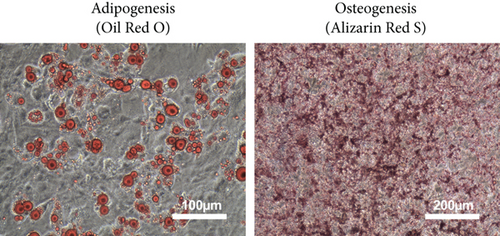
mMSC-ATs were cultured to 80% confluence using DMEM containing 10% FBS after isolation from AT. The whole medium was exchanged every two days. Microscopy was performed to confirm the absence of abnormalities with regard to the mMSC-AT (P0) size and shape and the culture state (Figure 1(a), left panel). Flow cytometry was performed using markers of mMSC-ATs (CD44, CD90.2), hematopoietic stem cells (CD34), and leukocytes (CD45). CD44 and CD90.2 were expressed in mMSC-ATs, while CD34 and CD45 were not detected (Figure 1(a), right panels). We induced differentiation into adipocytes (Figure 1(b), left panel) and osteoblasts (Figure 1(b), right panel) using mMSC-ATs. Mature adipocytes were stained with Oil Red O, and mature osteoblasts were stained with Alizarin Red S.
3.2. The Characteristics and Cell Quality of mMSC-ATs (P3)
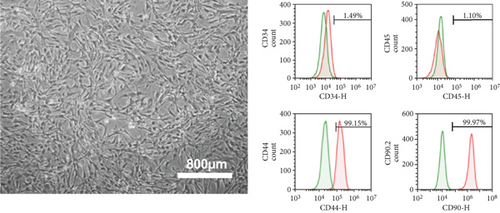
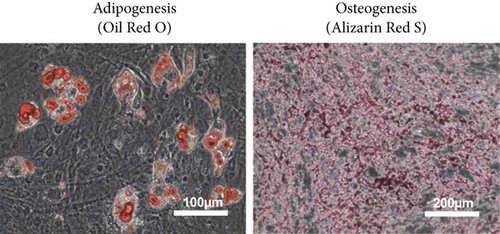
mMSC-ATs were cultured to 80% confluence using DMEM containing 10% FBS after isolation from adipose tissue. The whole medium was exchanged every two days. The passage of cells was performed every 3 to 4 days after reaching 80% confluence. Microscopy was performed to confirm the absence of abnormalities with regard to the mMSC-AT (P3) size and shape and the culture state (Figure 1(c), left panel). Flow cytometry was performed using markers of mMSC-ATs (CD44, CD90.2), hematopoietic stem cells (CD34) and leukocytes (CD45). CD44 and CD90.2 were expressed in mMSC-ATs while CD34 and CD45 were not detected (Figure 1(c), right panels). We induced differentiation into adipocytes (Figure 1(d), left panel) and osteoblasts (Figure 1(d), right panel) using mMSC-ATs. Mature adipocytes were stained with Oil Red O, and mature osteoblasts were stained with Alizarin Red S.
A proteome analysis using LC-MS/MS provided evidence supporting the safe application of cell therapy with MSCs and supplied information on the potential application of MSCs in various treatments. A protein analysis indicates the protein components present in the cell component. In this study, mMSC-ATs were used, but when human MSC-ATs are used, this analysis will show the protein components that should be administered to patients.
3.3. A Comprehensive Protein Expression Analysis of mMSC-ATs (P0 and P3)
We performed mMSC-AT isolation according to a protocol similar to that used in clinical studies at the University of the Ryukyus, and isolated mMSC-ATs were subjected to LC-MS/MS after 0 or 3 passages. The presence of a large amount of albumin in the medium reduces the accuracy in protein analyses. Therefore, the protein extracts obtained from the cells after washing with phosphate-buffered saline (PBS) were used in this study.
There were 1785 types of proteins identified from the mMSC-AT (P0) samples (Table 1) and 1825 types of proteins identified from the mMSC-AT (P3) samples (Table 2). Among the 1785 types of proteins in mouse P0 cells, there were 336 types of proteins unique to the primary cultured cells (group P0). A total of 1449 types of proteins in mouse P0 cells were also identified in mouse P3 cells (group P0&P3). Among the 1825 types of proteins in mouse P3 cells, there were 376 types of proteins unique to the cells passaged 3 times (group P3) (Figure 2). Therefore, the 336 types of proteins whose expression was eliminated by passage were deemed likely to have been derived from the different types of cells contained in the SVF.
| UniProt/Swiss-Prot ID | Description | Protein scorea | Protein mass (kDa) | pIb | Num. of matchesc | Num. of significant matchesd | Num. of sequencese | Num. of significant sequencesf | Num. of unique sequencesg | Sequence coverageh | emPAIi |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FLNA_MOUSE | Filamin-A | 2702 | 281046 | 5.68 | 220 | 138 | 87 | 56 | 81 | 0.54 | 1.34 |
| FLNB_MOUSE | Filamin-B | 1877 | 277651 | 5.46 | 164 | 94 | 81 | 51 | 73 | 0.52 | 1.23 |
| FLNC_MOUSE | Filamin-C | 1471 | 290937 | 5.63 | 115 | 68 | 57 | 33 | 11 | 0.39 | 0.68 |
| MYH9_MOUSE | Myosin-9 | 2689 | 226232 | 5.54 | 232 | 114 | 116 | 57 | 100 | 0.64 | 2.27 |
| TLN1_MOUSE | Talin-1 | 2390 | 269653 | 5.84 | 147 | 98 | 72 | 53 | 52 | 0.49 | 1.39 |
| IQGA1_MOUSE | Ras GTPase-activating-like protein IQGAP1 | 1740 | 188624 | 6.07 | 97 | 56 | 53 | 33 | 53 | 0.47 | 1.13 |
| CLH1_MOUSE | Clathrin heavy chain 1 | 1668 | 191435 | 5.48 | 114 | 80 | 64 | 49 | 64 | 0.6 | 2.26 |
| CH60_MOUSE | 60 kDa heat shock protein, mitochondrial | 1558 | 60917 | 5.91 | 75 | 58 | 29 | 22 | 29 | 0.66 | 5.35 |
| DYHC1_MOUSE | Cytoplasmic dynein 1 heavy chain 1 | 1460 | 531710 | 6.03 | 121 | 67 | 85 | 48 | 85 | 0.3 | 0.47 |
| ACTN1_MOUSE | Alpha-actinin-1 | 1455 | 103004 | 5.23 | 99 | 55 | 44 | 29 | 26 | 0.63 | 2.67 |
| ACTN4_MOUSE | Alpha-actinin-4 | 914 | 104911 | 5.25 | 76 | 36 | 39 | 23 | 21 | 0.56 | 1.5 |
| HSP7C_MOUSE | Heat shock cognate 71 kDa protein | 1427 | 70827 | 5.37 | 147 | 72 | 34 | 26 | 27 | 0.72 | 4.53 |
| GRP78_MOUSE | 78 kDa glucose-regulated protein | 1101 | 72377 | 5.07 | 114 | 54 | 35 | 23 | 32 | 0.64 | 2.77 |
| GRP75_MOUSE | Stress-70 protein, mitochondrial | 851 | 73416 | 5.81 | 65 | 36 | 22 | 17 | 21 | 0.46 | 1.79 |
| HS71A_MOUSE | Heat shock 70 kDa protein 1A | 246 | 70036 | 5.53 | 34 | 13 | 12 | 7 | 8 | 0.34 | 0.52 |
| NSF_MOUSE | Vesicle-fusing ATPase | 98 | 82561 | 6.52 | 8 | 4 | 7 | 4 | 7 | 0.15 | 0.22 |
| TBB6_MOUSE | Tubulin beta-6 chain | 517 | 50058 | 4.8 | 59 | 35 | 24 | 17 | 10 | 0.79 | 3.86 |
| VIME_MOUSE | Vimentin | 1321 | 53655 | 5.06 | 170 | 69 | 45 | 30 | 45 | 0.76 | 9.3 |
| ENPL_MOUSE | Endoplasmin | 1314 | 92418 | 4.74 | 108 | 60 | 44 | 32 | 20 | 0.58 | 4.09 |
| HS90A_MOUSE | Heat shock protein HSP 90-alpha | 1047 | 84735 | 4.93 | 93 | 47 | 38 | 22 | 25 | 0.56 | 2.27 |
| ENOA_MOUSE | Alpha-enolase | 1304 | 47111 | 6.37 | 95 | 58 | 23 | 17 | 23 | 0.74 | 6 |
| FAS_MOUSE | Fatty acid synthase | 1264 | 272257 | 6.13 | 89 | 59 | 61 | 44 | 61 | 0.45 | 1.03 |
| G3P_MOUSE | Glyceraldehyde-3-phosphate dehydrogenase | 1238 | 35787 | 8.44 | 94 | 50 | 19 | 13 | 15 | 0.76 | 9.22 |
| KPYM_MOUSE | Pyruvate kinase PKM | 1232 | 57808 | 7.18 | 98 | 58 | 37 | 30 | 12 | 0.74 | 13.41 |
| ACTBL_MOUSE | Beta-actin-like protein 2 | 361 | 41977 | 5.3 | 52 | 22 | 13 | 5 | 4 | 0.51 | 1.44 |
| TSP1_MOUSE | Thrombospondin-1 | 1160 | 129564 | 4.72 | 117 | 62 | 35 | 17 | 34 | 0.36 | 0.91 |
| ATPA_MOUSE | ATP synthase subunit alpha, mitochondrial | 1118 | 59716 | 9.22 | 64 | 44 | 24 | 18 | 16 | 0.58 | 3.34 |
| SPB6_MOUSE | Serpin B6 | 1096 | 42571 | 5.53 | 49 | 36 | 21 | 14 | 21 | 0.71 | 4.79 |
| ILEUA_MOUSE | Leukocyte elastase inhibitor A | 99 | 42548 | 5.85 | 4 | 4 | 3 | 3 | 3 | 0.11 | 0.34 |
| GDN_MOUSE | Glia-derived nexin | 84 | 44179 | 9.85 | 6 | 4 | 5 | 4 | 5 | 0.16 | 0.46 |
| FINC_MOUSE | Fibronectin | 1089 | 272368 | 5.39 | 97 | 52 | 57 | 34 | 57 | 0.4 | 0.69 |
| EF2_MOUSE | Elongation factor 2 | 1085 | 95253 | 6.41 | 91 | 53 | 47 | 28 | 46 | 0.66 | 2.42 |
| UBA1_MOUSE | Ubiquitin-like modifier-activating enzyme 1 | 1077 | 117734 | 5.43 | 55 | 39 | 26 | 19 | 26 | 0.45 | 1.11 |
| RINI_MOUSE | Ribonuclease inhibitor | 991 | 49784 | 4.69 | 48 | 34 | 23 | 17 | 19 | 0.81 | 3.14 |
| PDIA1_MOUSE | Protein disulfide-isomerase | 965 | 57023 | 4.77 | 91 | 48 | 34 | 21 | 34 | 0.62 | 3.99 |
| ATPB_MOUSE | ATP synthase subunit beta, mitochondrial | 853 | 56265 | 5.19 | 65 | 39 | 28 | 22 | 26 | 0.81 | 4.5 |
| TCPA_MOUSE | T-complex protein 1 subunit alpha | 833 | 60411 | 5.82 | 44 | 25 | 23 | 12 | 23 | 0.64 | 2.02 |
| 1433Z_MOUSE | 14-3-3 protein zeta/delta | 831 | 27754 | 4.73 | 46 | 29 | 15 | 11 | 12 | 0.6 | 4.16 |
| 1433B_MOUSE | 14-3-3 protein beta/alpha | 539 | 28069 | 4.77 | 28 | 19 | 13 | 10 | 3 | 0.63 | 3.37 |
| CO1A1_MOUSE | Collagen alpha-1(I) chain | 825 | 137948 | 5.65 | 141 | 65 | 42 | 24 | 25 | 0.53 | 1.27 |
| HSP74_MOUSE | Heat shock 70 kDa protein 4 | 817 | 94073 | 5.15 | 56 | 29 | 26 | 18 | 25 | 0.46 | 1.22 |
| HS105_MOUSE | Heat shock protein 105 kDa | 118 | 96346 | 5.39 | 14 | 6 | 12 | 5 | 11 | 0.25 | 0.24 |
| LRP1_MOUSE | Prolow-density lipoprotein receptor-related protein 1 | 815 | 504411 | 5.14 | 75 | 39 | 54 | 26 | 54 | 0.22 | 0.24 |
| WDR1_MOUSE | WD repeat-containing protein 1 | 796 | 66365 | 6.11 | 35 | 23 | 18 | 12 | 18 | 0.57 | 1.13 |
| LDHA_MOUSE | L-Lactate dehydrogenase A chain | 752 | 36475 | 7.62 | 81 | 28 | 19 | 11 | 19 | 0.79 | 3.92 |
| ANXA1_MOUSE | Annexin A1 | 750 | 38710 | 6.97 | 50 | 27 | 21 | 11 | 21 | 0.61 | 2.26 |
| MOES_MOUSE | Moesin | 737 | 67725 | 6.22 | 73 | 32 | 34 | 15 | 23 | 0.61 | 2.03 |
| TKT_MOUSE | Transketolase | 732 | 67588 | 7.23 | 53 | 32 | 26 | 16 | 26 | 0.69 | 2.04 |
| SERPH_MOUSE | Serpin H1 | 731 | 46504 | 8.88 | 56 | 35 | 23 | 16 | 23 | 0.68 | 4.01 |
| SC31A_MOUSE | Protein transport protein Sec31A | 707 | 133486 | 6.3 | 30 | 23 | 15 | 13 | 15 | 0.27 | 0.55 |
| ALDOA_MOUSE | Fructose-bisphosphate aldolase A | 674 | 39331 | 8.31 | 56 | 33 | 23 | 15 | 23 | 0.83 | 5.04 |
| VDAC1_MOUSE | Voltage-dependent anion-selective channel protein 1 | 667 | 32331 | 8.55 | 31 | 23 | 11 | 9 | 11 | 0.67 | 2.61 |
| VDAC2_MOUSE | Voltage-dependent anion-selective channel protein 2 | 454 | 31713 | 7.44 | 25 | 19 | 10 | 8 | 10 | 0.54 | 1.84 |
| RPN2_MOUSE | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2 | 666 | 69020 | 5.54 | 34 | 24 | 19 | 16 | 19 | 0.52 | 1.63 |
| ASSY_MOUSE | Argininosuccinate synthase | 660 | 46555 | 8.36 | 54 | 25 | 24 | 12 | 22 | 0.75 | 2.5 |
| SYAC_MOUSE | Alanine–tRNA ligase, cytoplasmic | 658 | 106841 | 5.45 | 46 | 25 | 29 | 18 | 29 | 0.55 | 1.02 |
| UGGG1_MOUSE | UDP-glucose:glycoprotein glucosyltransferase 1 | 656 | 176323 | 5.4 | 34 | 20 | 24 | 15 | 24 | 0.3 | 0.43 |
| ANXA5_MOUSE | Annexin A5 | 647 | 35730 | 4.83 | 65 | 34 | 19 | 12 | 18 | 0.71 | 4.71 |
| ANXA3_MOUSE | Annexin A3 | 313 | 36362 | 5.5 | 29 | 16 | 17 | 12 | 16 | 0.64 | 2.93 |
| ANXA6_MOUSE | Annexin A6 | 196 | 75837 | 5.34 | 17 | 11 | 12 | 8 | 12 | 0.27 | 0.55 |
| ANX11_MOUSE | Annexin A11 | 53 | 54045 | 7.53 | 4 | 3 | 3 | 2 | 3 | 0.11 | 0.17 |
| CO1A2_MOUSE | Collagen alpha-2(I) chain | 645 | 129478 | 9.27 | 61 | 38 | 29 | 17 | 29 | 0.44 | 0.85 |
| 6PGD_MOUSE | 6-Phosphogluconate dehydrogenase, decarboxylating | 642 | 53213 | 6.81 | 38 | 21 | 19 | 14 | 19 | 0.63 | 2 |
| SPTN1_MOUSE | Spectrin alpha chain, nonerythrocytic 1 | 639 | 284422 | 5.2 | 61 | 37 | 45 | 25 | 45 | 0.32 | 0.45 |
| AT1A1_MOUSE | Sodium/potassium-transporting ATPase subunit alpha-1 | 619 | 112910 | 5.3 | 47 | 26 | 25 | 15 | 25 | 0.34 | 0.74 |
| GDIB_MOUSE | Rab GDP dissociation inhibitor beta | 615 | 50505 | 5.93 | 40 | 22 | 18 | 10 | 13 | 0.58 | 1.48 |
| GDIA_MOUSE | Rab GDP dissociation inhibitor alpha | 274 | 50489 | 4.96 | 25 | 13 | 13 | 9 | 8 | 0.49 | 1.28 |
| EF1G_MOUSE | Elongation factor 1-gamma | 613 | 50029 | 6.31 | 43 | 29 | 21 | 14 | 21 | 0.67 | 2.48 |
| TCPQ_MOUSE | T-complex protein 1 subunit theta | 611 | 59518 | 5.44 | 35 | 19 | 20 | 12 | 20 | 0.53 | 1.32 |
| MDHM_MOUSE | Malate dehydrogenase, mitochondrial | 603 | 35589 | 8.93 | 39 | 27 | 16 | 11 | 16 | 0.58 | 3.55 |
| PDIA4_MOUSE | Protein disulfide-isomerase A4 | 602 | 71938 | 5.16 | 50 | 29 | 21 | 13 | 21 | 0.45 | 1.39 |
| CAP1_MOUSE | Adenylyl cyclase-associated protein 1 | 594 | 51532 | 7.16 | 58 | 28 | 24 | 14 | 24 | 0.73 | 2.1 |
| IMB1_MOUSE | Importin subunit beta-1 | 584 | 97122 | 4.68 | 33 | 24 | 24 | 17 | 24 | 0.41 | 1.08 |
| CAPG_MOUSE | Macrophage-capping protein | 263 | 39216 | 6.73 | 16 | 8 | 6 | 5 | 1 | 0.3 | 0.7 |
| ESTD_MOUSE | S-Formylglutathione hydrolase | 576 | 31299 | 6.7 | 50 | 28 | 17 | 12 | 17 | 0.87 | 5.39 |
| CAN2_MOUSE | Calpain-2 catalytic subunit | 566 | 79822 | 4.86 | 37 | 28 | 23 | 20 | 20 | 0.58 | 2 |
| VATB2_MOUSE | V-type proton ATPase subunit B, brain isoform | 562 | 56515 | 5.57 | 40 | 27 | 20 | 14 | 20 | 0.69 | 2.03 |
| VPS35_MOUSE | Vacuolar protein sorting-associated protein 35 | 561 | 91655 | 5.28 | 32 | 21 | 16 | 10 | 16 | 0.3 | 0.58 |
| LYAG_MOUSE | Lysosomal alpha-glucosidase | 561 | 106180 | 5.53 | 34 | 26 | 20 | 16 | 20 | 0.37 | 0.88 |
| ACLY_MOUSE | ATP-citrate synthase | 559 | 119651 | 7.13 | 34 | 23 | 27 | 18 | 27 | 0.39 | 0.94 |
| FPPS_MOUSE | Farnesyl pyrophosphate synthase | 554 | 40556 | 5.49 | 31 | 18 | 17 | 10 | 17 | 0.61 | 1.79 |
| COPG1_MOUSE | Coatomer subunit gamma-1 | 553 | 97450 | 5.23 | 35 | 22 | 24 | 18 | 20 | 0.46 | 1.16 |
| IDHC_MOUSE | Isocitrate dehydrogenase [NADP] cytoplasmic | 552 | 46644 | 6.73 | 35 | 25 | 17 | 10 | 16 | 0.57 | 1.92 |
| VIGLN_MOUSE | Vigilin | 550 | 141655 | 6.43 | 40 | 24 | 27 | 15 | 27 | 0.34 | 0.56 |
| P4HA1_MOUSE | Prolyl 4-hydroxylase subunit alpha-1 | 546 | 60872 | 5.62 | 33 | 18 | 17 | 12 | 17 | 0.5 | 1.44 |
| CATB_MOUSE | Cathepsin B | 542 | 37256 | 5.57 | 39 | 24 | 13 | 11 | 13 | 0.6 | 3.26 |
| VINC_MOUSE | Vinculin | 520 | 116644 | 5.77 | 53 | 24 | 33 | 19 | 33 | 0.41 | 0.98 |
| CNDP2_MOUSE | Cytosolic nonspecific dipeptidase | 516 | 52734 | 5.43 | 41 | 23 | 20 | 15 | 20 | 0.67 | 2.27 |
| SEPT2_MOUSE | Septin-2 | 515 | 41499 | 6.1 | 22 | 16 | 14 | 9 | 14 | 0.56 | 1.46 |
| DPYL2_MOUSE | Dihydropyrimidinase-related protein 2 | 509 | 62239 | 5.95 | 43 | 25 | 25 | 15 | 22 | 0.73 | 1.73 |
| DPYL3_MOUSE | Dihydropyrimidinase-related protein 3 | 203 | 61897 | 6.04 | 16 | 9 | 8 | 4 | 5 | 0.21 | 0.31 |
| TAGL2_MOUSE | Transgelin-2 | 509 | 22381 | 8.39 | 42 | 28 | 14 | 11 | 14 | 0.8 | 8.1 |
| TCPH_MOUSE | T-complex protein 1 subunit eta | 501 | 59614 | 7.95 | 34 | 22 | 18 | 15 | 18 | 0.53 | 2.06 |
| FRIL1_MOUSE | Ferritin light chain 1 | 500 | 20790 | 5.66 | 18 | 15 | 6 | 4 | 6 | 0.49 | 1.7 |
| NCPR_MOUSE | NADPH–cytochrome P450 reductase | 499 | 76995 | 5.34 | 19 | 15 | 12 | 8 | 12 | 0.33 | 0.54 |
| TCPZ_MOUSE | T-complex protein 1 subunit zeta | 491 | 57968 | 6.63 | 30 | 18 | 19 | 12 | 19 | 0.55 | 1.37 |
| VATA_MOUSE | V-type proton ATPase catalytic subunit A | 481 | 68283 | 5.42 | 43 | 23 | 27 | 14 | 27 | 0.6 | 1.35 |
| XDH_MOUSE | Xanthine dehydrogenase/oxidase | 475 | 146468 | 7.62 | 28 | 23 | 16 | 14 | 16 | 0.24 | 0.54 |
| PDC6I_MOUSE | Programmed cell death 6-interacting protein | 475 | 95964 | 6.15 | 25 | 17 | 17 | 12 | 17 | 0.35 | 0.76 |
| CAND1_MOUSE | Cullin-associated NEDD8-dissociated protein 1 | 473 | 136245 | 5.52 | 26 | 19 | 20 | 14 | 20 | 0.33 | 0.64 |
| COF1_MOUSE | Cofilin-1 | 473 | 18548 | 8.22 | 30 | 18 | 11 | 9 | 9 | 0.61 | 6.37 |
| COF2_MOUSE | Cofilin-2 | 182 | 18698 | 7.66 | 20 | 10 | 6 | 4 | 4 | 0.49 | 1.42 |
| LG3BP_MOUSE | Galectin-3-binding protein | 471 | 64450 | 5 | 17 | 12 | 10 | 6 | 9 | 0.3 | 0.47 |
| VAT1_MOUSE | Synaptic vesicle membrane protein VAT-1 homolog | 471 | 43069 | 5.95 | 44 | 25 | 21 | 14 | 21 | 0.65 | 3.69 |
| TCPD_MOUSE | T-complex protein 1 subunit delta | 467 | 58030 | 8.24 | 35 | 17 | 20 | 9 | 20 | 0.59 | 0.91 |
| AMPN_MOUSE | Aminopeptidase N | 465 | 109582 | 5.62 | 26 | 21 | 17 | 16 | 17 | 0.3 | 0.84 |
| PPIA_MOUSE | Peptidyl-prolyl cis-trans isomerase A | 465 | 17960 | 7.74 | 33 | 21 | 11 | 8 | 11 | 0.7 | 11.36 |
| MDHC_MOUSE | Malate dehydrogenase, cytoplasmic | 464 | 36488 | 6.16 | 21 | 14 | 10 | 6 | 10 | 0.52 | 1.22 |
| G6PI_MOUSE | Glucose-6-phosphate isomerase | 462 | 62727 | 8.14 | 29 | 18 | 15 | 10 | 8 | 0.4 | 1.08 |
| PDIA3_MOUSE | Protein disulfide-isomerase A3 | 449 | 56643 | 5.88 | 115 | 29 | 32 | 16 | 32 | 0.64 | 2.77 |
| THIO_MOUSE | Thioredoxin | 445 | 11668 | 4.8 | 24 | 16 | 7 | 6 | 7 | 0.78 | 7.1 |
| RLA0_MOUSE | 60S acidic ribosomal protein P0 | 441 | 34195 | 5.91 | 30 | 21 | 14 | 10 | 14 | 0.56 | 2.8 |
| ESYT1_MOUSE | Extended synaptotagmin-1 | 437 | 121478 | 5.63 | 20 | 17 | 13 | 11 | 13 | 0.22 | 0.46 |
| 2AAA_MOUSE | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | 437 | 65281 | 5 | 20 | 14 | 17 | 12 | 17 | 0.46 | 1.15 |
| PTBP1_MOUSE | Polypyrimidine tract-binding protein 1 | 437 | 56443 | 8.47 | 22 | 11 | 8 | 5 | 8 | 0.36 | 0.68 |
| AT2A2_MOUSE | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 436 | 114784 | 5.23 | 33 | 20 | 20 | 12 | 20 | 0.32 | 0.55 |
| SPTB2_MOUSE | Spectrin beta chain, nonerythrocytic 1 | 429 | 274052 | 5.4 | 52 | 21 | 39 | 20 | 39 | 0.25 | 0.38 |
| SCOT1_MOUSE | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial | 428 | 55953 | 8.73 | 29 | 20 | 14 | 12 | 14 | 0.5 | 1.63 |
| CALR_MOUSE | Calreticulin | 419 | 47965 | 4.33 | 43 | 26 | 19 | 11 | 19 | 0.6 | 1.84 |
| CKAP4_MOUSE | Cytoskeleton-associated protein 4 | 418 | 63654 | 5.46 | 48 | 27 | 25 | 13 | 25 | 0.54 | 1.86 |
| MYOF_MOUSE | Myoferlin | 417 | 233177 | 5.83 | 44 | 17 | 30 | 12 | 30 | 0.24 | 0.24 |
| GSTP1_MOUSE | Glutathione S-transferase P 1 | 417 | 23594 | 7.68 | 21 | 14 | 9 | 7 | 9 | 0.58 | 3.82 |
| PLEC_MOUSE | Plectin | 416 | 533861 | 5.74 | 66 | 21 | 52 | 16 | 52 | 0.18 | 0.13 |
| TCPB_MOUSE | T-complex protein 1 subunit beta | 414 | 57441 | 5.97 | 27 | 15 | 20 | 13 | 20 | 0.56 | 1.57 |
| KAD1_MOUSE | Adenylate kinase isoenzyme 1 | 410 | 21526 | 5.67 | 15 | 13 | 9 | 8 | 6 | 0.64 | 3.63 |
| PRDX6_MOUSE | Peroxiredoxin-6 | 405 | 24855 | 5.71 | 36 | 18 | 16 | 11 | 16 | 0.75 | 5.23 |
| RL9_MOUSE | 60S ribosomal protein L9 | 401 | 21868 | 9.96 | 12 | 10 | 5 | 4 | 5 | 0.48 | 1.13 |
| UGDH_MOUSE | UDP-glucose 6-dehydrogenase | 398 | 54797 | 7.49 | 33 | 18 | 22 | 14 | 22 | 0.66 | 1.9 |
| CATD_MOUSE | Cathepsin D | 389 | 44925 | 6.71 | 35 | 21 | 15 | 9 | 15 | 0.55 | 2.33 |
| NIBL1_MOUSE | Niban-like protein 1 | 388 | 84765 | 5.65 | 23 | 15 | 18 | 13 | 18 | 0.36 | 0.9 |
| TXND5_MOUSE | Thioredoxin domain-containing protein 5 | 386 | 46386 | 5.51 | 17 | 14 | 8 | 5 | 8 | 0.32 | 0.57 |
| TSP2_MOUSE | Thrombospondin-2 | 385 | 129798 | 4.61 | 44 | 19 | 22 | 10 | 21 | 0.28 | 0.38 |
| MAOX_MOUSE | NADP-dependent malic enzyme | 384 | 63913 | 7.16 | 15 | 11 | 14 | 10 | 14 | 0.49 | 0.92 |
| ARF4_MOUSE | ADP-ribosylation factor 4 | 382 | 20384 | 6.59 | 23 | 15 | 11 | 8 | 5 | 0.83 | 6.52 |
| COPB2_MOUSE | Coatomer subunit beta ′ | 381 | 102384 | 5.17 | 33 | 20 | 24 | 15 | 24 | 0.43 | 0.85 |
| PRDX1_MOUSE | Peroxiredoxin-1 | 379 | 22162 | 8.26 | 87 | 35 | 20 | 14 | 19 | 0.8 | 15.25 |
| PRDX2_MOUSE | Peroxiredoxin-2 | 316 | 21765 | 5.2 | 16 | 11 | 8 | 7 | 8 | 0.45 | 2.77 |
| PRDX4_MOUSE | Peroxiredoxin-4 | 152 | 31033 | 6.67 | 18 | 7 | 11 | 5 | 10 | 0.52 | 0.95 |
| PLSL_MOUSE | Plastin-2 | 378 | 70105 | 5.2 | 35 | 21 | 22 | 13 | 20 | 0.48 | 1.17 |
| GPNMB_MOUSE | Transmembrane glycoprotein NMB | 378 | 63635 | 7.55 | 27 | 16 | 10 | 5 | 10 | 0.22 | 0.48 |
| GELS_MOUSE | Gelsolin | 371 | 85888 | 5.83 | 32 | 21 | 17 | 13 | 17 | 0.45 | 0.98 |
| THIC_MOUSE | Acetyl-CoA acetyltransferase, cytosolic | 371 | 41271 | 7.16 | 20 | 12 | 12 | 8 | 12 | 0.59 | 1.24 |
| ECHA_MOUSE | Trifunctional enzyme subunit alpha, mitochondrial | 371 | 82617 | 9.24 | 24 | 16 | 16 | 10 | 16 | 0.44 | 0.66 |
| MYO1C_MOUSE | Unconventional myosin-Ic | 370 | 121868 | 9.41 | 34 | 17 | 29 | 14 | 29 | 0.4 | 0.62 |
| PDIA6_MOUSE | Protein disulfide-isomerase A6 | 369 | 48070 | 5 | 29 | 22 | 14 | 12 | 14 | 0.48 | 1.83 |
| EIF3L_MOUSE | Eukaryotic translation initiation factor 3 subunit L | 368 | 66570 | 6.01 | 16 | 11 | 7 | 5 | 7 | 0.21 | 0.37 |
| MVP_MOUSE | Major vault protein | 365 | 95865 | 5.43 | 30 | 16 | 17 | 10 | 17 | 0.35 | 0.55 |
| HYOU1_MOUSE | Hypoxia-upregulated protein 1 | 362 | 111112 | 5.12 | 18 | 10 | 15 | 8 | 15 | 0.27 | 0.35 |
| FABP5_MOUSE | Fatty acid-binding protein, epidermal | 361 | 15127 | 6.14 | 35 | 19 | 8 | 6 | 8 | 0.66 | 7.75 |
| IPO5_MOUSE | Importin-5 | 358 | 123511 | 4.82 | 42 | 28 | 25 | 19 | 25 | 0.44 | 1.11 |
| TCPE_MOUSE | T-complex protein 1 subunit epsilon | 356 | 59586 | 5.72 | 27 | 17 | 17 | 10 | 17 | 0.45 | 1.16 |
| OST48_MOUSE | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase 48 kDa subunit | 355 | 48997 | 5.52 | 21 | 13 | 11 | 5 | 11 | 0.38 | 0.81 |
| SYRC_MOUSE | Arginine–tRNA ligase, cytoplasmic | 354 | 75625 | 7.48 | 19 | 11 | 14 | 7 | 14 | 0.31 | 0.56 |
| NB5R3_MOUSE | NADH-cytochrome b5 reductase 3 | 354 | 34106 | 8.55 | 26 | 18 | 12 | 10 | 12 | 0.68 | 2.82 |
| NDKB_MOUSE | Nucleoside diphosphate kinase B | 353 | 17352 | 6.97 | 30 | 14 | 10 | 4 | 6 | 0.74 | 3.15 |
| NDKA_MOUSE | Nucleoside diphosphate kinase A | 171 | 17197 | 6.84 | 22 | 11 | 7 | 5 | 3 | 0.59 | 4.32 |
| CO5A1_MOUSE | Collagen alpha-1(V) chain | 351 | 183564 | 4.86 | 13 | 11 | 8 | 6 | 8 | 0.1 | 0.15 |
| RLA2_MOUSE | 60S acidic ribosomal protein P2 | 348 | 11644 | 4.42 | 13 | 9 | 5 | 5 | 5 | 0.7 | 4.72 |
| AK1A1_MOUSE | Alcohol dehydrogenase [NADP(+)] | 344 | 36564 | 6.9 | 23 | 16 | 16 | 11 | 16 | 0.64 | 2.92 |
| ALDR_MOUSE | Aldose reductase | 114 | 35709 | 6.71 | 14 | 6 | 7 | 4 | 7 | 0.26 | 0.59 |
| PCBP2_MOUSE | Poly(rC)-binding protein 2 | 343 | 38197 | 6.33 | 32 | 17 | 12 | 8 | 2 | 0.55 | 1.66 |
| NEDD4_MOUSE | E3 ubiquitin-protein ligase NEDD4 | 343 | 102642 | 5.12 | 29 | 17 | 18 | 11 | 18 | 0.33 | 0.57 |
| UAP1L_MOUSE | UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 | 342 | 56578 | 5.27 | 28 | 15 | 19 | 11 | 17 | 0.55 | 1.25 |
| UAP1_MOUSE | UDP-N-acetylhexosamine pyrophosphorylase | 87 | 58572 | 6.04 | 4 | 3 | 2 | 2 | 1 | 0.06 | 0.15 |
| CLIC1_MOUSE | Chloride intracellular channel protein 1 | 342 | 26996 | 5.09 | 24 | 13 | 14 | 8 | 13 | 0.72 | 2.41 |
| GSLG1_MOUSE | Golgi apparatus protein 1 | 342 | 133646 | 6.45 | 20 | 11 | 16 | 7 | 11 | 0.2 | 0.28 |
| DNJC3_MOUSE | DnaJ homolog subfamily C member 3 | 341 | 57428 | 5.61 | 9 | 5 | 6 | 3 | 6 | 0.29 | 0.24 |
| FSCN1_MOUSE | Fascin | 340 | 54474 | 6.44 | 24 | 10 | 15 | 8 | 15 | 0.51 | 0.84 |
| SODM_MOUSE | Superoxide dismutase [Mn], mitochondrial | 340 | 24588 | 8.8 | 12 | 9 | 5 | 4 | 5 | 0.46 | 0.96 |
| PROF1_MOUSE | Profilin-1 | 339 | 14948 | 8.46 | 65 | 22 | 11 | 7 | 11 | 0.78 | 10.68 |
| MBB1A_MOUSE | Myb-binding protein 1A | 339 | 151942 | 9.08 | 21 | 15 | 15 | 10 | 15 | 0.18 | 0.32 |
| CISY_MOUSE | Citrate synthase, mitochondrial | 338 | 51703 | 8.72 | 23 | 14 | 16 | 11 | 16 | 0.56 | 1.63 |
| SAP_MOUSE | Prosaposin | 338 | 61381 | 5.07 | 45 | 16 | 19 | 10 | 19 | 0.49 | 1.11 |
| ADT1_MOUSE | ADP/ATP translocase 1 | 278 | 32883 | 9.73 | 25 | 9 | 16 | 7 | 9 | 0.61 | 1.42 |
| CPNS1_MOUSE | Calpain small subunit 1 | 337 | 28445 | 5.41 | 14 | 9 | 7 | 5 | 7 | 0.41 | 1.07 |
| UGPA_MOUSE | UTP–glucose-1-phosphate uridylyltransferase | 335 | 56944 | 7.18 | 14 | 9 | 9 | 6 | 9 | 0.3 | 0.55 |
| NACAM_MOUSE | Nascent polypeptide-associated complex subunit alpha, muscle-specific form | 330 | 220364 | 9.39 | 20 | 12 | 8 | 3 | 8 | 0.06 | 0.06 |
| DLDH_MOUSE | Dihydrolipoyl dehydrogenase, mitochondrial | 329 | 54238 | 7.99 | 16 | 12 | 10 | 7 | 10 | 0.39 | 0.71 |
| HNRPF_MOUSE | Heterogeneous nuclear ribonucleoprotein F | 327 | 45701 | 5.31 | 17 | 10 | 10 | 6 | 8 | 0.42 | 0.73 |
| OAT_MOUSE | Ornithine aminotransferase, mitochondrial | 326 | 48324 | 6.19 | 25 | 14 | 9 | 6 | 9 | 0.4 | 0.68 |
| DHB12_MOUSE | Very-long-chain 3-oxoacyl-CoA reductase | 324 | 34719 | 9.55 | 18 | 13 | 12 | 9 | 12 | 0.56 | 1.93 |
| DHE3_MOUSE | Glutamate dehydrogenase 1, mitochondrial | 323 | 61298 | 8.05 | 36 | 16 | 16 | 9 | 16 | 0.44 | 0.85 |
| CATS_MOUSE | Cathepsin S | 323 | 38449 | 6.51 | 18 | 10 | 12 | 7 | 12 | 0.53 | 1.13 |
| ITB5_MOUSE | Integrin beta-5 | 322 | 87851 | 5.81 | 18 | 14 | 10 | 7 | 10 | 0.25 | 0.4 |
| CATL1_MOUSE | Cathepsin L1 | 321 | 37523 | 6.37 | 13 | 10 | 8 | 6 | 8 | 0.5 | 1.43 |
| PSMD1_MOUSE | 26S proteasome non-ATPase regulatory subunit 1 | 321 | 105663 | 5.25 | 20 | 12 | 15 | 10 | 15 | 0.29 | 0.49 |
| RPN1_MOUSE | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 | 320 | 68486 | 6.02 | 32 | 19 | 19 | 11 | 19 | 0.45 | 0.96 |
| LYOX_MOUSE | Protein-lysine 6-oxidase | 319 | 46671 | 8.73 | 20 | 13 | 8 | 5 | 8 | 0.37 | 0.56 |
| AOFA_MOUSE | Amine oxidase [flavin-containing] A | 319 | 59564 | 7.9 | 19 | 14 | 12 | 7 | 12 | 0.39 | 0.63 |
| PSD12_MOUSE | 26S proteasome non-ATPase regulatory subunit 12 | 315 | 52861 | 6.66 | 22 | 12 | 15 | 7 | 15 | 0.43 | 0.74 |
| COCA1_MOUSE | Collagen alpha-1(XII) chain | 315 | 340004 | 5.47 | 39 | 19 | 24 | 14 | 24 | 0.15 | 0.19 |
| ALDH2_MOUSE | Aldehyde dehydrogenase, mitochondrial | 315 | 56502 | 7.53 | 39 | 20 | 19 | 14 | 19 | 0.62 | 2.03 |
| PP1A_MOUSE | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 314 | 37516 | 5.94 | 15 | 12 | 10 | 7 | 10 | 0.37 | 1.17 |
| TPIS_MOUSE | Triosephosphate isomerase | 312 | 32171 | 5.56 | 36 | 14 | 14 | 9 | 14 | 0.64 | 2.63 |
| GSTM1_MOUSE | Glutathione S-transferase Mu 1 | 311 | 25953 | 7.71 | 32 | 13 | 12 | 6 | 9 | 0.61 | 2.05 |
| GSTM2_MOUSE | Glutathione S-transferase Mu 2 | 195 | 25700 | 6.9 | 17 | 8 | 11 | 6 | 8 | 0.6 | 1.62 |
| MPRI_MOUSE | Cation-independent mannose-6-phosphate receptor | 309 | 273639 | 5.47 | 30 | 18 | 23 | 16 | 23 | 0.18 | 0.28 |
| CALX_MOUSE | Calnexin | 307 | 67236 | 4.5 | 23 | 15 | 16 | 9 | 16 | 0.37 | 0.86 |
| EIF3D_MOUSE | Eukaryotic translation initiation factor 3 subunit D | 305 | 63948 | 5.79 | 11 | 8 | 7 | 5 | 7 | 0.25 | 0.39 |
| TPM4_MOUSE | Tropomyosin alpha-4 chain | 203 | 28450 | 4.65 | 21 | 9 | 11 | 7 | 10 | 0.46 | 1.77 |
| CATZ_MOUSE | Cathepsin Z | 303 | 33974 | 6.13 | 11 | 9 | 4 | 4 | 4 | 0.26 | 0.63 |
| GNAI2_MOUSE | Guanine nucleotide-binding protein G(i) subunit alpha-2 | 255 | 40463 | 5.28 | 19 | 9 | 11 | 6 | 5 | 0.38 | 0.85 |
| STOM_MOUSE | Erythrocyte band 7 integral membrane protein | 300 | 31355 | 6.45 | 18 | 10 | 7 | 5 | 7 | 0.38 | 0.94 |
| ITB1_MOUSE | Integrin beta-1 | 300 | 88173 | 5.68 | 29 | 17 | 18 | 10 | 18 | 0.41 | 0.61 |
| PSA7_MOUSE | Proteasome subunit alpha type-7 | 296 | 27838 | 8.59 | 15 | 6 | 8 | 4 | 8 | 0.48 | 0.81 |
| KCRB_MOUSE | Creatine kinase B-type | 295 | 42686 | 5.4 | 17 | 12 | 10 | 6 | 10 | 0.52 | 0.79 |
| COPB_MOUSE | Coatomer subunit beta | 295 | 106998 | 5.69 | 16 | 11 | 14 | 10 | 14 | 0.23 | 0.48 |
| LEG1_MOUSE | Galectin-1 | 294 | 14856 | 5.32 | 28 | 14 | 9 | 6 | 9 | 0.75 | 8.03 |
| ANXA2_MOUSE | Annexin A2 | 293 | 38652 | 7.55 | 24 | 19 | 14 | 11 | 14 | 0.49 | 2.27 |
| XPO2_MOUSE | Exportin-2 | 289 | 110384 | 5.52 | 10 | 9 | 6 | 5 | 6 | 0.12 | 0.26 |
| EF1D_MOUSE | Elongation factor 1-delta | 289 | 31274 | 4.91 | 22 | 11 | 11 | 7 | 10 | 0.5 | 1.53 |
| EF1B_MOUSE | Elongation factor 1-beta | 205 | 24678 | 4.53 | 16 | 9 | 6 | 6 | 5 | 0.42 | 1.74 |
| PAI1_MOUSE | Plasminogen activator inhibitor 1 | 288 | 45141 | 6.17 | 20 | 10 | 13 | 7 | 13 | 0.49 | 0.91 |
| APT_MOUSE | Adenine phosphoribosyltransferase | 287 | 19712 | 6.31 | 10 | 8 | 6 | 5 | 6 | 0.51 | 1.84 |
| AKA12_MOUSE | A-kinase anchor protein 12 | 286 | 180586 | 4.39 | 36 | 13 | 22 | 8 | 22 | 0.26 | 0.2 |
| IMA4_MOUSE | Importin subunit alpha-4 | 85 | 57737 | 4.8 | 7 | 4 | 5 | 3 | 2 | 0.22 | 0.24 |
| SERA_MOUSE | D-3-Phosphoglycerate dehydrogenase | 283 | 56549 | 6.12 | 21 | 14 | 14 | 9 | 14 | 0.34 | 0.94 |
| SND1_MOUSE | Staphylococcal nuclease domain-containing protein 1 | 282 | 102025 | 7.08 | 25 | 19 | 12 | 10 | 12 | 0.26 | 0.57 |
| ITA5_MOUSE | Integrin alpha-5 | 280 | 114971 | 5.65 | 23 | 14 | 13 | 9 | 13 | 0.2 | 0.44 |
| RRBP1_MOUSE | Ribosome-binding protein 1 | 280 | 172776 | 9.35 | 42 | 16 | 28 | 9 | 28 | 0.24 | 0.24 |
| PSB4_MOUSE | Proteasome subunit beta type-4 | 280 | 29097 | 5.47 | 9 | 5 | 7 | 3 | 7 | 0.45 | 0.53 |
| NUCL_MOUSE | Nucleolin | 278 | 76677 | 4.69 | 14 | 11 | 10 | 8 | 10 | 0.2 | 0.55 |
| SFXN3_MOUSE | Sideroflexin-3 | 277 | 35384 | 9.58 | 12 | 7 | 7 | 3 | 7 | 0.4 | 0.42 |
| ADK_MOUSE | Adenosine kinase | 276 | 40123 | 5.84 | 12 | 11 | 7 | 6 | 7 | 0.35 | 0.86 |
| HNRPU_MOUSE | Heterogeneous nuclear ribonucleoprotein U | 275 | 87863 | 5.92 | 25 | 15 | 17 | 12 | 17 | 0.27 | 0.77 |
| HXK3_MOUSE | Hexokinase-3 | 270 | 100037 | 5.63 | 20 | 12 | 13 | 7 | 13 | 0.26 | 0.34 |
| PGM1_MOUSE | Phosphoglucomutase-1 | 270 | 61380 | 6.14 | 15 | 11 | 8 | 6 | 8 | 0.27 | 0.61 |
| PSB2_MOUSE | Proteasome subunit beta type-2 | 269 | 22892 | 6.52 | 24 | 14 | 10 | 5 | 10 | 0.64 | 1.95 |
| ICAL_MOUSE | Calpastatin | 269 | 84871 | 5.37 | 10 | 7 | 5 | 4 | 5 | 0.15 | 0.22 |
| CD81_MOUSE | CD81 antigen | 268 | 25797 | 5.54 | 5 | 5 | 2 | 2 | 2 | 0.18 | 0.62 |
| PPCE_MOUSE | Prolyl endopeptidase | 267 | 80700 | 5.44 | 17 | 8 | 12 | 5 | 12 | 0.31 | 0.3 |
| PEBP1_MOUSE | Phosphatidylethanolamine-binding protein 1 | 267 | 20817 | 5.19 | 16 | 11 | 9 | 7 | 9 | 0.78 | 2.99 |
| MRC2_MOUSE | C-type mannose receptor 2 | 265 | 166968 | 5.65 | 18 | 11 | 14 | 8 | 14 | 0.19 | 0.22 |
| AP2A2_MOUSE | AP-2 complex subunit alpha-2 | 264 | 103951 | 6.51 | 24 | 11 | 16 | 7 | 10 | 0.28 | 0.33 |
| AP2A1_MOUSE | AP-2 complex subunit alpha-1 | 140 | 107596 | 6.63 | 18 | 7 | 17 | 6 | 11 | 0.31 | 0.26 |
| DDB1_MOUSE | DNA damage-binding protein 1 | 262 | 126772 | 5.14 | 19 | 11 | 9 | 4 | 9 | 0.16 | 0.18 |
| ITAV_MOUSE | Integrin alpha-V | 262 | 115287 | 5.41 | 28 | 10 | 15 | 5 | 15 | 0.18 | 0.2 |
| CH10_MOUSE | 10 kDa heat shock protein, mitochondrial | 260 | 10956 | 7.93 | 18 | 10 | 5 | 2 | 5 | 0.52 | 1.09 |
| COMT_MOUSE | Catechol O-methyltransferase | 258 | 29467 | 5.52 | 8 | 6 | 4 | 2 | 4 | 0.29 | 0.33 |
| DDX3X_MOUSE | ATP-dependent RNA helicase DDX3X | 257 | 73056 | 6.73 | 21 | 11 | 14 | 8 | 14 | 0.36 | 0.58 |
| PSB7_MOUSE | Proteasome subunit beta type-7 | 257 | 29872 | 8.14 | 8 | 6 | 5 | 3 | 5 | 0.32 | 0.52 |
| PNPH_MOUSE | Purine nucleoside phosphorylase | 257 | 32256 | 5.78 | 17 | 10 | 11 | 9 | 11 | 0.67 | 2.18 |
| PCNA_MOUSE | Proliferating cell nuclear antigen | 256 | 28766 | 4.66 | 15 | 10 | 9 | 5 | 9 | 0.66 | 1.06 |
| SYSC_MOUSE | Serine–tRNA ligase, cytoplasmic | 255 | 58352 | 5.95 | 17 | 12 | 13 | 8 | 13 | 0.41 | 0.77 |
| AATM_MOUSE | Aspartate aminotransferase, mitochondrial | 255 | 47381 | 9.13 | 32 | 14 | 18 | 10 | 18 | 0.54 | 1.41 |
| RL3_MOUSE | 60S ribosomal protein L3 | 254 | 46081 | 10.22 | 30 | 20 | 11 | 8 | 11 | 0.36 | 1.26 |
| FUMH_MOUSE | Fumarate hydratase, mitochondrial | 254 | 54322 | 9.12 | 10 | 8 | 8 | 6 | 8 | 0.36 | 0.59 |
| MAP4_MOUSE | Microtubule-associated protein 4 | 254 | 117357 | 4.9 | 24 | 13 | 14 | 8 | 14 | 0.23 | 0.33 |
| MAP1B_MOUSE | Microtubule-associated protein 1B | 254 | 270089 | 4.76 | 14 | 8 | 9 | 3 | 9 | 0.06 | 0.05 |
| SEC13_MOUSE | Protein SEC13 homolog | 253 | 35543 | 5.15 | 10 | 6 | 4 | 3 | 4 | 0.19 | 0.42 |
| LAMP1_MOUSE | Lysosome-associated membrane glycoprotein 1 | 253 | 43837 | 8.66 | 23 | 14 | 8 | 4 | 8 | 0.35 | 0.61 |
| FBLN2_MOUSE | Fibulin-2 | 253 | 131746 | 4.58 | 27 | 12 | 17 | 9 | 17 | 0.23 | 0.33 |
| GORS2_MOUSE | Golgi reassembly-stacking protein 2 | 252 | 47009 | 4.68 | 10 | 8 | 4 | 4 | 4 | 0.15 | 0.56 |
| PSB6_MOUSE | Proteasome subunit beta type-6 | 250 | 25362 | 4.97 | 14 | 10 | 9 | 7 | 9 | 0.67 | 2.14 |
| SERC_MOUSE | Phosphoserine aminotransferase | 249 | 40447 | 8.15 | 26 | 13 | 13 | 8 | 13 | 0.55 | 1.28 |
| ERP29_MOUSE | Endoplasmic reticulum resident protein 29 | 248 | 28805 | 5.9 | 22 | 13 | 9 | 6 | 9 | 0.5 | 1.37 |
| EHD1_MOUSE | EH domain-containing protein 1 | 247 | 60565 | 6.35 | 22 | 9 | 15 | 9 | 15 | 0.47 | 0.86 |
| RCN3_MOUSE | Reticulocalbin-3 | 246 | 37978 | 4.74 | 7 | 6 | 4 | 4 | 4 | 0.27 | 0.55 |
| COPE_MOUSE | Coatomer subunit epsilon | 245 | 34545 | 4.94 | 10 | 7 | 7 | 6 | 7 | 0.38 | 1.06 |
| PSME1_MOUSE | Proteasome activator complex subunit 1 | 244 | 28655 | 5.73 | 13 | 12 | 5 | 4 | 5 | 0.34 | 0.78 |
| TADBP_MOUSE | TAR DNA-binding protein 43 | 244 | 44519 | 6.26 | 9 | 9 | 7 | 7 | 7 | 0.34 | 0.92 |
| C1QBP_MOUSE | Complement component 1 Q subcomponent-binding Protein, mitochondrial | 206 | 30994 | 4.82 | 11 | 7 | 7 | 4 | 1 | 0.45 | 0.71 |
| PUR9_MOUSE | Bifunctional purine biosynthesis protein PURH | 242 | 64177 | 6.3 | 27 | 14 | 18 | 12 | 13 | 0.52 | 1.18 |
| RCN2_MOUSE | Reticulocalbin-2 | 242 | 37248 | 4.28 | 10 | 8 | 7 | 5 | 7 | 0.39 | 0.75 |
| PPIB_MOUSE | Peptidyl-prolyl cis-trans isomerase B | 241 | 23699 | 9.56 | 20 | 9 | 12 | 4 | 12 | 0.54 | 1.39 |
| PPIC_MOUSE | Peptidyl-prolyl cis-trans isomerase C | 140 | 22780 | 6.96 | 5 | 5 | 4 | 4 | 4 | 0.26 | 1.07 |
| RLA1_MOUSE | 60S acidic ribosomal protein P1 | 240 | 11468 | 4.28 | 6 | 6 | 3 | 3 | 3 | 0.53 | 1.9 |
| SNAA_MOUSE | Alpha-soluble NSF attachment protein | 239 | 33168 | 5.3 | 15 | 11 | 11 | 8 | 11 | 0.56 | 1.72 |
| KAD2_MOUSE | Adenylate kinase 2, mitochondrial | 239 | 26452 | 6.96 | 8 | 6 | 5 | 4 | 5 | 0.3 | 0.87 |
| GDIR1_MOUSE | Rho GDP-dissociation inhibitor 1 | 238 | 23393 | 5.12 | 16 | 12 | 10 | 8 | 10 | 0.51 | 3.1 |
| CNN2_MOUSE | Calponin-2 | 237 | 33134 | 7.53 | 20 | 7 | 11 | 5 | 4 | 0.65 | 0.87 |
| SNX9_MOUSE | Sorting nexin-9 | 236 | 66504 | 5.35 | 17 | 10 | 11 | 6 | 11 | 0.36 | 0.46 |
| CO3A1_MOUSE | Collagen alpha-1(III) chain | 236 | 138858 | 6.11 | 28 | 13 | 12 | 6 | 8 | 0.15 | 0.23 |
| IMA5_MOUSE | Importin subunit alpha-5 | 195 | 60144 | 4.93 | 8 | 7 | 4 | 3 | 3 | 0.18 | 0.32 |
| AMPL_MOUSE | Cytosol aminopeptidase | 234 | 56106 | 7.62 | 22 | 11 | 13 | 10 | 13 | 0.4 | 1.1 |
| PABP1_MOUSE | Polyadenylate-binding protein 1 | 233 | 70626 | 9.52 | 19 | 10 | 16 | 8 | 16 | 0.32 | 0.7 |
| SPA3N_MOUSE | Serine protease inhibitor A3N | 233 | 46688 | 5.59 | 4 | 4 | 3 | 3 | 3 | 0.13 | 0.31 |
| GARS_MOUSE | Glycine–tRNA ligase | 233 | 81826 | 6.24 | 15 | 8 | 12 | 6 | 12 | 0.33 | 0.36 |
| AP1B1_MOUSE | AP-1 complex subunit beta-1 | 231 | 103869 | 5.04 | 17 | 11 | 13 | 8 | 9 | 0.26 | 0.44 |
| NP1L1_MOUSE | Nucleosome assembly protein 1-like 1 | 231 | 45317 | 4.36 | 19 | 10 | 12 | 6 | 3 | 0.5 | 0.9 |
| MTPN_MOUSE | Myotrophin | 227 | 12853 | 5.27 | 12 | 7 | 6 | 4 | 6 | 0.68 | 2.55 |
| GRN_MOUSE | Granulins | 227 | 63413 | 6.42 | 21 | 15 | 5 | 4 | 5 | 0.14 | 0.3 |
| CTNA1_MOUSE | Catenin alpha-1 | 226 | 100044 | 5.91 | 14 | 10 | 9 | 7 | 9 | 0.19 | 0.34 |
| ECHM_MOUSE | Enoyl-CoA hydratase, mitochondrial | 226 | 31454 | 8.76 | 10 | 9 | 6 | 5 | 6 | 0.29 | 1.21 |
| ITB2_MOUSE | Integrin beta-2 | 226 | 84970 | 7.02 | 15 | 8 | 8 | 5 | 8 | 0.17 | 0.28 |
| LKHA4_MOUSE | Leukotriene A-4 hydrolase | 226 | 69007 | 5.98 | 13 | 10 | 7 | 6 | 7 | 0.21 | 0.44 |
| MYADM_MOUSE | Myeloid-associated differentiation marker | 225 | 35261 | 8.69 | 18 | 10 | 4 | 3 | 4 | 0.19 | 0.42 |
| ARC1B_MOUSE | Actin-related protein 2/3 complex subunit 1B | 224 | 41037 | 8.69 | 12 | 9 | 8 | 6 | 8 | 0.38 | 0.84 |
| COPD_MOUSE | Coatomer subunit delta | 222 | 57193 | 5.89 | 28 | 14 | 12 | 6 | 12 | 0.33 | 0.55 |
| DUS3_MOUSE | Dual specificity protein phosphatase 3 | 222 | 20459 | 6.07 | 6 | 4 | 5 | 4 | 5 | 0.46 | 1.24 |
| SYEP_MOUSE | Bifunctional glutamate/proline–tRNA ligase | 222 | 169972 | 7.75 | 29 | 11 | 21 | 8 | 21 | 0.24 | 0.22 |
| SC23A_MOUSE | Protein transport protein Sec23A | 221 | 86106 | 6.64 | 28 | 14 | 18 | 8 | 17 | 0.46 | 0.55 |
| NDUS1_MOUSE | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | 220 | 79726 | 5.51 | 12 | 6 | 9 | 5 | 9 | 0.22 | 0.37 |
| DDX1_MOUSE | ATP-dependent RNA helicase DDX1 | 220 | 82448 | 6.8 | 16 | 10 | 9 | 4 | 9 | 0.16 | 0.22 |
| SYVC_MOUSE | Valine–tRNA ligase | 220 | 140127 | 7.9 | 26 | 20 | 15 | 13 | 15 | 0.24 | 0.52 |
| RS3A_MOUSE | 40S ribosomal protein S3a | 219 | 29866 | 9.75 | 31 | 11 | 17 | 9 | 17 | 0.55 | 2.49 |
| PLOD3_MOUSE | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 | 216 | 84869 | 5.81 | 23 | 14 | 14 | 8 | 14 | 0.32 | 0.48 |
| F10A1_MOUSE | Hsc70-interacting protein | 216 | 41630 | 5.19 | 10 | 7 | 4 | 3 | 4 | 0.15 | 0.35 |
| LYZ2_MOUSE | Lysozyme C-2 | 214 | 16678 | 9.11 | 15 | 6 | 8 | 4 | 8 | 0.63 | 3.39 |
| MIF_MOUSE | Macrophage migration inhibitory factor | 214 | 12496 | 6.79 | 12 | 8 | 6 | 4 | 6 | 0.6 | 4.07 |
| DEST_MOUSE | Destrin | 214 | 18509 | 8.14 | 13 | 10 | 10 | 7 | 10 | 0.62 | 4.9 |
| HCD2_MOUSE | 3-Hydroxyacyl-CoA dehydrogenase type-2 | 214 | 27402 | 8.53 | 11 | 7 | 6 | 4 | 6 | 0.39 | 0.83 |
| DNJA2_MOUSE | DnaJ homolog subfamily A member 2 | 213 | 45717 | 6.06 | 7 | 7 | 3 | 3 | 3 | 0.18 | 0.31 |
| MARCS_MOUSE | Myristoylated alanine-rich C-kinase substrate | 213 | 29644 | 4.34 | 13 | 9 | 3 | 3 | 3 | 0.27 | 0.52 |
| LICH_MOUSE | Lysosomal acid lipase/cholesteryl ester hydrolase | 212 | 45296 | 8.16 | 11 | 10 | 6 | 6 | 6 | 0.29 | 0.9 |
| SAE2_MOUSE | SUMO-activating enzyme subunit 2 | 211 | 70525 | 5.09 | 9 | 8 | 5 | 4 | 5 | 0.2 | 0.27 |
| RL6_MOUSE | 60S ribosomal protein L6 | 210 | 33489 | 10.69 | 17 | 8 | 10 | 5 | 10 | 0.37 | 0.86 |
| FKB10_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP10 | 208 | 64656 | 5.38 | 21 | 8 | 12 | 6 | 12 | 0.3 | 0.47 |
| IPO4_MOUSE | Importin-4 | 208 | 119198 | 4.92 | 15 | 7 | 12 | 6 | 12 | 0.19 | 0.23 |
| RL14_MOUSE | 60S ribosomal protein L14 | 206 | 23549 | 11.03 | 12 | 6 | 6 | 2 | 6 | 0.33 | 0.42 |
| PLD3_MOUSE | Phospholipase D3 | 205 | 54354 | 6.07 | 15 | 10 | 9 | 7 | 9 | 0.37 | 0.85 |
| LRC59_MOUSE | Leucine-rich repeat-containing protein 59 | 205 | 34856 | 9.57 | 15 | 5 | 9 | 3 | 9 | 0.35 | 0.43 |
| NIBAN_MOUSE | Protein Niban | 204 | 102585 | 4.72 | 15 | 10 | 9 | 6 | 9 | 0.17 | 0.28 |
| CATK_MOUSE | Cathepsin K | 203 | 36865 | 8.61 | 5 | 4 | 4 | 3 | 4 | 0.25 | 0.4 |
| HMCS1_MOUSE | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 201 | 57532 | 5.65 | 17 | 10 | 12 | 6 | 12 | 0.39 | 0.66 |
| GANAB_MOUSE | Neutral alpha-glucosidase AB | 201 | 106844 | 5.67 | 16 | 11 | 12 | 9 | 12 | 0.24 | 0.42 |
| PSB5_MOUSE | Proteasome subunit beta type-5 | 200 | 28514 | 6.52 | 10 | 7 | 7 | 4 | 7 | 0.36 | 0.79 |
| LEG3_MOUSE | Galectin-3 | 200 | 27498 | 8.46 | 27 | 10 | 14 | 6 | 14 | 0.42 | 1.47 |
| SYNC_MOUSE | Asparagine–tRNA ligase, cytoplasmic | 197 | 64238 | 5.62 | 19 | 11 | 15 | 9 | 15 | 0.41 | 0.79 |
| DCTN2_MOUSE | Dynactin subunit 2 | 197 | 44090 | 5.14 | 8 | 5 | 8 | 5 | 8 | 0.34 | 0.6 |
| ZYX_MOUSE | Zyxin | 197 | 60507 | 5.99 | 15 | 7 | 7 | 3 | 7 | 0.22 | 0.23 |
| LTOR3_MOUSE | Ragulator complex protein LAMTOR3 | 195 | 13544 | 6.73 | 7 | 7 | 4 | 4 | 4 | 0.63 | 2.34 |
| S10AB_MOUSE | Protein S100-A11 | 195 | 11075 | 5.28 | 8 | 6 | 4 | 3 | 4 | 0.77 | 3.32 |
| VATL_MOUSE | V-type proton ATPase 16 kDa proteolipid subunit | 195 | 15798 | 9.1 | 4 | 3 | 2 | 1 | 2 | 0.32 | 0.68 |
| VMA5A_MOUSE | von Willebrand factor A domain-containing protein 5A | 194 | 87087 | 6.15 | 10 | 5 | 9 | 4 | 9 | 0.23 | 0.21 |
| IBP7_MOUSE | Insulin-like growth factor-binding protein 7 | 194 | 28951 | 8.71 | 16 | 7 | 8 | 3 | 8 | 0.46 | 0.54 |
| EIF3H_MOUSE | Eukaryotic translation initiation factor 3 subunit H | 192 | 39807 | 6.2 | 8 | 7 | 6 | 5 | 6 | 0.34 | 0.68 |
| FHL2_MOUSE | Four and a half LIM domains protein 2 | 192 | 32051 | 7.31 | 14 | 6 | 6 | 3 | 6 | 0.28 | 0.47 |
| PRS6B_MOUSE | 26S proteasome regulatory subunit 6B | 192 | 47379 | 5.09 | 9 | 5 | 8 | 4 | 8 | 0.4 | 0.42 |
| ETFA_MOUSE | Electron transfer flavoprotein subunit alpha, mitochondrial | 190 | 34988 | 8.62 | 17 | 8 | 8 | 5 | 7 | 0.47 | 0.81 |
| CSF1_MOUSE | Macrophage colony-stimulating factor 1 | 190 | 60611 | 5.1 | 9 | 7 | 4 | 3 | 4 | 0.13 | 0.32 |
| P4HA2_MOUSE | Prolyl 4-hydroxylase subunit alpha-2 | 189 | 60964 | 5.55 | 24 | 10 | 12 | 9 | 12 | 0.35 | 0.85 |
| LAMB1_MOUSE | Laminin subunit beta-1 | 188 | 196961 | 4.82 | 17 | 5 | 12 | 3 | 12 | 0.12 | 0.07 |
| TCPG_MOUSE | T-complex protein 1 subunit gamma | 188 | 60591 | 6.28 | 25 | 8 | 17 | 6 | 17 | 0.42 | 0.51 |
| SQSTM_MOUSE | Sequestosome-1 | 186 | 48132 | 5.09 | 10 | 7 | 6 | 4 | 6 | 0.28 | 0.41 |
| DC1I2_MOUSE | Cytoplasmic dynein 1 intermediate chain 2 | 185 | 68352 | 5.16 | 5 | 4 | 4 | 3 | 4 | 0.19 | 0.2 |
| CATA_MOUSE | Catalase | 184 | 59758 | 7.72 | 15 | 6 | 11 | 4 | 11 | 0.35 | 0.32 |
| VATC1_MOUSE | V-type proton ATPase subunit C 1 | 183 | 43860 | 7.02 | 8 | 5 | 5 | 3 | 5 | 0.25 | 0.33 |
| ECHB_MOUSE | Trifunctional enzyme subunit beta, mitochondrial | 183 | 51353 | 9.43 | 10 | 6 | 8 | 5 | 8 | 0.32 | 0.5 |
| STIP1_MOUSE | Stress-induced phosphoprotein 1 | 183 | 62542 | 6.4 | 15 | 6 | 11 | 4 | 11 | 0.35 | 0.31 |
| THY1_MOUSE | Thy-1 membrane glycoprotein | 182 | 18069 | 9.16 | 14 | 8 | 5 | 2 | 5 | 0.49 | 0.58 |
| PYRG1_MOUSE | CTP synthase 1 | 182 | 66640 | 6.14 | 17 | 10 | 11 | 7 | 11 | 0.33 | 0.65 |
| GSTO1_MOUSE | Glutathione S-transferase omega-1 | 181 | 27480 | 6.92 | 36 | 11 | 19 | 7 | 19 | 0.83 | 1.87 |
| HNRPM_MOUSE | Heterogeneous nuclear ribonucleoprotein M | 180 | 77597 | 8.8 | 16 | 8 | 11 | 6 | 11 | 0.23 | 0.38 |
| PCOC1_MOUSE | Procollagen C-endopeptidase enhancer 1 | 180 | 50136 | 8.73 | 14 | 6 | 9 | 4 | 9 | 0.38 | 0.39 |
| ATPO_MOUSE | ATP synthase subunit O, mitochondrial | 180 | 23349 | 10 | 10 | 7 | 6 | 5 | 6 | 0.38 | 1.43 |
| IDH3A_MOUSE | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | 179 | 39613 | 6.27 | 9 | 5 | 4 | 1 | 4 | 0.15 | 0.23 |
| POSTN_MOUSE | Periostin | 179 | 93085 | 7.27 | 15 | 7 | 10 | 4 | 10 | 0.25 | 0.2 |
| HCDH_MOUSE | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 179 | 34442 | 8.76 | 10 | 6 | 6 | 4 | 6 | 0.5 | 0.62 |
| DHB4_MOUSE | Peroxisomal multifunctional enzyme type 2 | 179 | 79432 | 8.76 | 22 | 13 | 13 | 8 | 13 | 0.24 | 0.52 |
| PTGIS_MOUSE | Prostacyclin synthase | 178 | 57011 | 6.26 | 21 | 9 | 18 | 8 | 18 | 0.53 | 0.79 |
| TPP1_MOUSE | Tripeptidyl-peptidase 1 | 177 | 61304 | 6.1 | 8 | 7 | 6 | 5 | 6 | 0.21 | 0.5 |
| SAE1_MOUSE | SUMO-activating enzyme subunit 1 | 177 | 38596 | 5.24 | 7 | 5 | 5 | 3 | 5 | 0.24 | 0.38 |
| 6PGL_MOUSE | 6-Phosphogluconolactonase | 177 | 27237 | 5.55 | 9 | 6 | 5 | 2 | 5 | 0.35 | 0.36 |
| SUCB1_MOUSE | Succinate–CoA ligase [ADP-forming] subunit beta, mitochondrial | 176 | 50082 | 6.57 | 8 | 5 | 6 | 3 | 6 | 0.27 | 0.28 |
| IDI1_MOUSE | Isopentenyl-diphosphate delta-isomerase 1 | 175 | 26272 | 5.79 | 14 | 5 | 6 | 4 | 6 | 0.49 | 0.88 |
| PLIN3_MOUSE | Perilipin-3 | 175 | 47233 | 5.45 | 12 | 9 | 10 | 8 | 10 | 0.43 | 1.02 |
| RL7_MOUSE | 60S ribosomal protein L7 | 174 | 31400 | 10.89 | 19 | 9 | 13 | 5 | 13 | 0.46 | 0.93 |
| RAB1B_MOUSE | Ras-related protein Rab-1B | 130 | 22173 | 5.55 | 15 | 7 | 10 | 5 | 4 | 0.52 | 1.53 |
| FERM2_MOUSE | Fermitin family homolog 2 | 174 | 77750 | 6.26 | 9 | 4 | 6 | 3 | 6 | 0.18 | 0.18 |
| CAZA2_MOUSE | F-actin-capping protein subunit alpha-2 | 173 | 32947 | 5.57 | 19 | 10 | 11 | 7 | 4 | 0.65 | 1.41 |
| CAZA1_MOUSE | F-actin-capping protein subunit alpha-1 | 61 | 32919 | 5.34 | 9 | 3 | 7 | 3 | 5 | 0.44 | 0.46 |
| EIF3K_MOUSE | Eukaryotic translation initiation factor 3 subunit K | 173 | 25070 | 4.81 | 10 | 3 | 8 | 2 | 8 | 0.61 | 0.39 |
| G6PD1_MOUSE | Glucose-6-phosphate 1-dehydrogenase X | 172 | 59225 | 6.06 | 22 | 11 | 14 | 7 | 14 | 0.4 | 0.64 |
| ASAH1_MOUSE | Acid ceramidase | 172 | 44641 | 8.68 | 10 | 7 | 9 | 6 | 9 | 0.3 | 0.75 |
| ADHX_MOUSE | Alcohol dehydrogenase class-3 | 172 | 39522 | 6.97 | 14 | 7 | 9 | 7 | 9 | 0.51 | 1.09 |
| IF4G1_MOUSE | Eukaryotic translation initiation factor 4 gamma 1 | 172 | 175967 | 5.3 | 16 | 10 | 11 | 7 | 11 | 0.11 | 0.18 |
| IL6RB_MOUSE | Interleukin-6 receptor subunit beta | 171 | 102387 | 5.35 | 14 | 8 | 4 | 2 | 4 | 0.06 | 0.09 |
| NMT1_MOUSE | Glycylpeptide N-tetradecanoyltransferase 1 | 171 | 56852 | 8.04 | 8 | 5 | 6 | 3 | 6 | 0.24 | 0.25 |
| ANXA4_MOUSE | Annexin A4 | 170 | 35893 | 5.43 | 10 | 7 | 8 | 5 | 8 | 0.3 | 0.78 |
| PCYOX_MOUSE | Prenylcysteine oxidase | 169 | 56459 | 6.44 | 8 | 5 | 7 | 4 | 7 | 0.24 | 0.34 |
| BASP1_MOUSE | Brain acid soluble protein 1 | 168 | 22074 | 4.5 | 21 | 9 | 5 | 3 | 5 | 0.42 | 0.75 |
| PSMD5_MOUSE | 26S proteasome non-ATPase regulatory subunit 5 | 168 | 55937 | 5.13 | 10 | 5 | 7 | 3 | 7 | 0.28 | 0.25 |
| IPYR_MOUSE | Inorganic pyrophosphatase | 165 | 32646 | 5.37 | 6 | 4 | 4 | 2 | 4 | 0.26 | 0.29 |
| ISG15_MOUSE | Ubiquitin-like protein ISG15 | 164 | 17886 | 7.74 | 6 | 5 | 3 | 2 | 3 | 0.5 | 0.58 |
| ACO13_MOUSE | Acyl-coenzyme A thioesterase 13 | 164 | 15173 | 8.95 | 8 | 7 | 5 | 5 | 5 | 0.56 | 2.84 |
| CP51A_MOUSE | Lanosterol 14-alpha demethylase | 164 | 56739 | 8.6 | 12 | 4 | 8 | 3 | 5 | 0.3 | 0.25 |
| IMPA1_MOUSE | Inositol monophosphatase 1 | 163 | 30416 | 5.08 | 13 | 4 | 5 | 2 | 5 | 0.21 | 0.31 |
| P3H1_MOUSE | Prolyl 3-hydroxylase 1 | 162 | 83598 | 5.03 | 12 | 6 | 8 | 4 | 8 | 0.22 | 0.22 |
| SDHA_MOUSE | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 161 | 72539 | 7.06 | 12 | 9 | 5 | 4 | 5 | 0.15 | 0.26 |
| VPS25_MOUSE | Vacuolar protein-sorting-associated protein 25 | 161 | 20735 | 5.97 | 4 | 4 | 1 | 1 | 1 | 0.19 | 0.49 |
| PHB_MOUSE | Prohibitin | 159 | 29802 | 5.57 | 17 | 10 | 11 | 7 | 11 | 0.65 | 1.65 |
| PSA6_MOUSE | Proteasome subunit alpha type-6 | 158 | 27355 | 6.34 | 14 | 5 | 8 | 3 | 8 | 0.52 | 0.58 |
| TOM40_MOUSE | Mitochondrial import receptor subunit TOM40 homolog | 156 | 37871 | 7.64 | 10 | 8 | 6 | 4 | 6 | 0.32 | 0.73 |
| UBP5_MOUSE | Ubiquitin carboxyl-terminal hydrolase 5 | 156 | 95772 | 4.89 | 18 | 10 | 10 | 7 | 10 | 0.23 | 0.36 |
| EFTU_MOUSE | Elongation factor Tu, mitochondrial | 156 | 49477 | 7.23 | 16 | 5 | 12 | 3 | 11 | 0.44 | 0.29 |
| DHB7_MOUSE | 3-Keto-steroid reductase | 154 | 37293 | 6.25 | 9 | 5 | 6 | 4 | 6 | 0.26 | 0.75 |
| CAVN1_MOUSE | Caveolae-associated protein 1 | 153 | 43927 | 5.43 | 7 | 6 | 3 | 2 | 3 | 0.15 | 0.21 |
| ODPB_MOUSE | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | 152 | 38912 | 6.41 | 14 | 7 | 6 | 4 | 6 | 0.32 | 0.53 |
| DNPEP_MOUSE | Aspartyl aminopeptidase | 152 | 52174 | 6.82 | 18 | 12 | 10 | 7 | 10 | 0.41 | 0.75 |
| PRDX5_MOUSE | Peroxiredoxin-5, mitochondrial | 151 | 21884 | 9.1 | 20 | 5 | 10 | 4 | 10 | 0.54 | 1.13 |
| ARPC2_MOUSE | Actin-related protein 2/3 complex subunit 2 | 150 | 34336 | 6.84 | 13 | 4 | 10 | 2 | 10 | 0.39 | 0.27 |
| PCKGM_MOUSE | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | 150 | 70482 | 6.92 | 4 | 3 | 3 | 2 | 3 | 0.08 | 0.13 |
| GLCM_MOUSE | Glucosylceramidase | 150 | 57585 | 7.64 | 14 | 10 | 10 | 8 | 10 | 0.37 | 0.78 |
| NB5R1_MOUSE | NADH-cytochrome b5 reductase 1 | 150 | 34113 | 8.97 | 4 | 3 | 4 | 3 | 4 | 0.26 | 0.44 |
| PUR6_MOUSE | Multifunctional protein ADE2 | 149 | 46976 | 6.94 | 11 | 6 | 8 | 5 | 8 | 0.37 | 0.56 |
| LPPRC_MOUSE | Leucine-rich PPR motif-containing protein, mitochondrial | 149 | 156516 | 6.42 | 18 | 4 | 14 | 4 | 14 | 0.16 | 0.11 |
| SAC1_MOUSE | Phosphatidylinositide phosphatase SAC1 | 148 | 66901 | 6.85 | 9 | 4 | 4 | 2 | 4 | 0.12 | 0.13 |
| GFPT2_MOUSE | Glutamine–fructose-6-phosphate aminotransferase [isomerizing] 2 | 147 | 76960 | 6.72 | 10 | 4 | 9 | 4 | 9 | 0.21 | 0.24 |
| MTAP_MOUSE | S-Methyl-5 ′-thioadenosine phosphorylase | 146 | 31042 | 6.71 | 7 | 4 | 5 | 3 | 5 | 0.29 | 0.49 |
| IKIP_MOUSE | Inhibitor of nuclear factor kappa-B kinase-interacting protein | 146 | 42505 | 5.03 | 13 | 10 | 4 | 2 | 4 | 0.29 | 0.22 |
| ADPGK_MOUSE | ADP-dependent glucokinase | 146 | 53869 | 5.37 | 9 | 7 | 4 | 3 | 4 | 0.15 | 0.26 |
| MA2A1_MOUSE | Alpha-mannosidase 2 | 145 | 131548 | 8.17 | 12 | 7 | 10 | 6 | 10 | 0.15 | 0.21 |
| SYLC_MOUSE | Leucine–tRNA ligase, cytoplasmic | 145 | 134106 | 6.64 | 13 | 6 | 10 | 4 | 10 | 0.16 | 0.13 |
| SYIC_MOUSE | Isoleucine–tRNA ligase, cytoplasmic | 145 | 144179 | 6.14 | 18 | 8 | 12 | 5 | 12 | 0.19 | 0.16 |
| PARVA_MOUSE | Alpha-parvin | 144 | 42304 | 5.69 | 9 | 5 | 5 | 3 | 5 | 0.26 | 0.34 |
| DYN2_MOUSE | Dynamin-2 | 144 | 98084 | 7.02 | 12 | 5 | 9 | 3 | 7 | 0.19 | 0.14 |
| IMA1_MOUSE | Importin subunit alpha-1 | 144 | 57892 | 5.49 | 13 | 4 | 8 | 3 | 8 | 0.27 | 0.24 |
| CYGB_MOUSE | Cytoglobin | 144 | 21452 | 6.32 | 8 | 4 | 3 | 2 | 3 | 0.33 | 0.47 |
| ODO1_MOUSE | 2-Oxoglutarate dehydrogenase, mitochondrial | 143 | 116375 | 6.36 | 34 | 6 | 9 | 4 | 9 | 0.14 | 0.15 |
| MK01_MOUSE | Mitogen-activated protein kinase 1 | 143 | 41249 | 6.5 | 8 | 5 | 7 | 4 | 7 | 0.38 | 0.5 |
| UD17C_MOUSE | UDP-glucuronosyltransferase 1-7C | 143 | 59719 | 8.64 | 14 | 5 | 8 | 3 | 8 | 0.27 | 0.23 |
| HMOX1_MOUSE | Heme oxygenase 1 | 142 | 32908 | 6.08 | 16 | 7 | 10 | 6 | 10 | 0.57 | 1.13 |
| USO1_MOUSE | General vesicular transport factor p115 | 142 | 106917 | 4.85 | 11 | 7 | 10 | 6 | 10 | 0.17 | 0.26 |
| RTN4_MOUSE | Reticulon-4 | 142 | 126535 | 4.47 | 23 | 9 | 10 | 5 | 10 | 0.16 | 0.18 |
| THIKA_MOUSE | 3-Ketoacyl-CoA thiolase A, peroxisomal | 141 | 43926 | 8.74 | 14 | 8 | 10 | 5 | 10 | 0.47 | 0.61 |
| MGST1_MOUSE | Microsomal glutathione S-transferase 1 | 141 | 17540 | 9.67 | 6 | 5 | 2 | 2 | 2 | 0.22 | 0.6 |
| HYEP_MOUSE | Epoxide hydrolase 1 | 141 | 52543 | 8.43 | 18 | 5 | 14 | 4 | 14 | 0.41 | 0.37 |
| GUAD_MOUSE | Guanine deaminase | 141 | 50981 | 5.36 | 6 | 5 | 4 | 3 | 4 | 0.24 | 0.28 |
| LASP1_MOUSE | LIM and SH3 domain protein 1 | 140 | 29975 | 6.61 | 23 | 7 | 7 | 2 | 7 | 0.29 | 0.51 |
| 4F2_MOUSE | 4F2 cell-surface antigen heavy chain | 139 | 58300 | 5.62 | 12 | 8 | 7 | 5 | 7 | 0.26 | 0.43 |
| PPAC_MOUSE | Low molecular weight phosphotyrosine protein phosphatase | 139 | 18180 | 6.3 | 6 | 3 | 4 | 3 | 4 | 0.45 | 0.97 |
| CD36_MOUSE | Platelet glycoprotein 4 | 139 | 52664 | 8.6 | 6 | 5 | 6 | 5 | 6 | 0.19 | 0.49 |
| ATPG_MOUSE | ATP synthase subunit gamma, mitochondrial | 139 | 32865 | 9.06 | 9 | 6 | 6 | 3 | 6 | 0.3 | 0.46 |
| AL9A1_MOUSE | 4-Trimethylaminobutyraldehyde dehydrogenase | 138 | 53480 | 6.63 | 7 | 6 | 4 | 3 | 4 | 0.18 | 0.26 |
| ASPH_MOUSE | Aspartyl/asparaginyl beta-hydroxylase | 138 | 82991 | 4.97 | 6 | 4 | 5 | 3 | 5 | 0.1 | 0.16 |
| ATPD_MOUSE | ATP synthase subunit delta, mitochondrial | 138 | 17589 | 5.03 | 14 | 9 | 5 | 2 | 5 | 0.63 | 0.6 |
| IRGM1_MOUSE | Immunity-related GTPase family M protein 1 | 137 | 46522 | 8.56 | 7 | 5 | 4 | 4 | 4 | 0.2 | 0.43 |
| PON3_MOUSE | Serum paraoxonase/lactonase 3 | 137 | 39326 | 5.44 | 9 | 5 | 5 | 4 | 5 | 0.26 | 0.53 |
| UMPS_MOUSE | Uridine 5 ′-monophosphate synthase | 137 | 52259 | 6.17 | 7 | 5 | 6 | 4 | 6 | 0.23 | 0.38 |
| CMTD1_MOUSE | Catechol O-methyltransferase domain-containing protein 1 | 137 | 28943 | 8.61 | 7 | 4 | 6 | 3 | 6 | 0.51 | 0.54 |
| SYUG_MOUSE | Gamma-synuclein | 137 | 13152 | 4.68 | 6 | 5 | 3 | 3 | 3 | 0.4 | 1.54 |
| CLIC4_MOUSE | Chloride intracellular channel protein 4 | 137 | 28711 | 5.44 | 10 | 7 | 8 | 5 | 7 | 0.41 | 1.06 |
| CRIP2_MOUSE | Cysteine-rich protein 2 | 136 | 22712 | 8.94 | 8 | 5 | 3 | 3 | 3 | 0.34 | 1.07 |
| PLOD1_MOUSE | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | 136 | 83542 | 6.08 | 12 | 5 | 10 | 4 | 10 | 0.2 | 0.22 |
| CPNE1_MOUSE | Copine-1 | 136 | 58849 | 5.4 | 7 | 4 | 5 | 3 | 5 | 0.18 | 0.24 |
| RL4_MOUSE | 60S ribosomal protein L4 | 136 | 47124 | 11.01 | 40 | 10 | 20 | 9 | 20 | 0.45 | 1.21 |
| RN213_MOUSE | E3 ubiquitin-protein ligase RNF213 | 135 | 584411 | 6.35 | 29 | 8 | 22 | 6 | 22 | 0.09 | 0.04 |
| CD109_MOUSE | CD109 antigen | 134 | 161557 | 5.37 | 5 | 5 | 4 | 4 | 4 | 0.05 | 0.11 |
| LAMP2_MOUSE | Lysosome-associated membrane glycoprotein 2 | 134 | 45652 | 7.05 | 11 | 4 | 7 | 3 | 7 | 0.16 | 0.31 |
| ARL8A_MOUSE | ADP-ribosylation factor-like protein 8A | 134 | 21376 | 7.63 | 14 | 8 | 6 | 4 | 6 | 0.4 | 1.16 |
| NCEH1_MOUSE | Neutral cholesterol ester hydrolase 1 | 133 | 45711 | 6.56 | 12 | 7 | 7 | 4 | 7 | 0.34 | 0.44 |
| GNPI1_MOUSE | Glucosamine-6-phosphate isomerase 1 | 133 | 32528 | 6.13 | 8 | 5 | 7 | 4 | 7 | 0.36 | 0.67 |
| NOMO1_MOUSE | Nodal modulator 1 | 133 | 133336 | 5.75 | 13 | 8 | 10 | 7 | 10 | 0.19 | 0.25 |
| RRAS_MOUSE | Ras-related protein R-Ras | 133 | 23749 | 6.32 | 6 | 4 | 4 | 3 | 2 | 0.23 | 0.68 |
| RCN1_MOUSE | Reticulocalbin-1 | 133 | 38090 | 4.7 | 12 | 6 | 6 | 4 | 6 | 0.25 | 0.55 |
| PFKAP_MOUSE | ATP-dependent 6-phosphofructokinase, platelet type | 132 | 85400 | 6.73 | 12 | 4 | 9 | 2 | 9 | 0.24 | 0.1 |
| BLVRB_MOUSE | Flavin reductase (NADPH) | 132 | 22183 | 6.49 | 15 | 6 | 9 | 5 | 9 | 0.69 | 1.53 |
| TMM43_MOUSE | Transmembrane protein 43 | 132 | 44755 | 6.85 | 6 | 5 | 6 | 5 | 6 | 0.31 | 0.59 |
| P5CS_MOUSE | Delta-1-pyrroline-5-carboxylate synthase | 131 | 87212 | 7.18 | 12 | 5 | 9 | 4 | 9 | 0.19 | 0.21 |
| MATR3_MOUSE | Matrin-3 | 131 | 94572 | 5.87 | 6 | 4 | 5 | 3 | 5 | 0.14 | 0.14 |
| PTGR1_MOUSE | Prostaglandin reductase 1 | 131 | 35537 | 8.09 | 5 | 5 | 5 | 5 | 5 | 0.26 | 0.79 |
| COTL1_MOUSE | Coactosin-like protein | 131 | 15934 | 5.28 | 10 | 3 | 6 | 2 | 6 | 0.55 | 0.67 |
| ACAD9_MOUSE | Acyl-CoA dehydrogenase family member 9, mitochondrial | 131 | 68679 | 7.16 | 6 | 6 | 3 | 3 | 3 | 0.11 | 0.2 |
| IFM3_MOUSE | Interferon-induced transmembrane protein 3 | 130 | 14945 | 6.89 | 14 | 6 | 3 | 3 | 2 | 0.32 | 1.98 |
| IFM2_MOUSE | Interferon-induced transmembrane protein 2 | 59 | 15733 | 6.81 | 9 | 3 | 3 | 2 | 2 | 0.17 | 0.68 |
| DHX9_MOUSE | ATP-dependent RNA helicase A | 130 | 149381 | 6.39 | 17 | 9 | 11 | 5 | 11 | 0.13 | 0.15 |
| SDCB1_MOUSE | Syntenin-1 | 130 | 32359 | 6.66 | 9 | 4 | 4 | 3 | 4 | 0.29 | 0.47 |
| ORN_MOUSE | Oligoribonuclease, mitochondrial | 129 | 26722 | 6.67 | 8 | 4 | 6 | 3 | 6 | 0.38 | 0.59 |
| ITM2B_MOUSE | Integral membrane protein 2B | 128 | 30240 | 5.14 | 16 | 7 | 7 | 5 | 7 | 0.45 | 0.99 |
| EIF3I_MOUSE | Eukaryotic translation initiation factor 3 subunit I | 128 | 36438 | 5.38 | 15 | 6 | 9 | 5 | 9 | 0.47 | 0.77 |
| FIS1_MOUSE | Mitochondrial fission 1 protein | 128 | 16998 | 8.56 | 5 | 4 | 4 | 3 | 4 | 0.31 | 1.07 |
| SAHH_MOUSE | Adenosylhomocysteinase | 128 | 47657 | 6.08 | 15 | 8 | 11 | 7 | 11 | 0.43 | 0.84 |
| TFR1_MOUSE | Transferrin receptor protein 1 | 127 | 85677 | 6.13 | 4 | 4 | 3 | 3 | 3 | 0.05 | 0.16 |
| STML2_MOUSE | Stomatin-like protein 2, mitochondrial | 127 | 38361 | 8.95 | 13 | 8 | 9 | 6 | 9 | 0.52 | 0.91 |
| IF6_MOUSE | Eukaryotic translation initiation factor 6 | 127 | 26494 | 4.63 | 7 | 6 | 5 | 4 | 5 | 0.56 | 1.18 |
| VPP1_MOUSE | V-type proton ATPase 116 kDa subunit a isoform 1 | 126 | 96404 | 6.29 | 10 | 4 | 7 | 3 | 7 | 0.13 | 0.14 |
| PREP_MOUSE | Presequence protease, mitochondrial | 126 | 117297 | 6.76 | 10 | 6 | 10 | 6 | 10 | 0.17 | 0.24 |
| BIN1_MOUSE | Myc box-dependent-interacting protein 1 | 125 | 64430 | 4.95 | 16 | 6 | 10 | 5 | 10 | 0.32 | 0.38 |
| CSN7A_MOUSE | COP9 signalosome complex subunit 7a | 125 | 30206 | 7.68 | 8 | 6 | 4 | 3 | 4 | 0.26 | 0.51 |
| SSRD_MOUSE | Translocon-associated protein subunit delta | 125 | 18924 | 5.5 | 7 | 6 | 3 | 2 | 3 | 0.25 | 0.55 |
| COR1B_MOUSE | Coronin-1B | 124 | 53878 | 5.54 | 15 | 7 | 7 | 3 | 7 | 0.29 | 0.26 |
| VKOR1_MOUSE | Vitamin K epoxide reductase complex subunit 1 | 124 | 17756 | 9.37 | 6 | 4 | 2 | 2 | 2 | 0.19 | 0.59 |
| FUBP2_MOUSE | Far upstream element-binding protein 2 | 124 | 76728 | 6.9 | 11 | 4 | 7 | 3 | 7 | 0.12 | 0.18 |
| MFGM_MOUSE | Lactadherin | 122 | 51208 | 6.1 | 22 | 9 | 13 | 7 | 13 | 0.45 | 0.77 |
| PSME2_MOUSE | Proteasome activator complex subunit 2 | 122 | 27040 | 5.54 | 10 | 6 | 7 | 4 | 7 | 0.46 | 0.84 |
| C1TC_MOUSE | C-1-tetrahydrofolate synthase, cytoplasmic | 122 | 101136 | 6.7 | 12 | 5 | 8 | 3 | 8 | 0.12 | 0.13 |
| RISC_MOUSE | Retinoid-inducible serine carboxypeptidase | 122 | 50932 | 5.48 | 11 | 5 | 8 | 4 | 8 | 0.21 | 0.39 |
| ETFB_MOUSE | Electron transfer flavoprotein subunit beta | 122 | 27606 | 8.24 | 16 | 4 | 8 | 2 | 8 | 0.43 | 0.35 |
| PRRC1_MOUSE | Protein PRRC1 | 122 | 46268 | 5.62 | 10 | 5 | 8 | 5 | 8 | 0.31 | 0.57 |
| MAOM_MOUSE | NAD-dependent malic enzyme, mitochondrial | 121 | 65757 | 7.53 | 6 | 5 | 3 | 3 | 3 | 0.15 | 0.21 |
| SH3L1_MOUSE | SH3 domain-binding glutamic acid-rich-like protein | 120 | 12803 | 4.87 | 4 | 3 | 3 | 2 | 3 | 0.38 | 0.89 |
| PSB1_MOUSE | Proteasome subunit beta type-1 | 120 | 26355 | 7.67 | 9 | 5 | 7 | 4 | 7 | 0.55 | 0.88 |
| LY6E_MOUSE | Lymphocyte antigen 6E | 120 | 13791 | 6.67 | 4 | 3 | 2 | 2 | 2 | 0.29 | 1.44 |
| SPRC_MOUSE | SPARC | 120 | 34428 | 4.77 | 8 | 5 | 6 | 4 | 6 | 0.33 | 0.62 |
| ECI2_MOUSE | Enoyl-CoA delta isomerase 2, mitochondrial | 119 | 43240 | 9.08 | 9 | 5 | 4 | 1 | 4 | 0.16 | 0.1 |
| AIMP2_MOUSE | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | 119 | 35355 | 7.7 | 9 | 6 | 5 | 4 | 5 | 0.34 | 0.6 |
| TOM1_MOUSE | Target of Myb protein 1 | 119 | 54291 | 4.83 | 5 | 4 | 5 | 4 | 5 | 0.22 | 0.36 |
| TALDO_MOUSE | Transaldolase | 119 | 37363 | 6.57 | 16 | 5 | 10 | 4 | 10 | 0.31 | 0.56 |
| SAMH1_MOUSE | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | 118 | 72604 | 8.17 | 8 | 4 | 6 | 2 | 6 | 0.17 | 0.12 |
| LPP_MOUSE | Lipoma-preferred partner homolog | 118 | 65848 | 7.19 | 11 | 7 | 6 | 4 | 6 | 0.22 | 0.29 |
| STX12_MOUSE | Syntaxin-12 | 118 | 31176 | 5.33 | 9 | 5 | 8 | 4 | 8 | 0.45 | 0.7 |
| CYB5B_MOUSE | Cytochrome b5 type B | 118 | 16308 | 4.79 | 5 | 4 | 4 | 3 | 4 | 0.51 | 1.13 |
| PDLI1_MOUSE | PDZ and LIM domain protein 1 | 117 | 35752 | 6.38 | 9 | 4 | 7 | 4 | 7 | 0.38 | 0.59 |
| FABP4_MOUSE | Fatty acid-binding protein, adipocyte | 117 | 14641 | 8.53 | 6 | 3 | 5 | 3 | 5 | 0.47 | 1.31 |
| BACH_MOUSE | Cytosolic acyl coenzyme A thioester hydrolase | 117 | 42510 | 8.9 | 6 | 3 | 5 | 3 | 5 | 0.24 | 0.34 |
| NSF1C_MOUSE | NSFL1 cofactor p47 | 117 | 40685 | 5.04 | 9 | 7 | 2 | 2 | 2 | 0.11 | 0.23 |
| CMC1_MOUSE | Calcium-binding mitochondrial carrier protein Aralar1 | 117 | 74523 | 8.43 | 8 | 2 | 7 | 2 | 7 | 0.18 | 0.12 |
| S10A1_MOUSE | Protein S100-A1 | 117 | 10498 | 4.37 | 2 | 2 | 1 | 1 | 1 | 0.16 | 0.47 |
| ACDSB_MOUSE | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial | 116 | 47843 | 8 | 7 | 4 | 4 | 2 | 4 | 0.19 | 0.19 |
| CO4A1_MOUSE | Collagen alpha-1(IV) chain | 116 | 160579 | 8.51 | 16 | 3 | 8 | 2 | 8 | 0.13 | 0.08 |
| SPEE_MOUSE | Spermidine synthase | 116 | 33973 | 5.31 | 10 | 4 | 10 | 4 | 10 | 0.65 | 0.63 |
| EDC4_MOUSE | Enhancer of mRNA-decapping protein 4 | 115 | 152389 | 5.51 | 6 | 4 | 3 | 2 | 3 | 0.03 | 0.06 |
| SGT1_MOUSE | Protein SGT1 homolog | 115 | 38135 | 5.32 | 4 | 3 | 3 | 2 | 3 | 0.16 | 0.24 |
| SQOR_MOUSE | Sulfide:quinone oxidoreductase, mitochondrial | 115 | 50250 | 9.2 | 6 | 4 | 4 | 2 | 4 | 0.12 | 0.18 |
| SYWC_MOUSE | Tryptophan–tRNA ligase, cytoplasmic | 114 | 54323 | 6.44 | 9 | 5 | 7 | 4 | 7 | 0.26 | 0.36 |
| PDXD1_MOUSE | Pyridoxal-dependent decarboxylase domain-containing protein 1 | 114 | 87281 | 5.31 | 12 | 6 | 7 | 4 | 7 | 0.18 | 0.21 |
| BROX_MOUSE | BRO1 domain-containing protein BROX | 114 | 46172 | 7.59 | 6 | 4 | 2 | 2 | 2 | 0.11 | 0.2 |
| SCFD1_MOUSE | Sec1 family domain-containing protein 1 | 112 | 72277 | 5.98 | 5 | 2 | 5 | 2 | 5 | 0.19 | 0.12 |
| DHPR_MOUSE | Dihydropteridine reductase | 113 | 25554 | 7.67 | 4 | 2 | 2 | 1 | 2 | 0.11 | 0.18 |
| NDUS3_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | 113 | 30131 | 6.67 | 4 | 3 | 2 | 1 | 2 | 0.11 | 0.15 |
| TAGL_MOUSE | Transgelin | 113 | 22561 | 8.85 | 12 | 6 | 8 | 5 | 8 | 0.51 | 2 |
| PALLD_MOUSE | Palladin | 113 | 152037 | 5.87 | 11 | 4 | 9 | 3 | 9 | 0.11 | 0.09 |
| RL18_MOUSE | 60S ribosomal protein L18 | 112 | 21631 | 11.79 | 7 | 6 | 4 | 3 | 4 | 0.25 | 0.77 |
| MPCP_MOUSE | Phosphate carrier protein, mitochondrial | 112 | 39606 | 9.36 | 39 | 5 | 9 | 3 | 9 | 0.28 | 0.37 |
| CYFP1_MOUSE | Cytoplasmic FMR1-interacting protein 1 | 112 | 145148 | 6.46 | 6 | 3 | 6 | 3 | 6 | 0.07 | 0.09 |
| G3BP1_MOUSE | Ras GTPase-activating protein-binding protein 1 | 112 | 51797 | 5.41 | 6 | 4 | 6 | 4 | 6 | 0.17 | 0.38 |
| PYGB_MOUSE | Glycogen phosphorylase, brain form | 112 | 96668 | 6.28 | 8 | 3 | 7 | 2 | 7 | 0.15 | 0.09 |
| S10A6_MOUSE | Protein S100-A6 | 112 | 10044 | 5.3 | 12 | 5 | 6 | 3 | 6 | 0.66 | 4.05 |
| TCTP_MOUSE | Translationally-controlled tumor protein | 110 | 19450 | 4.76 | 9 | 4 | 5 | 3 | 5 | 0.31 | 0.89 |
| RL5_MOUSE | 60S ribosomal protein L5 | 110 | 34379 | 9.78 | 12 | 4 | 9 | 3 | 9 | 0.43 | 0.44 |
| SYHC_MOUSE | Histidine–tRNA ligase, cytoplasmic | 110 | 57396 | 5.79 | 10 | 5 | 6 | 4 | 6 | 0.22 | 0.34 |
| CLPT1_MOUSE | Cleft lip and palate transmembrane protein 1 homolog | 109 | 75243 | 5.88 | 13 | 9 | 5 | 4 | 5 | 0.18 | 0.25 |
| ACSF2_MOUSE | Acyl-CoA synthetase family member 2, mitochondrial | 109 | 67907 | 8.44 | 7 | 4 | 6 | 3 | 6 | 0.2 | 0.2 |
| THIL_MOUSE | Acetyl-CoA acetyltransferase, mitochondrial | 108 | 44787 | 8.71 | 15 | 9 | 6 | 4 | 6 | 0.23 | 0.45 |
| USMG5_MOUSE | Upregulated during skeletal muscle growth protein 5 | 108 | 6377 | 9.84 | 2 | 2 | 1 | 1 | 1 | 0.26 | 0.87 |
| ISC2A_MOUSE | Isochorismatase domain-containing protein 2A | 108 | 22403 | 8.25 | 2 | 2 | 1 | 1 | 1 | 0.09 | 0.2 |
| HNRPL_MOUSE | Heterogeneous nuclear ribonucleoprotein L | 107 | 63923 | 8.33 | 14 | 6 | 6 | 2 | 6 | 0.24 | 0.14 |
| PP1R7_MOUSE | Protein phosphatase 1 regulatory subunit 7 | 106 | 41266 | 4.85 | 3 | 3 | 2 | 2 | 2 | 0.11 | 0.22 |
| SE1L1_MOUSE | Protein sel-1 homolog 1 | 106 | 88285 | 5.36 | 4 | 3 | 3 | 2 | 3 | 0.1 | 0.1 |
| CCD47_MOUSE | Coiled-coil domain-containing protein 47 | 106 | 55808 | 4.74 | 6 | 5 | 3 | 2 | 1 | 0.14 | 0.16 |
| HUWE1_MOUSE | E3 ubiquitin-protein ligase HUWE1 | 105 | 482332 | 5.1 | 15 | 2 | 13 | 1 | 13 | 0.06 | 0.01 |
| GDIR2_MOUSE | Rho GDP-dissociation inhibitor 2 | 104 | 22836 | 4.97 | 5 | 4 | 4 | 3 | 4 | 0.45 | 0.72 |
| SGPL1_MOUSE | Sphingosine-1-phosphate lyase 1 | 104 | 63636 | 9.2 | 12 | 5 | 8 | 4 | 8 | 0.29 | 0.3 |
| PLIN4_MOUSE | Perilipin-4 | 104 | 139328 | 8.81 | 7 | 4 | 6 | 3 | 6 | 0.07 | 0.09 |
| UBA6_MOUSE | Ubiquitin-like modifier-activating enzyme 6 | 104 | 117891 | 5.75 | 6 | 2 | 5 | 1 | 5 | 0.07 | 0.04 |
| ARL1_MOUSE | ADP-ribosylation factor-like protein 1 | 104 | 20398 | 5.63 | 8 | 5 | 6 | 3 | 6 | 0.57 | 0.83 |
| G6PE_MOUSE | GDH/6PGL endoplasmic bifunctional protein | 103 | 88872 | 6.44 | 6 | 4 | 5 | 3 | 5 | 0.11 | 0.15 |
| UBP14_MOUSE | Ubiquitin carboxyl-terminal hydrolase 14 | 103 | 55966 | 5.15 | 8 | 4 | 7 | 3 | 7 | 0.25 | 0.25 |
| FSTL1_MOUSE | Follistatin-related protein 1 | 103 | 34532 | 5.58 | 6 | 2 | 5 | 1 | 5 | 0.21 | 0.13 |
| PGFRB_MOUSE | Platelet-derived growth factor receptor beta | 103 | 122728 | 4.99 | 10 | 7 | 7 | 4 | 7 | 0.12 | 0.15 |
| CD180_MOUSE | CD180 antigen | 103 | 74255 | 5.55 | 4 | 2 | 3 | 1 | 3 | 0.12 | 0.06 |
| GLRX1_MOUSE | Glutaredoxin-1 | 103 | 11863 | 8.67 | 4 | 3 | 2 | 1 | 2 | 0.49 | 0.41 |
| GCN1_MOUSE | eIF-2-alpha kinase activator GCN1 | 102 | 292834 | 7.14 | 18 | 5 | 16 | 3 | 16 | 0.12 | 0.04 |
| PLPP3_MOUSE | Phospholipid phosphatase 3 | 102 | 35193 | 9.25 | 6 | 4 | 4 | 2 | 4 | 0.23 | 0.27 |
| NU155_MOUSE | Nuclear pore complex protein Nup155 | 101 | 155019 | 5.77 | 9 | 4 | 7 | 2 | 7 | 0.09 | 0.06 |
| ACADV_MOUSE | Very-long-chain-specific acyl-CoA dehydrogenase, mitochondrial | 95 | 70831 | 8.91 | 3 | 2 | 3 | 2 | 3 | 0.08 | 0.13 |
| PFD5_MOUSE | Prefoldin subunit 5 | 101 | 17345 | 5.93 | 8 | 2 | 5 | 2 | 5 | 0.43 | 0.61 |
| PSA2_MOUSE | Proteasome subunit alpha type-2 | 101 | 25910 | 6.92 | 9 | 5 | 8 | 5 | 8 | 0.52 | 1.22 |
| ZWINT_MOUSE | ZW10 interactor | 101 | 28695 | 8.56 | 4 | 2 | 3 | 1 | 3 | 0.24 | 0.33 |
| ERG7_MOUSE | Lanosterol synthase | 101 | 83088 | 5.96 | 13 | 4 | 10 | 4 | 10 | 0.21 | 0.22 |
| EMC1_MOUSE | ER membrane protein complex subunit 1 | 100 | 111535 | 7 | 12 | 6 | 11 | 5 | 11 | 0.19 | 0.21 |
| PYC_MOUSE | Pyruvate carboxylase, mitochondrial | 100 | 129602 | 6.25 | 4 | 2 | 4 | 2 | 4 | 0.07 | 0.07 |
| SRPRB_MOUSE | Signal recognition particle receptor subunit beta | 99 | 29561 | 9.34 | 7 | 5 | 4 | 2 | 4 | 0.26 | 0.32 |
| CD34_MOUSE | Hematopoietic progenitor cell antigen CD34 | 99 | 40957 | 5.2 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.11 |
| AIFM1_MOUSE | Apoptosis-inducing factor 1, mitochondrial | 98 | 66724 | 9.23 | 5 | 3 | 4 | 3 | 4 | 0.14 | 0.21 |
| 3HIDH_MOUSE | 3-Hydroxyisobutyrate dehydrogenase, mitochondrial | 98 | 35417 | 8.37 | 6 | 3 | 5 | 2 | 5 | 0.31 | 0.26 |
| ATLA3_MOUSE | Atlastin-3 | 98 | 60537 | 5.73 | 7 | 5 | 5 | 4 | 5 | 0.18 | 0.32 |
| TXTP_MOUSE | Tricarboxylate transport protein, mitochondrial | 98 | 33910 | 9.91 | 6 | 2 | 4 | 2 | 4 | 0.27 | 0.28 |
| IF1A_MOUSE | Eukaryotic translation initiation factor 1A | 98 | 16492 | 5.07 | 8 | 4 | 3 | 1 | 3 | 0.21 | 0.28 |
| TRI25_MOUSE | E3 ubiquitin/ISG15 ligase TRIM25 | 98 | 71680 | 8.62 | 9 | 2 | 7 | 2 | 7 | 0.21 | 0.12 |
| QCR1_MOUSE | Cytochrome b-c1 complex subunit 1, mitochondrial | 97 | 52818 | 5.81 | 11 | 3 | 6 | 2 | 6 | 0.26 | 0.17 |
| ACACA_MOUSE | Acetyl-CoA carboxylase 1 | 96 | 265088 | 5.97 | 11 | 6 | 10 | 5 | 10 | 0.11 | 0.08 |
| DYLT1_MOUSE | Dynein light chain Tctex-type 1 | 95 | 12475 | 5 | 1 | 1 | 1 | 1 | 1 | 0.16 | 0.39 |
| ATOX1_MOUSE | Copper transport protein ATOX1 | 95 | 7334 | 6.04 | 5 | 3 | 3 | 2 | 3 | 0.72 | 4.12 |
| RENBP_MOUSE | N-Acylglucosamine 2-epimerase | 94 | 49739 | 5.69 | 4 | 3 | 3 | 2 | 3 | 0.13 | 0.18 |
| ROAA_MOUSE | Heterogeneous nuclear ribonucleoprotein A/B | 94 | 30812 | 7.68 | 15 | 6 | 7 | 5 | 6 | 0.29 | 0.96 |
| GNS_MOUSE | N-Acetylglucosamine-6-sulfatase | 94 | 61136 | 8.52 | 13 | 5 | 8 | 3 | 8 | 0.22 | 0.23 |
| TMX1_MOUSE | Thioredoxin-related transmembrane protein 1 | 93 | 31376 | 5.19 | 3 | 3 | 1 | 1 | 1 | 0.04 | 0.14 |
| OCAD1_MOUSE | OCIA domain-containing protein 1 | 90 | 27593 | 7.66 | 4 | 3 | 3 | 2 | 3 | 0.2 | 0.35 |
| STS_MOUSE | Steryl-sulfatase | 93 | 66549 | 8.83 | 2 | 2 | 1 | 1 | 1 | 0.03 | 0.06 |
| TIF1B_MOUSE | Transcription intermediary factor 1-beta | 93 | 88791 | 5.52 | 11 | 4 | 7 | 4 | 7 | 0.12 | 0.21 |
| LMAN2_MOUSE | Vesicular integral-membrane protein VIP36 | 92 | 40404 | 6.46 | 10 | 4 | 8 | 3 | 8 | 0.32 | 0.36 |
| SH3L3_MOUSE | SH3 domain-binding glutamic acid-rich-like protein 3 | 92 | 10470 | 5.02 | 10 | 6 | 4 | 2 | 4 | 0.68 | 3.72 |
| PTGR3_MOUSE | Prostaglandin reductase-3 | 92 | 40503 | 7.01 | 4 | 3 | 4 | 3 | 4 | 0.25 | 0.36 |
| KIME_MOUSE | Mevalonate kinase | 91 | 41851 | 6.22 | 5 | 2 | 5 | 2 | 5 | 0.3 | 0.22 |
| PAPS2_MOUSE | Bifunctional 3 ′-phosphoadenosine 5 ′-phosphosulfate synthase 2 | 91 | 70306 | 7.31 | 8 | 3 | 7 | 3 | 7 | 0.24 | 0.2 |
| PGRC1_MOUSE | Membrane-associated progesterone receptor component 1 | 91 | 21681 | 4.57 | 8 | 5 | 4 | 3 | 4 | 0.4 | 0.77 |
| FRIH_MOUSE | Ferritin heavy chain | 90 | 21053 | 5.53 | 16 | 6 | 9 | 4 | 9 | 0.73 | 1.19 |
| THIM_MOUSE | 3-Ketoacyl-CoA thiolase, mitochondrial | 90 | 41803 | 8.33 | 13 | 8 | 8 | 4 | 8 | 0.34 | 0.49 |
| LGMN_MOUSE | Legumain | 90 | 49341 | 5.92 | 6 | 3 | 5 | 2 | 5 | 0.22 | 0.18 |
| CD9_MOUSE | CD9 antigen | 90 | 25241 | 6.88 | 4 | 3 | 1 | 1 | 1 | 0.11 | 0.39 |
| HEBP1_MOUSE | Heme-binding protein 1 | 90 | 21053 | 5.18 | 2 | 1 | 2 | 1 | 2 | 0.29 | 0.22 |
| SCPDL_MOUSE | Saccharopine dehydrogenase-like oxidoreductase | 90 | 47099 | 8.86 | 4 | 4 | 2 | 2 | 2 | 0.13 | 0.19 |
| TOIP1_MOUSE | Torsin-1A-interacting protein 1 | 90 | 66740 | 6.58 | 2 | 1 | 2 | 1 | 2 | 0.04 | 0.06 |
| ACADL_MOUSE | Long-chain specific acyl-CoA dehydrogenase, mitochondrial | 89 | 47877 | 8.53 | 15 | 5 | 8 | 4 | 8 | 0.35 | 0.42 |
| PTGES_MOUSE | Prostaglandin E synthase | 89 | 17274 | 9.47 | 2 | 2 | 1 | 1 | 1 | 0.17 | 0.27 |
| PSA1_MOUSE | Proteasome subunit alpha type-1 | 89 | 29528 | 6 | 11 | 5 | 7 | 4 | 7 | 0.32 | 0.75 |
| DX39A_MOUSE | ATP-dependent RNA helicase DDX39A | 89 | 49036 | 5.46 | 5 | 2 | 5 | 2 | 5 | 0.23 | 0.19 |
| ATP5H_MOUSE | ATP synthase subunit d, mitochondrial | 89 | 18738 | 5.52 | 7 | 4 | 6 | 3 | 6 | 0.55 | 0.93 |
| CNPY2_MOUSE | Protein canopy homolog 2 | 88 | 20754 | 4.95 | 8 | 5 | 3 | 2 | 3 | 0.28 | 0.49 |
| AACS_MOUSE | Acetoacetyl-CoA synthetase | 88 | 75152 | 6.25 | 15 | 5 | 11 | 5 | 11 | 0.28 | 0.32 |
| GNAS1_MOUSE | Guanine nucleotide-binding protein G(s) subunit alpha isoforms Xlas | 88 | 121429 | 4.72 | 15 | 5 | 8 | 3 | 7 | 0.11 | 0.11 |
| HA11_MOUSE | H-2 class I histocompatibility antigen, D-B alpha chain | 88 | 40810 | 6.28 | 13 | 2 | 8 | 2 | 8 | 0.29 | 0.23 |
| GNA11_MOUSE | Guanine nucleotide-binding protein subunit alpha-11 | 88 | 41997 | 5.7 | 7 | 4 | 5 | 3 | 2 | 0.22 | 0.35 |
| VATG1_MOUSE | V-type proton ATPase subunit G 1 | 88 | 13716 | 7.77 | 4 | 3 | 2 | 1 | 2 | 0.24 | 0.35 |
| NDUS5_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 | 87 | 12639 | 9.1 | 1 | 1 | 1 | 1 | 1 | 0.11 | 0.38 |
| SYFB_MOUSE | Phenylalanine–tRNA ligase beta subunit | 87 | 65655 | 6.69 | 8 | 3 | 8 | 3 | 8 | 0.21 | 0.21 |
| THOP1_MOUSE | Thimet oligopeptidase | 87 | 77976 | 5.72 | 3 | 3 | 2 | 2 | 2 | 0.04 | 0.11 |
| HSPB1_MOUSE | Heat shock protein beta-1 | 86 | 23000 | 6.12 | 9 | 2 | 7 | 2 | 7 | 0.56 | 0.43 |
| IF2G_MOUSE | Eukaryotic translation initiation factor 2 subunit 3, X-linked | 86 | 51033 | 8.66 | 15 | 5 | 10 | 5 | 10 | 0.3 | 0.5 |
| PDIA5_MOUSE | Protein disulfide-isomerase A5 | 86 | 59229 | 7.25 | 6 | 2 | 5 | 1 | 5 | 0.22 | 0.15 |
| BAG3_MOUSE | BAG family molecular chaperone regulator 3 | 86 | 61822 | 6.78 | 12 | 4 | 7 | 3 | 7 | 0.23 | 0.22 |
| MRGRF_MOUSE | Mas-related G-protein coupled receptor member F | 86 | 38497 | 9 | 2 | 2 | 1 | 1 | 1 | 0.06 | 0.11 |
| SET_MOUSE | Protein SET | 85 | 33358 | 4.22 | 10 | 3 | 7 | 3 | 7 | 0.37 | 0.45 |
| GLTP_MOUSE | Glycolipid transfer protein | 85 | 23674 | 6.9 | 2 | 2 | 2 | 2 | 2 | 0.2 | 0.42 |
| DPP3_MOUSE | Dipeptidyl peptidase 3 | 85 | 82846 | 5.26 | 6 | 3 | 4 | 1 | 4 | 0.12 | 0.05 |
| CARM1_MOUSE | Histone-arginine methyltransferase CARM1 | 85 | 65811 | 6.28 | 5 | 3 | 4 | 3 | 4 | 0.12 | 0.21 |
| SERC1_MOUSE | Serine incorporator 1 | 85 | 50475 | 5.91 | 3 | 2 | 2 | 1 | 2 | 0.06 | 0.09 |
| DCUP_MOUSE | Uroporphyrinogen decarboxylase | 85 | 40666 | 6.21 | 4 | 3 | 3 | 2 | 3 | 0.21 | 0.23 |
| KINH_MOUSE | Kinesin-1 heavy chain | 85 | 109484 | 6.06 | 11 | 5 | 8 | 3 | 8 | 0.13 | 0.12 |
| KANK2_MOUSE | KN motif and ankyrin repeat domain-containing protein 2 | 85 | 90190 | 5.38 | 6 | 2 | 3 | 1 | 3 | 0.06 | 0.05 |
| AGM1_MOUSE | Phosphoacetylglucosamine mutase | 84 | 59415 | 5.8 | 3 | 2 | 3 | 2 | 3 | 0.14 | 0.15 |
| USP9X_MOUSE | Probable ubiquitin carboxyl-terminal hydrolase FAF-X | 84 | 290526 | 5.57 | 8 | 1 | 5 | 1 | 5 | 0.04 | 0.01 |
| ADDA_MOUSE | Alpha-adducin | 84 | 80596 | 5.62 | 6 | 2 | 4 | 1 | 4 | 0.11 | 0.05 |
| FA98B_MOUSE | Protein FAM98B | 84 | 45321 | 8.77 | 3 | 2 | 3 | 2 | 3 | 0.15 | 0.2 |
| GALK2_MOUSE | N-Acetylgalactosamine kinase | 84 | 50470 | 6.47 | 10 | 4 | 6 | 3 | 6 | 0.38 | 0.39 |
| UB2V1_MOUSE | Ubiquitin-conjugating enzyme E2 variant 1 | 84 | 16344 | 7.74 | 11 | 3 | 6 | 2 | 6 | 0.58 | 0.65 |
| PPGB_MOUSE | Lysosomal protective protein | 83 | 53809 | 5.56 | 9 | 3 | 8 | 3 | 8 | 0.27 | 0.26 |
| EIF2A_MOUSE | Eukaryotic translation initiation factor 2A | 83 | 64363 | 9.04 | 11 | 6 | 5 | 4 | 5 | 0.14 | 0.3 |
| CATH_MOUSE | Pro-cathepsin H | 83 | 37146 | 8.68 | 4 | 3 | 3 | 3 | 3 | 0.21 | 0.4 |
| CD97_MOUSE | CD97 antigen | 83 | 90354 | 7.38 | 2 | 2 | 1 | 1 | 1 | 0.03 | 0.05 |
| RDH11_MOUSE | Retinol dehydrogenase 11 | 83 | 35125 | 9.1 | 10 | 6 | 5 | 3 | 5 | 0.24 | 0.43 |
| QCR2_MOUSE | Cytochrome b-c1 complex subunit 2, mitochondrial | 82 | 48205 | 9.26 | 14 | 5 | 7 | 2 | 7 | 0.37 | 0.19 |
| ITPA_MOUSE | Inosine triphosphate pyrophosphatase | 82 | 21883 | 5.6 | 5 | 2 | 2 | 1 | 2 | 0.15 | 0.21 |
| TWF1_MOUSE | Twinfilin-1 | 82 | 40054 | 6.21 | 7 | 4 | 4 | 3 | 4 | 0.19 | 0.37 |
| SEPT9_MOUSE | Septin-9 | 82 | 65534 | 9.01 | 16 | 5 | 9 | 3 | 9 | 0.23 | 0.21 |
| PGFS_MOUSE | Prostamide/prostaglandin F synthase | 81 | 21656 | 6.31 | 2 | 2 | 1 | 1 | 1 | 0.15 | 0.21 |
| ECI1_MOUSE | Enoyl-CoA delta isomerase 1, mitochondrial | 81 | 32230 | 9.12 | 3 | 3 | 2 | 2 | 2 | 0.13 | 0.29 |
| RPE_MOUSE | Ribulose-phosphate 3-epimerase | 81 | 24928 | 5.2 | 3 | 3 | 2 | 2 | 2 | 0.24 | 0.39 |
| RL13_MOUSE | 60S ribosomal protein L13 | 80 | 24290 | 11.54 | 18 | 6 | 7 | 4 | 6 | 0.32 | 0.97 |
| RWDD1_MOUSE | RWD domain-containing protein 1 | 80 | 27768 | 4.18 | 4 | 2 | 2 | 1 | 2 | 0.23 | 0.16 |
| CAN1_MOUSE | Calpain-1 catalytic subunit | 80 | 82054 | 5.62 | 3 | 2 | 3 | 2 | 3 | 0.09 | 0.11 |
| ECM29_MOUSE | Proteasome-associated protein ECM29 homolog | 80 | 203573 | 6.68 | 10 | 4 | 7 | 4 | 7 | 0.08 | 0.09 |
| ADA_MOUSE | Adenosine deaminase | 80 | 39966 | 5.48 | 3 | 2 | 3 | 2 | 3 | 0.27 | 0.23 |
| PSMD7_MOUSE | 26S proteasome non-ATPase regulatory subunit 7 | 80 | 36517 | 6.29 | 5 | 3 | 5 | 3 | 5 | 0.34 | 0.41 |
| RAGP1_MOUSE | Ran GTPase-activating protein 1 | 80 | 63491 | 4.59 | 8 | 5 | 5 | 4 | 5 | 0.15 | 0.3 |
| ACON_MOUSE | Aconitate hydratase, mitochondrial | 79 | 85410 | 8.08 | 10 | 4 | 9 | 4 | 9 | 0.2 | 0.22 |
| NEB2_MOUSE | Neurabin-2 | 79 | 89466 | 4.86 | 5 | 2 | 4 | 2 | 4 | 0.13 | 0.1 |
| IF2B_MOUSE | Eukaryotic translation initiation factor 2 subunit 2 | 79 | 38068 | 5.61 | 6 | 4 | 4 | 3 | 4 | 0.18 | 0.39 |
| NXP20_MOUSE | Protein Noxp20 | 78 | 60975 | 4.49 | 6 | 3 | 4 | 2 | 4 | 0.11 | 0.15 |
| ABCF1_MOUSE | ATP-binding cassette sub-family F member 1 | 78 | 94887 | 6.15 | 9 | 3 | 6 | 3 | 6 | 0.11 | 0.14 |
| HEM3_MOUSE | Porphobilinogen deaminase | 78 | 39320 | 6.41 | 4 | 2 | 3 | 1 | 3 | 0.17 | 0.11 |
| CSN4_MOUSE | COP9 signalosome complex subunit 4 | 78 | 46256 | 5.57 | 6 | 4 | 5 | 3 | 5 | 0.2 | 0.31 |
| C1TM_MOUSE | Monofunctional C1-tetrahydrofolate synthase, mitochondrial | 78 | 105662 | 6.58 | 10 | 2 | 8 | 1 | 8 | 0.15 | 0.04 |
| SPRE_MOUSE | Sepiapterin reductase | 78 | 27865 | 5.58 | 2 | 2 | 2 | 2 | 2 | 0.12 | 0.35 |
| PLCD1_MOUSE | 1-Phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | 77 | 85819 | 5.82 | 3 | 2 | 1 | 1 | 1 | 0.02 | 0.05 |
| IF2B2_MOUSE | Insulin-like growth factor 2 mRNA-binding protein 2 | 77 | 65543 | 7.81 | 3 | 2 | 3 | 2 | 3 | 0.07 | 0.14 |
| MCAT_MOUSE | Mitochondrial carnitine/acylcarnitine carrier protein | 77 | 33005 | 9.24 | 6 | 3 | 4 | 2 | 4 | 0.31 | 0.29 |
| GSH0_MOUSE | Glutamate–cysteine ligase regulatory subunit | 76 | 30516 | 5.35 | 6 | 2 | 5 | 2 | 5 | 0.32 | 0.31 |
| HA1B_MOUSE | H-2 class I histocompatibility antigen, K-B alpha chain | 76 | 41276 | 5.96 | 6 | 3 | 3 | 3 | 3 | 0.14 | 0.35 |
| AR6P1_MOUSE | ADP-ribosylation factor-like protein 6-interacting protein 1 | 76 | 23421 | 9.38 | 3 | 2 | 2 | 1 | 2 | 0.19 | 0.19 |
| TM9S2_MOUSE | Transmembrane 9 superfamily member 2 | 76 | 75280 | 7.21 | 6 | 3 | 5 | 2 | 5 | 0.14 | 0.12 |
| SFXN1_MOUSE | Sideroflexin-1 | 76 | 35626 | 9.35 | 4 | 1 | 4 | 1 | 4 | 0.26 | 0.12 |
| RS21_MOUSE | 40S ribosomal protein S21 | 76 | 9136 | 8.71 | 7 | 3 | 4 | 1 | 4 | 0.54 | 0.56 |
| IL4RA_MOUSE | Interleukin-4 receptor subunit alpha | 76 | 87571 | 4.97 | 3 | 2 | 2 | 1 | 2 | 0.04 | 0.05 |
| EIF1_MOUSE | Eukaryotic translation initiation factor 1 | 75 | 12739 | 6.89 | 3 | 2 | 2 | 2 | 2 | 0.24 | 0.89 |
| NUCB1_MOUSE | Nucleobindin-1 | 75 | 53376 | 4.99 | 17 | 4 | 11 | 2 | 11 | 0.34 | 0.17 |
| MMS19_MOUSE | MMS19 nucleotide excision repair protein homolog | 75 | 113017 | 5.81 | 4 | 2 | 3 | 2 | 3 | 0.07 | 0.08 |
| GCP60_MOUSE | Golgi resident protein GCP60 | 75 | 60144 | 5.07 | 12 | 5 | 5 | 2 | 5 | 0.2 | 0.15 |
| FDFT_MOUSE | Squalene synthase | 75 | 48123 | 5.91 | 9 | 3 | 6 | 2 | 6 | 0.27 | 0.3 |
| TPD52_MOUSE | Tumor protein D52 | 75 | 24298 | 4.69 | 4 | 3 | 3 | 2 | 3 | 0.25 | 0.4 |
| RBM3_MOUSE | RNA-binding protein 3 | 75 | 16595 | 6.84 | 14 | 7 | 5 | 4 | 5 | 0.57 | 1.68 |
| RBMS1_MOUSE | RNA-binding motif, single-stranded-interacting protein 1 | 75 | 43963 | 8.79 | 3 | 2 | 3 | 2 | 3 | 0.21 | 0.21 |
| NDUAA_MOUSE | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | 75 | 40578 | 7.63 | 5 | 3 | 5 | 3 | 5 | 0.23 | 0.36 |
| SRC8_MOUSE | Src substrate cortactin | 75 | 61212 | 5.24 | 10 | 2 | 8 | 2 | 8 | 0.15 | 0.15 |
| HNRPC_MOUSE | Heterogeneous nuclear ribonucleoproteins C1/C2 | 74 | 34364 | 4.92 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.13 |
| CP20A_MOUSE | Cytochrome P450 20A1 | 74 | 52116 | 6.48 | 5 | 3 | 4 | 2 | 4 | 0.19 | 0.17 |
| LDLR_MOUSE | Low-density lipoprotein receptor | 74 | 94885 | 4.82 | 5 | 2 | 5 | 2 | 5 | 0.13 | 0.09 |
| MARC2_MOUSE | Mitochondrial amidoxime reducing component 2 | 73 | 38170 | 8.95 | 2 | 2 | 2 | 2 | 2 | 0.15 | 0.24 |
| ERF3A_MOUSE | Eukaryotic peptide chain release factor GTP-binding subunit ERF3A | 73 | 68582 | 5.12 | 13 | 6 | 9 | 4 | 9 | 0.22 | 0.28 |
| ALD2_MOUSE | Aldose reductase-related protein 2 | 73 | 36098 | 5.97 | 4 | 2 | 4 | 2 | 4 | 0.2 | 0.26 |
| ADPRH_MOUSE | [Protein ADP-ribosylarginine] hydrolase | 73 | 40042 | 5.46 | 3 | 2 | 3 | 2 | 3 | 0.22 | 0.23 |
| ATP5I_MOUSE | ATP synthase subunit e, mitochondrial | 72 | 8230 | 9.34 | 9 | 4 | 4 | 2 | 4 | 0.62 | 1.66 |
| GLU2B_MOUSE | Glucosidase 2 subunit beta | 72 | 58756 | 4.41 | 12 | 5 | 8 | 4 | 8 | 0.24 | 0.33 |
| ACSL4_MOUSE | Long-chain-fatty-acid–CoA ligase 4 | 72 | 79026 | 8.57 | 12 | 5 | 8 | 3 | 6 | 0.19 | 0.17 |
| PUF60_MOUSE | Poly(U)-binding-splicing factor PUF60 | 72 | 60211 | 5.2 | 6 | 2 | 5 | 1 | 5 | 0.24 | 0.07 |
| PTPA_MOUSE | Serine/threonine-protein phosphatase 2A activator | 72 | 36687 | 5.95 | 5 | 2 | 4 | 1 | 4 | 0.29 | 0.12 |
| PDXK_MOUSE | Pyridoxal kinase | 72 | 34993 | 5.88 | 4 | 2 | 4 | 2 | 4 | 0.19 | 0.27 |
| AP1G1_MOUSE | AP-1 complex subunit gamma-1 | 71 | 91292 | 6.36 | 6 | 4 | 5 | 3 | 5 | 0.16 | 0.15 |
| ATG3_MOUSE | Ubiquitin-like-conjugating enzyme ATG3 | 71 | 35773 | 4.63 | 4 | 2 | 3 | 1 | 3 | 0.28 | 0.26 |
| KAP0_MOUSE | cAMP-dependent protein kinase type I-alpha regulatory subunit | 71 | 43158 | 5.27 | 9 | 2 | 8 | 2 | 8 | 0.28 | 0.21 |
| IF4B_MOUSE | Eukaryotic translation initiation factor 4B | 71 | 68799 | 5.47 | 8 | 6 | 2 | 2 | 2 | 0.07 | 0.13 |
| NPL4_MOUSE | Nuclear protein localization protein 4 homolog | 71 | 67974 | 6.01 | 3 | 1 | 3 | 1 | 3 | 0.11 | 0.06 |
| CX6A1_MOUSE | Cytochrome c oxidase subunit 6A1, mitochondrial | 71 | 12344 | 9.97 | 3 | 2 | 2 | 1 | 2 | 0.45 | 0.39 |
| CDC37_MOUSE | Hsp90 co-chaperone Cdc37 | 71 | 44565 | 5.24 | 6 | 4 | 5 | 3 | 5 | 0.21 | 0.32 |
| AN32A_MOUSE | Acidic leucine-rich nuclear phosphoprotein 32 family member A | 71 | 28520 | 3.99 | 4 | 2 | 4 | 2 | 4 | 0.22 | 0.34 |
| PA24A_MOUSE | Cytosolic phospholipase A2 | 71 | 85168 | 5.28 | 6 | 2 | 6 | 2 | 6 | 0.18 | 0.1 |
| CO5A2_MOUSE | Collagen alpha-2(V) chain | 70 | 144929 | 6.28 | 17 | 7 | 9 | 5 | 9 | 0.14 | 0.16 |
| ANXA7_MOUSE | Annexin A7 | 70 | 49893 | 5.91 | 10 | 3 | 8 | 2 | 8 | 0.22 | 0.18 |
| AIF1L_MOUSE | Allograft inflammatory factor 1-like | 70 | 17013 | 6.63 | 3 | 2 | 2 | 1 | 2 | 0.16 | 0.27 |
| MECR_MOUSE | Enoyl-[acyl-carrier-protein] reductase, mitochondrial | 70 | 40317 | 9.17 | 3 | 3 | 1 | 1 | 1 | 0.09 | 0.11 |
| MPI_MOUSE | Mannose-6-phosphate isomerase | 70 | 46545 | 5.62 | 2 | 1 | 2 | 1 | 2 | 0.1 | 0.09 |
| SUCB2_MOUSE | Succinate–CoA ligase [GDP-forming] subunit beta, mitochondrial | 70 | 46811 | 6.58 | 4 | 1 | 4 | 1 | 4 | 0.14 | 0.09 |
| MMAB_MOUSE | Cob(I)yrinic acid a,c-diamide adenosyltransferase, mitochondrial | 69 | 26256 | 9.32 | 2 | 1 | 2 | 1 | 2 | 0.16 | 0.17 |
| AGAL_MOUSE | Alpha-galactosidase A | 69 | 47611 | 5.44 | 4 | 2 | 4 | 2 | 4 | 0.14 | 0.19 |
| SP16H_MOUSE | FACT complex subunit SPT16 | 69 | 119749 | 5.5 | 4 | 1 | 4 | 1 | 4 | 0.07 | 0.04 |
| NP1L4_MOUSE | Nucleosome assembly protein 1-like 4 | 69 | 42653 | 4.56 | 9 | 3 | 7 | 3 | 5 | 0.32 | 0.34 |
| UBQL1_MOUSE | Ubiquilin-1 | 69 | 61937 | 4.86 | 10 | 4 | 6 | 3 | 6 | 0.24 | 0.22 |
| RET1_MOUSE | Retinol-binding protein 1 | 69 | 15836 | 5.1 | 4 | 2 | 4 | 2 | 4 | 0.44 | 0.68 |
| ABHEB_MOUSE | Protein ABHD14B | 69 | 22437 | 5.82 | 2 | 1 | 2 | 1 | 2 | 0.1 | 0.2 |
| HM13_MOUSE | Minor histocompatibility antigen H13 | 69 | 41721 | 5.7 | 4 | 2 | 2 | 1 | 2 | 0.1 | 0.1 |
| GALM_MOUSE | Aldose 1-epimerase | 68 | 37775 | 6.26 | 4 | 2 | 3 | 2 | 3 | 0.13 | 0.25 |
| AP3B1_MOUSE | AP-3 complex subunit beta-1 | 68 | 122664 | 5.49 | 13 | 3 | 9 | 3 | 9 | 0.11 | 0.11 |
| LEG9_MOUSE | Galectin-9 | 68 | 40010 | 9.41 | 9 | 3 | 6 | 3 | 6 | 0.24 | 0.37 |
| SPART_MOUSE | Spartin | 68 | 72610 | 5.64 | 3 | 2 | 3 | 2 | 3 | 0.09 | 0.12 |
| ERAP1_MOUSE | Endoplasmic reticulum aminopeptidase 1 | 67 | 106531 | 5.82 | 7 | 3 | 5 | 3 | 5 | 0.09 | 0.12 |
| PIEZ1_MOUSE | Piezo-type mechanosensitive ion channel component 1 | 67 | 291813 | 7.41 | 2 | 1 | 2 | 1 | 2 | 0.01 | 0.01 |
| AL1L2_MOUSE | Mitochondrial 10-formyltetrahydrofolate dehydrogenase | 67 | 101526 | 5.93 | 8 | 3 | 7 | 3 | 6 | 0.13 | 0.13 |
| PDLI7_MOUSE | PDZ and LIM domain protein 7 | 67 | 50087 | 8.82 | 5 | 1 | 5 | 1 | 5 | 0.23 | 0.09 |
| ADA15_MOUSE | Disintegrin and metalloproteinase domain-containing protein 15 | 67 | 92604 | 6.09 | 5 | 2 | 3 | 1 | 3 | 0.09 | 0.05 |
| PGPI_MOUSE | Pyroglutamyl-peptidase 1 | 66 | 22919 | 5.21 | 4 | 3 | 4 | 3 | 4 | 0.43 | 0.72 |
| ANTR1_MOUSE | Anthrax toxin receptor 1 | 66 | 62269 | 7.53 | 3 | 1 | 3 | 1 | 3 | 0.08 | 0.07 |
| OSTF1_MOUSE | Osteoclast-stimulating factor 1 | 66 | 23768 | 5.46 | 4 | 2 | 4 | 2 | 4 | 0.3 | 0.42 |
| IL1RA_MOUSE | Interleukin-1 receptor antagonist protein | 66 | 20261 | 5.82 | 3 | 2 | 3 | 2 | 3 | 0.42 | 0.5 |
| CC90B_MOUSE | Coiled-coil domain-containing protein 90B, mitochondrial | 66 | 29578 | 8.54 | 3 | 1 | 3 | 1 | 3 | 0.25 | 0.15 |
| PON2_MOUSE | Serum paraoxonase/arylesterase 2 | 66 | 39592 | 5.49 | 5 | 3 | 3 | 1 | 3 | 0.18 | 0.11 |
| PGH1_MOUSE | Prostaglandin G/H synthase 1 | 66 | 68998 | 6.36 | 7 | 5 | 6 | 5 | 6 | 0.12 | 0.35 |
| NRADD_MOUSE | Death domain-containing membrane protein NRADD | 66 | 24711 | 4.89 | 2 | 2 | 1 | 1 | 1 | 0.14 | 0.18 |
| PEA15_MOUSE | Astrocytic phosphoprotein PEA-15 | 66 | 15045 | 4.94 | 9 | 3 | 4 | 3 | 4 | 0.45 | 1.26 |
| B2MG_MOUSE | Beta-2-microglobulin | 66 | 13770 | 8.55 | 3 | 2 | 3 | 2 | 3 | 0.35 | 0.81 |
| ACADM_MOUSE | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 66 | 46452 | 8.6 | 4 | 2 | 4 | 2 | 4 | 0.18 | 0.2 |
| SCRB2_MOUSE | Lysosome membrane protein 2 | 65 | 54009 | 4.99 | 5 | 2 | 5 | 2 | 5 | 0.12 | 0.17 |
| AATC_MOUSE | Aspartate aminotransferase, cytoplasmic | 65 | 46219 | 6.68 | 14 | 3 | 10 | 3 | 10 | 0.34 | 0.31 |
| ERO1A_MOUSE | ERO1-like protein alpha | 65 | 54050 | 6.12 | 6 | 3 | 4 | 2 | 4 | 0.14 | 0.17 |
| PUR8_MOUSE | Adenylosuccinate lyase | 65 | 54831 | 6.9 | 6 | 3 | 5 | 2 | 5 | 0.19 | 0.16 |
| LSG1_MOUSE | Large subunit GTPase 1 homolog | 64 | 73111 | 6.08 | 5 | 2 | 4 | 1 | 4 | 0.1 | 0.06 |
| NAGAB_MOUSE | Alpha-N-acetylgalactosaminidase | 64 | 47204 | 6.02 | 4 | 2 | 3 | 1 | 3 | 0.14 | 0.09 |
| IDHG1_MOUSE | Isocitrate dehydrogenase [NAD] subunit gamma 1, mitochondrial | 64 | 42758 | 9.17 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.1 |
| CTGF_MOUSE | Connective tissue growth factor | 64 | 37798 | 8.22 | 14 | 6 | 8 | 3 | 8 | 0.33 | 0.39 |
| EFTS_MOUSE | Elongation factor Ts, mitochondrial | 64 | 35312 | 6.62 | 5 | 3 | 4 | 2 | 4 | 0.25 | 0.27 |
| DBNL_MOUSE | Drebrin-like protein | 64 | 48670 | 4.9 | 5 | 1 | 4 | 1 | 4 | 0.13 | 0.09 |
| TENA_MOUSE | Tenascin | 64 | 231659 | 4.77 | 11 | 3 | 8 | 2 | 8 | 0.09 | 0.04 |
| SYTC_MOUSE | Threonine–tRNA ligase, cytoplasmic | 64 | 83303 | 7.03 | 6 | 4 | 5 | 4 | 5 | 0.12 | 0.22 |
| TMM59_MOUSE | Transmembrane protein 59 | 64 | 36290 | 4.8 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.12 |
| ARK72_MOUSE | Aflatoxin B1 aldehyde reductase member 2 | 63 | 40586 | 8.36 | 2 | 2 | 1 | 1 | 1 | 0.1 | 0.11 |
| PR2C3_MOUSE | Prolactin-2C3 | 63 | 25322 | 5.61 | 9 | 2 | 5 | 1 | 5 | 0.55 | 0.18 |
| AL7A1_MOUSE | Alpha-aminoadipic semialdehyde dehydrogenase | 62 | 58824 | 7.16 | 5 | 4 | 5 | 4 | 5 | 0.15 | 0.33 |
| STAT1_MOUSE | Signal transducer and activator of transcription 1 | 49 | 87142 | 5.42 | 8 | 4 | 6 | 3 | 3 | 0.14 | 0.15 |
| FMR1_MOUSE | Synaptic functional regulator FMR1 | 62 | 68947 | 7.27 | 7 | 2 | 6 | 2 | 5 | 0.17 | 0.13 |
| EGFR_MOUSE | Epidermal growth factor receptor | 59 | 134766 | 6.46 | 7 | 2 | 7 | 2 | 7 | 0.09 | 0.06 |
| DCAKD_MOUSE | Dephospho-CoA kinase domain-containing protein | 61 | 26459 | 9.61 | 4 | 2 | 3 | 1 | 3 | 0.2 | 0.17 |
| TIM13_MOUSE | Mitochondrial import inner membrane translocase subunit Tim13 | 61 | 10451 | 8.42 | 6 | 2 | 2 | 1 | 2 | 0.26 | 0.47 |
| ITAM_MOUSE | Integrin alpha-M | 61 | 127400 | 6.87 | 4 | 2 | 3 | 1 | 3 | 0.04 | 0.03 |
| ARP10_MOUSE | Actin-related protein 10 | 61 | 46178 | 7.54 | 2 | 2 | 2 | 2 | 2 | 0.05 | 0.2 |
| TMED5_MOUSE | Transmembrane emp24 domain-containing protein 5 | 61 | 26155 | 4.81 | 5 | 2 | 4 | 2 | 4 | 0.31 | 0.37 |
| TRXR1_MOUSE | Thioredoxin reductase 1, cytoplasmic | 61 | 67042 | 7.42 | 9 | 6 | 7 | 5 | 7 | 0.22 | 0.37 |
| NAGK_MOUSE | N-acetyl-D-glucosamine kinase | 60 | 37245 | 5.43 | 5 | 3 | 3 | 2 | 3 | 0.23 | 0.25 |
| GAPR1_MOUSE | Golgi-associated plant pathogenesis-related protein 1 | 60 | 17080 | 9.54 | 5 | 2 | 3 | 1 | 3 | 0.32 | 0.27 |
| ODO2_MOUSE | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | 60 | 48963 | 9.11 | 8 | 2 | 7 | 2 | 7 | 0.2 | 0.19 |
| STX7_MOUSE | Syntaxin-7 | 60 | 29802 | 5.6 | 9 | 4 | 5 | 2 | 5 | 0.15 | 0.32 |
| GID8_MOUSE | Glucose-induced degradation protein 8 homolog | 60 | 26762 | 4.92 | 4 | 2 | 3 | 1 | 3 | 0.3 | 0.17 |
| PGBM_MOUSE | Basement membrane-specific heparan sulfate proteoglycan core protein | 60 | 398039 | 5.88 | 10 | 4 | 8 | 3 | 8 | 0.05 | 0.04 |
| MDR1A_MOUSE | Multidrug resistance protein 1A | 60 | 140558 | 8.94 | 4 | 1 | 3 | 1 | 3 | 0.04 | 0.03 |
| CASP1_MOUSE | Caspase-1 | 59 | 45611 | 5.73 | 4 | 2 | 3 | 1 | 3 | 0.15 | 0.1 |
| SYCC_MOUSE | Cysteine–tRNA ligase, cytoplasmic | 59 | 94800 | 6.32 | 13 | 3 | 8 | 3 | 8 | 0.15 | 0.14 |
| PRRX1_MOUSE | Paired mesoderm homeobox protein 1 | 59 | 27253 | 9.48 | 2 | 2 | 1 | 1 | 1 | 0.12 | 0.16 |
| LY6A_MOUSE | Lymphocyte antigen 6A-2/6E-1 | 59 | 14367 | 4.75 | 4 | 3 | 1 | 1 | 1 | 0.16 | 0.33 |
| AMPD3_MOUSE | AMP deaminase 3 | 59 | 88596 | 6.87 | 4 | 1 | 4 | 1 | 4 | 0.08 | 0.05 |
| NDRG2_MOUSE | Protein NDRG2 | 59 | 40763 | 5.23 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.11 |
| S39AE_MOUSE | Zinc transporter ZIP14 | 59 | 53927 | 5.11 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.08 |
| PLPHP_MOUSE | Pyridoxal phosphate homeostasis protein | 59 | 30030 | 8.37 | 4 | 4 | 4 | 4 | 4 | 0.33 | 0.74 |
| PSMD3_MOUSE | 26S proteasome non-ATPase regulatory subunit 3 | 59 | 60680 | 8.48 | 7 | 2 | 6 | 2 | 6 | 0.18 | 0.15 |
| TRADD_MOUSE | Tumor necrosis factor receptor type 1-associated DEATH domain protein | 59 | 34556 | 5.11 | 5 | 1 | 3 | 1 | 3 | 0.12 | 0.13 |
| ANM5_MOUSE | Protein arginine N-methyltransferase 5 | 59 | 72634 | 5.99 | 5 | 3 | 3 | 2 | 3 | 0.13 | 0.12 |
| IPO9_MOUSE | Importin-9 | 58 | 115978 | 4.71 | 6 | 2 | 4 | 2 | 4 | 0.09 | 0.07 |
| VPS52_MOUSE | Vacuolar protein sorting-associated protein 52 homolog | 58 | 81993 | 5.65 | 7 | 2 | 3 | 1 | 3 | 0.07 | 0.05 |
| LRRF1_MOUSE | Leucine-rich repeat flightless-interacting protein 1 | 58 | 79201 | 4.75 | 2 | 1 | 2 | 1 | 2 | 0.08 | 0.05 |
| E41L3_MOUSE | Band 4.1-like protein 3 | 58 | 103274 | 5.2 | 5 | 2 | 4 | 2 | 4 | 0.05 | 0.08 |
| EMIL1_MOUSE | EMILIN-1 | 58 | 107518 | 5.21 | 3 | 1 | 3 | 1 | 3 | 0.06 | 0.04 |
| EEA1_MOUSE | Early endosome antigen 1 | 58 | 160817 | 5.59 | 6 | 2 | 5 | 2 | 5 | 0.06 | 0.05 |
| MESD_MOUSE | LRP chaperone MESD | 58 | 25191 | 6.06 | 2 | 2 | 1 | 1 | 1 | 0.06 | 0.18 |
| IMPCT_MOUSE | Protein IMPACT | 57 | 36253 | 4.97 | 3 | 2 | 3 | 2 | 3 | 0.15 | 0.26 |
| GUAA_MOUSE | GMP synthase [glutamine-hydrolyzing] | 57 | 76675 | 6.29 | 4 | 2 | 3 | 1 | 3 | 0.11 | 0.06 |
| GCSH_MOUSE | Glycine cleavage system H protein, mitochondrial | 57 | 18625 | 4.78 | 3 | 3 | 1 | 1 | 1 | 0.22 | 0.55 |
| VA0D1_MOUSE | V-type proton ATPase subunit d 1 | 57 | 40275 | 4.89 | 9 | 4 | 8 | 4 | 8 | 0.34 | 0.51 |
| PUR2_MOUSE | Trifunctional purine biosynthetic protein adenosine-3 | 57 | 107436 | 6.25 | 6 | 3 | 5 | 2 | 5 | 0.1 | 0.08 |
| GLOD4_MOUSE | Glyoxalase domain-containing protein 4 | 57 | 33296 | 5.28 | 2 | 2 | 2 | 2 | 2 | 0.17 | 0.28 |
| WDR61_MOUSE | WD repeat-containing protein 61 | 57 | 33752 | 5.1 | 3 | 2 | 3 | 2 | 3 | 0.18 | 0.28 |
| CUL5_MOUSE | Cullin-5 | 57 | 90916 | 7.86 | 10 | 2 | 6 | 1 | 6 | 0.17 | 0.05 |
| NICA_MOUSE | Nicastrin | 57 | 78443 | 5.75 | 5 | 2 | 4 | 2 | 4 | 0.06 | 0.11 |
| FKB14_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP14 | 57 | 24236 | 5.66 | 4 | 2 | 3 | 2 | 3 | 0.24 | 0.41 |
| PLEK_MOUSE | Pleckstrin | 57 | 39876 | 8.53 | 3 | 2 | 3 | 2 | 3 | 0.19 | 0.23 |
| LA_MOUSE | Lupus La protein homolog | 57 | 47727 | 9.77 | 5 | 2 | 5 | 2 | 5 | 0.13 | 0.19 |
| TPBG_MOUSE | Trophoblast glycoprotein | 56 | 46422 | 6.36 | 5 | 2 | 5 | 2 | 5 | 0.18 | 0.2 |
| ETHE1_MOUSE | Persulfide dioxygenase ETHE1, mitochondrial | 56 | 27721 | 6.78 | 2 | 1 | 2 | 1 | 2 | 0.23 | 0.16 |
| UBR4_MOUSE | E3 ubiquitin-protein ligase UBR4 | 56 | 571927 | 5.72 | 20 | 2 | 16 | 2 | 16 | 0.07 | 0.01 |
| ACSA_MOUSE | Acetyl-coenzyme A synthetase, cytoplasmic | 56 | 78811 | 6.19 | 4 | 2 | 3 | 1 | 3 | 0.09 | 0.05 |
| PSA_MOUSE | Puromycin-sensitive aminopeptidase | 56 | 103260 | 5.61 | 7 | 4 | 6 | 3 | 6 | 0.14 | 0.13 |
| S10A4_MOUSE | Protein S100-A4 | 56 | 11714 | 5.23 | 4 | 2 | 2 | 1 | 2 | 0.23 | 0.41 |
| P3H3_MOUSE | Prolyl 3-hydroxylase 3 | 55 | 81650 | 6.21 | 3 | 1 | 3 | 1 | 3 | 0.08 | 0.05 |
| LMNA_MOUSE | Prelamin-A/C | 55 | 74193 | 6.54 | 10 | 1 | 5 | 1 | 5 | 0.11 | 0.06 |
| VAC14_MOUSE | Protein VAC14 homolog | 54 | 87992 | 5.75 | 5 | 1 | 4 | 1 | 4 | 0.09 | 0.05 |
| PYR1_MOUSE | CAD protein | 54 | 243084 | 6 | 13 | 5 | 11 | 4 | 11 | 0.11 | 0.07 |
| ABRAL_MOUSE | Costars family protein ABRACL | 54 | 9025 | 5.52 | 4 | 1 | 3 | 1 | 3 | 0.57 | 0.56 |
| CTND1_MOUSE | Catenin delta-1 | 54 | 104860 | 6.41 | 8 | 2 | 8 | 2 | 8 | 0.12 | 0.08 |
| FKB15_MOUSE | FK506-binding protein 15 | 54 | 132878 | 4.98 | 4 | 2 | 3 | 1 | 3 | 0.06 | 0.03 |
| ECE1_MOUSE | Endothelin-converting enzyme 1 | 54 | 87017 | 5.64 | 5 | 5 | 5 | 5 | 5 | 0.17 | 0.27 |
| AT5F1_MOUSE | ATP synthase F(0) complex subunit B1, mitochondrial | 54 | 28930 | 9.11 | 8 | 2 | 6 | 2 | 6 | 0.27 | 0.33 |
| LDAH_MOUSE | Lipid droplet-associated hydrolase | 54 | 37349 | 8.5 | 3 | 1 | 2 | 1 | 2 | 0.18 | 0.12 |
| TNIK_MOUSE | Traf2 and NCK-interacting protein kinase | 54 | 150274 | 6.82 | 9 | 2 | 7 | 1 | 7 | 0.07 | 0.03 |
| EI2BA_MOUSE | Translation initiation factor eIF-2B subunit alpha | 54 | 33795 | 8.48 | 4 | 3 | 2 | 1 | 2 | 0.1 | 0.13 |
| VAPA_MOUSE | Vesicle-associated membrane protein-associated protein A | 53 | 27837 | 8.59 | 9 | 2 | 6 | 2 | 6 | 0.36 | 0.35 |
| DYLT3_MOUSE | Dynein light chain Tctex-type 3 | 53 | 12949 | 4.98 | 1 | 1 | 1 | 1 | 1 | 0.29 | 0.37 |
| ZZEF1_MOUSE | Zinc finger ZZ-type and EF-hand domain-containing protein 1 | 53 | 328102 | 5.74 | 6 | 2 | 4 | 1 | 4 | 0.03 | 0.01 |
| NDRG1_MOUSE | Protein NDRG1 | 53 | 42981 | 5.69 | 5 | 2 | 4 | 2 | 4 | 0.21 | 0.21 |
| PDLI5_MOUSE | PDZ and LIM domain protein 5 | 53 | 63259 | 8.61 | 15 | 3 | 7 | 3 | 7 | 0.27 | 0.22 |
| LIMA1_MOUSE | LIM domain and actin-binding protein 1 | 53 | 84008 | 6.18 | 10 | 2 | 7 | 2 | 7 | 0.12 | 0.1 |
| NPC2_MOUSE | Epididymal secretory protein E1 | 53 | 16432 | 7.59 | 13 | 3 | 8 | 2 | 8 | 0.7 | 0.65 |
| AN32B_MOUSE | Acidic leucine-rich nuclear phosphoprotein 32 family member B | 53 | 31060 | 3.89 | 2 | 1 | 2 | 1 | 2 | 0.11 | 0.14 |
| ERP44_MOUSE | Endoplasmic reticulum resident protein 44 | 53 | 46823 | 5.09 | 6 | 2 | 6 | 2 | 6 | 0.24 | 0.19 |
| CBR2_MOUSE | Carbonyl reductase [NADPH] 2 | 53 | 25942 | 9.1 | 2 | 2 | 2 | 2 | 2 | 0.23 | 0.38 |
| TNR12_MOUSE | Tumor necrosis factor receptor superfamily member 12A | 53 | 13632 | 8.18 | 2 | 1 | 2 | 1 | 2 | 0.43 | 0.35 |
| GGT5_MOUSE | Glutathione hydrolase 5 proenzyme | 53 | 61635 | 8.76 | 3 | 1 | 3 | 1 | 3 | 0.13 | 0.07 |
| AL3A2_MOUSE | Fatty aldehyde dehydrogenase | 52 | 53936 | 8.59 | 2 | 2 | 2 | 2 | 2 | 0.09 | 0.17 |
| MYDGF_MOUSE | Myeloid-derived growth factor | 52 | 17971 | 6.3 | 5 | 2 | 2 | 1 | 2 | 0.12 | 0.26 |
| FKBP4_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP4 | 52 | 51540 | 5.54 | 4 | 2 | 3 | 1 | 3 | 0.13 | 0.08 |
| AL1L1_MOUSE | Cytosolic 10-formyltetrahydrofolate dehydrogenase | 52 | 98647 | 5.64 | 9 | 2 | 7 | 2 | 6 | 0.17 | 0.09 |
| TCP4_MOUSE | Activated RNA polymerase II transcriptional coactivator p15 | 52 | 14418 | 9.6 | 5 | 2 | 3 | 1 | 3 | 0.28 | 0.33 |
| GLO2_MOUSE | Hydroxyacylglutathione hydrolase, mitochondrial | 51 | 34062 | 7.66 | 4 | 2 | 2 | 1 | 2 | 0.14 | 0.28 |
| PARK7_MOUSE | Protein/nucleic acid deglycase DJ-1 | 51 | 20008 | 6.32 | 12 | 5 | 6 | 4 | 6 | 0.6 | 1.28 |
| DHSO_MOUSE | Sorbitol dehydrogenase | 51 | 38225 | 6.56 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.11 |
| TENS3_MOUSE | Tensin-3 | 51 | 155491 | 6.19 | 6 | 1 | 4 | 1 | 4 | 0.07 | 0.03 |
| RSU1_MOUSE | Ras suppressor protein 1 | 51 | 31531 | 8.86 | 5 | 1 | 5 | 1 | 5 | 0.35 | 0.14 |
| PAK2_MOUSE | Serine/threonine-protein kinase PAK 2 | 49 | 57894 | 5.57 | 5 | 3 | 4 | 2 | 4 | 0.14 | 0.15 |
| HSBP1_MOUSE | Heat shock factor-binding protein 1 | 51 | 8605 | 4.11 | 3 | 2 | 2 | 1 | 2 | 0.58 | 0.59 |
| TMX3_MOUSE | Protein disulfide-isomerase TMX3 | 51 | 51815 | 5.02 | 4 | 2 | 4 | 2 | 4 | 0.17 | 0.17 |
| CREG1_MOUSE | Protein CREG1 | 51 | 24436 | 5.96 | 4 | 3 | 3 | 3 | 3 | 0.23 | 0.66 |
| MRP1_MOUSE | Multidrug resistance-associated protein 1 | 50 | 171075 | 7.03 | 2 | 1 | 2 | 1 | 2 | 0.04 | 0.02 |
| NU4M_MOUSE | NADH-ubiquinone oxidoreductase chain 4 | 50 | 51847 | 9.42 | 4 | 3 | 3 | 2 | 3 | 0.11 | 0.17 |
| TIMP2_MOUSE | Metalloproteinase inhibitor 2 | 50 | 24312 | 7.45 | 6 | 2 | 6 | 2 | 6 | 0.5 | 0.4 |
| NUP93_MOUSE | Nuclear pore complex protein Nup93 | 50 | 93222 | 5.5 | 5 | 2 | 5 | 2 | 5 | 0.09 | 0.09 |
| TGBR3_MOUSE | Transforming growth factor beta receptor type 3 | 50 | 93769 | 5.65 | 7 | 4 | 4 | 2 | 4 | 0.1 | 0.14 |
| RM46_MOUSE | 39S ribosomal protein L46, mitochondrial | 50 | 32112 | 6.93 | 4 | 1 | 3 | 1 | 3 | 0.14 | 0.14 |
| SRXN1_MOUSE | Sulfiredoxin-1 | 50 | 14140 | 7.82 | 4 | 1 | 2 | 1 | 2 | 0.26 | 0.34 |
| SYQ_MOUSE | Glutamine–tRNA ligase | 50 | 87621 | 6.93 | 9 | 2 | 6 | 1 | 6 | 0.16 | 0.05 |
| SORCN_MOUSE | Sorcin | 50 | 21613 | 5.32 | 10 | 2 | 4 | 1 | 4 | 0.26 | 0.21 |
| NDUA2_MOUSE | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 | 50 | 10909 | 10.02 | 3 | 2 | 1 | 1 | 1 | 0.14 | 0.45 |
| NASP_MOUSE | Nuclear autoantigenic sperm protein | 50 | 83903 | 4.35 | 4 | 2 | 4 | 2 | 4 | 0.1 | 0.1 |
| A16A1_MOUSE | Aldehyde dehydrogenase family 16 member A1 | 49 | 84703 | 5.9 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.05 |
| NQO1_MOUSE | NAD(P)H dehydrogenase [quinone] 1 | 49 | 30940 | 8.74 | 12 | 4 | 6 | 2 | 6 | 0.36 | 0.31 |
| NRBP_MOUSE | Nuclear receptor-binding protein | 49 | 59828 | 5.02 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.07 |
| NIF3L_MOUSE | NIF3-like protein 1 | 49 | 41719 | 6.28 | 5 | 2 | 3 | 1 | 3 | 0.12 | 0.1 |
| NDUS4_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | 49 | 19772 | 10 | 3 | 1 | 3 | 1 | 2 | 0.26 | 0.23 |
| CERS2_MOUSE | Ceramide synthase 2 | 49 | 44995 | 8.85 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.1 |
| GT251_MOUSE | Procollagen galactosyltransferase 1 | 49 | 71015 | 6.83 | 11 | 4 | 9 | 4 | 9 | 0.28 | 0.27 |
| S23IP_MOUSE | SEC23-interacting protein | 49 | 110711 | 5.63 | 4 | 1 | 3 | 1 | 3 | 0.07 | 0.04 |
| DDRGK_MOUSE | DDRGK domain-containing protein 1 | 49 | 35956 | 5.32 | 3 | 1 | 2 | 1 | 2 | 0.13 | 0.12 |
| LMAN1_MOUSE | Protein ERGIC-53 | 49 | 57753 | 5.92 | 11 | 5 | 7 | 4 | 7 | 0.25 | 0.34 |
| AHSA1_MOUSE | Activator of 90 kDa heat shock protein ATPase homolog 1 | 49 | 38093 | 5.41 | 5 | 2 | 5 | 2 | 5 | 0.27 | 0.24 |
| HMGCL_MOUSE | Hydroxymethylglutaryl-CoA lyase, mitochondrial | 48 | 34217 | 8.7 | 3 | 2 | 3 | 2 | 3 | 0.2 | 0.27 |
| PURA2_MOUSE | Adenylosuccinate synthetase isozyme 2 | 48 | 49990 | 5.98 | 6 | 2 | 5 | 2 | 5 | 0.16 | 0.18 |
| SYK_MOUSE | Lysine–tRNA ligase | 48 | 67796 | 5.65 | 6 | 1 | 4 | 1 | 4 | 0.11 | 0.06 |
| CGAT1_MOUSE | Chondroitin sulfate N-acetylgalactosaminyltransferase 1 | 48 | 60878 | 9.03 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.07 |
| NSDHL_MOUSE | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | 48 | 40660 | 7.71 | 6 | 4 | 4 | 3 | 4 | 0.19 | 0.36 |
| KTN1_MOUSE | Kinectin | 48 | 152498 | 5.67 | 11 | 1 | 11 | 1 | 11 | 0.12 | 0.03 |
| BID_MOUSE | BH3-interacting domain death agonist | 48 | 21938 | 4.71 | 2 | 2 | 2 | 2 | 2 | 0.15 | 0.46 |
| DEGS1_MOUSE | Sphingolipid delta(4)-desaturase DES1 | 45 | 38216 | 7.33 | 2 | 2 | 2 | 2 | 2 | 0.1 | 0.24 |
| EP15R_MOUSE | Epidermal growth factor receptor substrate 15-like 1 | 47 | 99248 | 4.86 | 8 | 2 | 6 | 1 | 6 | 0.17 | 0.04 |
| ARL2_MOUSE | ADP-ribosylation factor-like protein 2 | 47 | 20851 | 5.67 | 1 | 1 | 1 | 1 | 1 | 0.14 | 0.22 |
| NPC1_MOUSE | Niemann-Pick C1 protein | 47 | 142791 | 5.44 | 5 | 2 | 4 | 2 | 4 | 0.06 | 0.06 |
| WASC2_MOUSE | WASH complex subunit 2 | 47 | 145224 | 4.63 | 4 | 2 | 4 | 2 | 4 | 0.05 | 0.06 |
| NPL_MOUSE | N-Acetylneuraminate lyase | 47 | 35108 | 7.74 | 3 | 2 | 1 | 1 | 1 | 0.09 | 0.27 |
| CD63_MOUSE | CD63 antigen | 47 | 25749 | 6.69 | 4 | 2 | 4 | 2 | 4 | 0.23 | 0.38 |
| GLRX3_MOUSE | Glutaredoxin-3 | 47 | 37754 | 5.42 | 12 | 3 | 11 | 3 | 11 | 0.6 | 0.39 |
| DNJB4_MOUSE | DnaJ homolog subfamily B member 4 | 47 | 37758 | 8.7 | 2 | 1 | 2 | 1 | 2 | 0.08 | 0.12 |
| DC1L2_MOUSE | Cytoplasmic dynein 1 light intermediate chain 2 | 46 | 54185 | 5.89 | 7 | 3 | 7 | 3 | 6 | 0.25 | 0.26 |
| ULA1_MOUSE | NEDD8-activating enzyme E1 regulatory subunit | 46 | 60236 | 5.34 | 6 | 3 | 4 | 2 | 4 | 0.16 | 0.23 |
| SETD3_MOUSE | Histone-lysine N-methyltransferase setd3 | 44 | 67134 | 5.47 | 2 | 2 | 2 | 2 | 2 | 0.08 | 0.13 |
| LXN_MOUSE | Latexin | 46 | 25476 | 5.48 | 2 | 1 | 2 | 1 | 2 | 0.14 | 0.18 |
| MPEG1_MOUSE | Macrophage-expressed gene 1 protein | 46 | 78340 | 5.64 | 4 | 2 | 3 | 1 | 3 | 0.1 | 0.05 |
| YKT6_MOUSE | Synaptobrevin homolog YKT6 | 46 | 22300 | 5.97 | 4 | 1 | 4 | 1 | 4 | 0.35 | 0.2 |
| GSHR_MOUSE | Glutathione reductase, mitochondrial | 46 | 53629 | 8.19 | 8 | 2 | 6 | 1 | 6 | 0.19 | 0.08 |
| CP062_MOUSE | UPF0505 protein C16orf62 homolog | 46 | 109007 | 6.97 | 3 | 1 | 3 | 1 | 3 | 0.06 | 0.04 |
| SCMC1_MOUSE | Calcium-binding mitochondrial carrier protein SCaMC-1 | 46 | 52868 | 7.02 | 7 | 2 | 6 | 2 | 6 | 0.23 | 0.17 |
| CO4A2_MOUSE | Collagen alpha-2(IV) chain | 46 | 167220 | 8.75 | 4 | 1 | 4 | 1 | 4 | 0.04 | 0.03 |
| RDH13_MOUSE | Retinol dehydrogenase 13 | 46 | 36441 | 9.02 | 2 | 1 | 2 | 1 | 2 | 0.08 | 0.12 |
| DPP2_MOUSE | Dipeptidyl peptidase 2 | 46 | 56218 | 5.17 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.08 |
| ATX10_MOUSE | Ataxin-10 | 46 | 53673 | 5.12 | 5 | 2 | 4 | 2 | 4 | 0.12 | 0.17 |
| APMAP_MOUSE | Adipocyte plasma membrane-associated protein | 45 | 46405 | 5.97 | 2 | 2 | 1 | 1 | 1 | 0.07 | 0.09 |
| EFHD2_MOUSE | EF-hand domain-containing protein D2 | 45 | 26775 | 5.01 | 8 | 1 | 3 | 1 | 3 | 0.12 | 0.17 |
| PEDF_MOUSE | Pigment epithelium-derived factor | 45 | 46205 | 6.48 | 3 | 1 | 2 | 1 | 2 | 0.1 | 0.09 |
| SODC_MOUSE | Superoxide dismutase [Cu-Zn] | 45 | 15933 | 6.02 | 9 | 3 | 5 | 2 | 5 | 0.39 | 0.67 |
| S38A2_MOUSE | Sodium-coupled neutral amino acid transporter 2 | 45 | 55467 | 8.05 | 4 | 2 | 4 | 2 | 4 | 0.16 | 0.16 |
| NUP50_MOUSE | Nuclear pore complex protein Nup50 | 45 | 49455 | 5.94 | 3 | 1 | 3 | 1 | 3 | 0.16 | 0.09 |
| CD44_MOUSE | CD44 antigen | 45 | 85565 | 4.82 | 6 | 1 | 5 | 1 | 5 | 0.11 | 0.05 |
| NDUS8_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial | 45 | 24023 | 5.89 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.19 |
| STX2_MOUSE | Syntaxin-2 | 45 | 33157 | 5.98 | 2 | 1 | 1 | 1 | 1 | 0.11 | 0.13 |
| TMED3_MOUSE | Transmembrane emp24 domain-containing protein 3 | 45 | 25449 | 5.62 | 3 | 2 | 2 | 2 | 2 | 0.16 | 0.38 |
| VMP1_MOUSE | Vacuole membrane protein 1 | 45 | 45931 | 6.47 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.09 |
| PRAF1_MOUSE | Prenylated Rab acceptor protein 1 | 45 | 20606 | 7.74 | 1 | 1 | 1 | 1 | 1 | 0.08 | 0.22 |
| ARMC6_MOUSE | Armadillo repeat-containing protein 6 | 44 | 50651 | 5.65 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.09 |
| GRHPR_MOUSE | Glyoxylate reductase/hydroxypyruvate reductase | 44 | 35306 | 7.57 | 3 | 3 | 2 | 2 | 2 | 0.16 | 0.27 |
| CRTAP_MOUSE | Cartilage-associated protein | 44 | 46140 | 5.46 | 3 | 3 | 3 | 3 | 3 | 0.14 | 0.31 |
| TM14C_MOUSE | Transmembrane protein 14C | 44 | 11634 | 9.7 | 2 | 2 | 2 | 2 | 2 | 0.51 | 1.01 |
| COX5B_MOUSE | Cytochrome c oxidase subunit 5B, mitochondrial | 44 | 13804 | 8.69 | 10 | 4 | 3 | 2 | 3 | 0.34 | 0.81 |
| NNRE_MOUSE | NAD(P)H-hydrate epimerase | 44 | 30953 | 7.59 | 2 | 2 | 2 | 2 | 2 | 0.1 | 0.31 |
| TPR_MOUSE | Nucleoprotein TPR | 44 | 273824 | 4.98 | 10 | 1 | 10 | 1 | 10 | 0.06 | 0.02 |
| SYFA_MOUSE | Phenylalanine–tRNA ligase alpha subunit | 44 | 57563 | 8.27 | 3 | 1 | 2 | 1 | 2 | 0.05 | 0.08 |
| E41L2_MOUSE | Band 4.1-like protein 2 | 43 | 109873 | 5.31 | 7 | 1 | 7 | 1 | 7 | 0.09 | 0.04 |
| CBR3_MOUSE | Carbonyl reductase [NADPH] 3 | 43 | 30934 | 6.15 | 4 | 2 | 3 | 2 | 3 | 0.19 | 0.31 |
| NDUB7_MOUSE | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 | 43 | 16320 | 8.35 | 5 | 2 | 2 | 2 | 2 | 0.26 | 0.65 |
| LRN4L_MOUSE | LRRN4 C-terminal-like protein | 43 | 25914 | 5.15 | 2 | 1 | 2 | 1 | 2 | 0.11 | 0.17 |
| CSTFT_MOUSE | Cleavage stimulation factor subunit 2 tau variant | 43 | 65820 | 6.78 | 4 | 1 | 3 | 1 | 3 | 0.13 | 0.07 |
| RFA2_MOUSE | Replication protein A 32 kDa subunit | 43 | 29700 | 5.76 | 1 | 1 | 1 | 1 | 1 | 0.09 | 0.15 |
| NPS3B_MOUSE | Protein NipSnap homolog 3B | 43 | 28290 | 9.51 | 2 | 2 | 1 | 1 | 1 | 0.1 | 0.16 |
| HDAC7_MOUSE | Histone deacetylase 7 | 43 | 101224 | 7.11 | 2 | 1 | 2 | 1 | 2 | 0.07 | 0.04 |
| PEX19_MOUSE | Peroxisomal biogenesis factor 19 | 43 | 32713 | 4.26 | 4 | 1 | 4 | 1 | 4 | 0.34 | 0.14 |
| GALK1_MOUSE | Galactokinase | 43 | 42268 | 5.17 | 7 | 2 | 5 | 2 | 5 | 0.21 | 0.22 |
| DCTN1_MOUSE | Dynactin subunit 1 | 43 | 141588 | 5.66 | 7 | 2 | 6 | 2 | 6 | 0.08 | 0.06 |
| OAS1A_MOUSE | 2 ′-5 ′-Oligoadenylate synthase 1A | 43 | 42402 | 8.3 | 2 | 1 | 2 | 1 | 2 | 0.08 | 0.1 |
| FKBP3_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP3 | 43 | 25132 | 9.29 | 3 | 2 | 3 | 2 | 3 | 0.21 | 0.39 |
| GIPC1_MOUSE | PDZ domain-containing protein GIPC1 | 43 | 36107 | 5.65 | 3 | 2 | 3 | 2 | 3 | 0.17 | 0.26 |
| EMC3_MOUSE | ER membrane protein complex subunit 3 | 43 | 29960 | 6.33 | 3 | 1 | 3 | 1 | 3 | 0.17 | 0.15 |
| MMSA_MOUSE | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial | 43 | 57878 | 8.29 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.07 |
| PLXB2_MOUSE | Plexin-B2 | 43 | 206099 | 5.58 | 12 | 1 | 9 | 1 | 9 | 0.09 | 0.02 |
| FADS3_MOUSE | Fatty acid desaturase 3 | 42 | 51436 | 7 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.08 |
| FUND2_MOUSE | FUN14 domain-containing protein 2 | 42 | 16554 | 9.71 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.28 |
| GPX7_MOUSE | Glutathione peroxidase 7 | 42 | 21048 | 8.42 | 3 | 1 | 3 | 1 | 3 | 0.28 | 0.22 |
| MTX2_MOUSE | Metaxin-2 | 42 | 29739 | 5.44 | 2 | 1 | 2 | 1 | 2 | 0.16 | 0.15 |
| HTR5A_MOUSE | HEAT repeat-containing protein 5A | 42 | 219754 | 6.31 | 4 | 1 | 4 | 1 | 4 | 0.05 | 0.02 |
| ALG11_MOUSE | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase | 42 | 55234 | 8.58 | 3 | 1 | 2 | 1 | 2 | 0.09 | 0.08 |
| DHSD_MOUSE | Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial | 42 | 17003 | 9.3 | 4 | 2 | 2 | 1 | 2 | 0.23 | 0.27 |
| UBP19_MOUSE | Ubiquitin carboxyl-terminal hydrolase 19 | 42 | 150454 | 5.99 | 3 | 1 | 3 | 1 | 3 | 0.04 | 0.03 |
| OSMR_MOUSE | Oncostatin-M-specific receptor subunit beta | 42 | 110160 | 6.16 | 3 | 1 | 1 | 1 | 1 | 0.04 | 0.04 |
| PGP_MOUSE | Glycerol-3-phosphate phosphatase | 42 | 34519 | 5.21 | 3 | 1 | 2 | 1 | 2 | 0.2 | 0.13 |
| DDR2_MOUSE | Discoidin domain-containing receptor 2 | 38 | 96420 | 5.31 | 3 | 2 | 2 | 2 | 2 | 0.05 | 0.09 |
| F162A_MOUSE | Protein FAM162A | 42 | 17713 | 9.9 | 3 | 1 | 3 | 1 | 3 | 0.25 | 0.26 |
| ACOT9_MOUSE | Acyl-coenzyme A thioesterase 9, mitochondrial | 41 | 50528 | 8.74 | 8 | 3 | 5 | 2 | 5 | 0.16 | 0.18 |
| UCHL1_MOUSE | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 41 | 24822 | 5.14 | 4 | 2 | 3 | 1 | 3 | 0.22 | 0.18 |
| TPP2_MOUSE | Tripeptidyl-peptidase 2 | 41 | 139790 | 6.13 | 9 | 3 | 7 | 3 | 7 | 0.11 | 0.09 |
| HEXB_MOUSE | Beta-hexosaminidase subunit beta | 41 | 61077 | 8.29 | 5 | 1 | 3 | 1 | 3 | 0.14 | 0.07 |
| CDIPT_MOUSE | CDP-diacylglycerol–inositol 3-phosphatidyltransferase | 41 | 23583 | 8.56 | 2 | 1 | 2 | 1 | 2 | 0.21 | 0.19 |
| GAS6_MOUSE | Growth arrest-specific protein 6 | 41 | 74561 | 5.34 | 5 | 2 | 4 | 1 | 4 | 0.09 | 0.06 |
| PIN1_MOUSE | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | 41 | 18359 | 8.93 | 2 | 1 | 2 | 1 | 2 | 0.19 | 0.25 |
| APOE_MOUSE | Apolipoprotein E | 41 | 35844 | 5.56 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.12 |
| SGMR1_MOUSE | Sigma nonopioid intracellular receptor 1 | 41 | 25234 | 5.56 | 1 | 1 | 1 | 1 | 1 | 0.15 | 0.18 |
| RTRAF_MOUSE | RNA transcription, translation and transport factor protein | 41 | 28135 | 6.4 | 4 | 1 | 3 | 1 | 3 | 0.21 | 0.16 |
| TF_MOUSE | Tissue factor | 41 | 32914 | 9.4 | 5 | 2 | 5 | 2 | 5 | 0.26 | 0.29 |
| ERGI3_MOUSE | Endoplasmic reticulum-Golgi intermediate compartment protein 3 | 40 | 43181 | 6.02 | 3 | 2 | 2 | 1 | 2 | 0.17 | 0.21 |
| LONM_MOUSE | Lon protease homolog, mitochondrial | 40 | 105776 | 6.15 | 6 | 1 | 5 | 1 | 5 | 0.14 | 0.04 |
| TXD17_MOUSE | Thioredoxin domain-containing protein 17 | 40 | 14006 | 4.72 | 3 | 1 | 2 | 1 | 2 | 0.19 | 0.34 |
| DJC10_MOUSE | DnaJ homolog subfamily C member 10 | 40 | 90525 | 6.53 | 7 | 2 | 4 | 2 | 4 | 0.09 | 0.1 |
| PML_MOUSE | Protein PML | 40 | 98180 | 5.42 | 4 | 1 | 4 | 1 | 4 | 0.09 | 0.04 |
| NAGA_MOUSE | N-Acetylglucosamine-6-phosphate deacetylase | 40 | 43473 | 5.78 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.1 |
| HGH1_MOUSE | Protein HGH1 homolog | 40 | 42889 | 4.66 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.1 |
| GLRX5_MOUSE | Glutaredoxin-related protein 5, mitochondrial | 40 | 16282 | 6.1 | 2 | 1 | 2 | 1 | 2 | 0.14 | 0.29 |
| IPO11_MOUSE | Importin-11 | 40 | 112344 | 5.14 | 3 | 1 | 3 | 1 | 3 | 0.07 | 0.04 |
| T176A_MOUSE | Transmembrane protein 176A | 40 | 26579 | 7.62 | 3 | 1 | 2 | 1 | 2 | 0.16 | 0.17 |
| S10AD_MOUSE | Protein S100-A13 | 40 | 11151 | 5.89 | 2 | 1 | 2 | 1 | 2 | 0.23 | 0.44 |
| PEX14_MOUSE | Peroxisomal membrane protein PEX14 | 40 | 41183 | 5.03 | 2 | 1 | 2 | 1 | 2 | 0.15 | 0.11 |
| DHRS1_MOUSE | Dehydrogenase/reductase SDR family member 1 | 40 | 33983 | 8.66 | 6 | 2 | 5 | 2 | 5 | 0.28 | 0.28 |
| MVD1_MOUSE | Diphosphomevalonate decarboxylase | 40 | 44044 | 5.89 | 3 | 2 | 3 | 2 | 3 | 0.19 | 0.21 |
| VCAM1_MOUSE | Vascular cell adhesion protein 1 | 40 | 81265 | 5.21 | 5 | 2 | 3 | 1 | 3 | 0.09 | 0.05 |
| PRDX3_MOUSE | Thioredoxin-dependent peroxide reductase, mitochondrial | 40 | 28109 | 7.15 | 7 | 2 | 6 | 2 | 6 | 0.39 | 0.34 |
| PP14B_MOUSE | Protein phosphatase 1 regulatory subunit 14B | 40 | 15947 | 4.74 | 2 | 1 | 2 | 1 | 2 | 0.33 | 0.29 |
| BPNT1_MOUSE | 3 ′(2 ′),5 ′-Bisphosphate nucleotidase 1 | 39 | 33175 | 5.54 | 3 | 1 | 3 | 1 | 3 | 0.15 | 0.13 |
| FAF2_MOUSE | FAS-associated factor 2 | 39 | 52438 | 5.35 | 6 | 1 | 4 | 1 | 4 | 0.23 | 0.08 |
| TI8AB_MOUSE | Putative mitochondrial import inner membrane translocase subunit Tim8 A-B | 39 | 11276 | 6.11 | 4 | 1 | 3 | 1 | 3 | 0.39 | 0.43 |
| GBP2_MOUSE | Guanylate-binding protein 2 | 39 | 66697 | 5.56 | 3 | 1 | 3 | 1 | 3 | 0.14 | 0.06 |
| ABCD3_MOUSE | ATP-binding cassette sub-family D member 3 | 39 | 75426 | 9.32 | 7 | 2 | 7 | 2 | 7 | 0.18 | 0.12 |
| DPP9_MOUSE | Dipeptidyl peptidase 9 | 39 | 97939 | 6.18 | 3 | 2 | 2 | 1 | 2 | 0.05 | 0.04 |
| MSMO1_MOUSE | Methylsterol monooxygenase 1 | 39 | 34750 | 7.29 | 2 | 1 | 1 | 1 | 1 | 0.1 | 0.13 |
| TRNT1_MOUSE | CCA tRNA nucleotidyltransferase 1, mitochondrial | 39 | 49864 | 8.65 | 3 | 2 | 2 | 1 | 2 | 0.07 | 0.09 |
| RIPK3_MOUSE | Receptor-interacting serine/threonine-protein kinase 3 | 39 | 53289 | 7.22 | 8 | 3 | 6 | 3 | 6 | 0.17 | 0.26 |
| ERGI1_MOUSE | Endoplasmic reticulum-Golgi intermediate compartment protein 1 | 39 | 32541 | 6.59 | 4 | 2 | 3 | 1 | 3 | 0.14 | 0.14 |
| P2RX4_MOUSE | P2X purinoceptor 4 | 38 | 43410 | 8.29 | 2 | 2 | 1 | 1 | 1 | 0.07 | 0.1 |
| FCGRN_MOUSE | IgG receptor FcRn large subunit p51 | 38 | 40067 | 5.2 | 2 | 2 | 2 | 2 | 2 | 0.1 | 0.23 |
| NCF2_MOUSE | Neutrophil cytosol factor 2 | 38 | 59448 | 6.18 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.07 |
| TOIP2_MOUSE | Torsin-1A-interacting protein 2 | 38 | 54463 | 4.75 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.08 |
| DHB11_MOUSE | Estradiol 17-beta-dehydrogenase 11 | 38 | 32860 | 8.85 | 2 | 2 | 1 | 1 | 1 | 0.06 | 0.13 |
| SGTA_MOUSE | Small glutamine-rich tetratricopeptide repeat-containing protein alpha | 38 | 34301 | 4.99 | 4 | 2 | 3 | 2 | 3 | 0.12 | 0.27 |
| HEM2_MOUSE | Delta-aminolevulinic acid dehydratase | 38 | 36000 | 6.32 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.12 |
| CX7A2_MOUSE | Cytochrome c oxidase subunit 7A2, mitochondrial | 38 | 9285 | 10.28 | 3 | 1 | 3 | 1 | 3 | 0.53 | 0.54 |
| PSB8_MOUSE | Proteasome subunit beta type-8 | 38 | 30241 | 6.22 | 4 | 1 | 4 | 1 | 4 | 0.19 | 0.15 |
| DECR_MOUSE | 2,4-Dienoyl-CoA reductase, mitochondrial | 36 | 36191 | 9.1 | 4 | 2 | 4 | 2 | 4 | 0.2 | 0.26 |
| XPP1_MOUSE | Xaa-Pro aminopeptidase 1 | 37 | 69547 | 5.33 | 3 | 2 | 3 | 2 | 3 | 0.09 | 0.13 |
| SYYC_MOUSE | Tyrosine–tRNA ligase, cytoplasmic | 37 | 59068 | 6.57 | 6 | 1 | 6 | 1 | 6 | 0.16 | 0.07 |
| FLOT1_MOUSE | Flotillin-1 | 37 | 47484 | 6.71 | 12 | 2 | 4 | 1 | 4 | 0.13 | 0.09 |
| BAP31_MOUSE | B cell receptor-associated protein 31 | 37 | 27939 | 8.73 | 5 | 1 | 3 | 1 | 3 | 0.16 | 0.16 |
| NECP2_MOUSE | Adaptin ear-binding coat-associated protein 2 | 37 | 28580 | 7.71 | 9 | 4 | 4 | 2 | 4 | 0.28 | 0.34 |
| GINM1_MOUSE | Glycoprotein integral membrane protein 1 | 37 | 36056 | 5.45 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.12 |
| ATG7_MOUSE | Ubiquitin-like modifier-activating enzyme ATG7 | 37 | 77470 | 5.97 | 5 | 1 | 5 | 1 | 5 | 0.12 | 0.06 |
| MA2B2_MOUSE | Epididymis-specific alpha-mannosidase | 37 | 115537 | 6.9 | 1 | 1 | 1 | 1 | 1 | 0.02 | 0.04 |
| TB182_MOUSE | 182 kDa tankyrase-1-binding protein | 37 | 181714 | 4.8 | 5 | 1 | 4 | 1 | 4 | 0.04 | 0.02 |
| PICAL_MOUSE | Phosphatidylinositol-binding clathrin assembly protein | 37 | 71498 | 7.71 | 4 | 1 | 4 | 1 | 4 | 0.1 | 0.06 |
| PELP1_MOUSE | Proline-, glutamic acid-, and leucine-rich protein 1 | 37 | 117995 | 4.31 | 2 | 1 | 2 | 1 | 2 | 0.05 | 0.04 |
| ARHG7_MOUSE | Rho guanine nucleotide exchange factor 7 | 37 | 96995 | 6.35 | 5 | 1 | 3 | 1 | 3 | 0.09 | 0.04 |
| RT11_MOUSE | 28S ribosomal protein S11, mitochondrial | 37 | 20196 | 10.77 | 2 | 1 | 2 | 1 | 2 | 0.18 | 0.23 |
| ODP2_MOUSE | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | 37 | 67899 | 8.81 | 7 | 1 | 5 | 1 | 5 | 0.13 | 0.06 |
| NT5C_MOUSE | 5 ′(3 ′)-Deoxyribonucleotidase, cytosolic type | 37 | 23062 | 5.31 | 2 | 1 | 2 | 1 | 2 | 0.22 | 0.2 |
| INF2_MOUSE | Inverted formin-2 | 37 | 138474 | 5.09 | 5 | 1 | 4 | 1 | 4 | 0.07 | 0.03 |
| WIPI1_MOUSE | WD repeat domain phosphoinositide-interacting protein 1 | 37 | 48727 | 6.03 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.09 |
| GPDM_MOUSE | Glycerol-3-phosphate dehydrogenase, mitochondrial | 37 | 80902 | 6.17 | 7 | 2 | 5 | 2 | 5 | 0.11 | 0.11 |
| ATP8_MOUSE | ATP synthase protein 8 | 36 | 7761 | 9.88 | 2 | 1 | 1 | 1 | 1 | 0.15 | 0.67 |
| BST2_MOUSE | Bone marrow stromal antigen 2 | 36 | 19140 | 6.82 | 2 | 2 | 1 | 1 | 1 | 0.05 | 0.24 |
| DERL2_MOUSE | Derlin-2 | 36 | 27621 | 6.73 | 1 | 1 | 1 | 1 | 1 | 0.13 | 0.16 |
| IF4G2_MOUSE | Eukaryotic translation initiation factor 4 gamma 2 | 36 | 102041 | 6.7 | 6 | 1 | 6 | 1 | 6 | 0.1 | 0.04 |
| FKBP7_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP7 | 36 | 24897 | 5.69 | 3 | 2 | 3 | 2 | 3 | 0.17 | 0.39 |
| PLP2_MOUSE | Proteolipid protein 2 | 36 | 16597 | 6.69 | 4 | 3 | 1 | 1 | 1 | 0.08 | 0.28 |
| RL29_MOUSE | 60S ribosomal protein L29 | 36 | 17576 | 11.84 | 5 | 1 | 5 | 1 | 5 | 0.34 | 0.26 |
| WASH1_MOUSE | WASH complex subunit 1 | 36 | 51627 | 5.33 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.08 |
| SIAS_MOUSE | Sialic acid synthase | 36 | 39998 | 6.61 | 6 | 1 | 5 | 1 | 5 | 0.23 | 0.11 |
| RT28_MOUSE | 28S ribosomal protein S28, mitochondrial | 36 | 20508 | 9.1 | 1 | 1 | 1 | 1 | 1 | 0.13 | 0.22 |
| FACE1_MOUSE | CAAX prenyl protease 1 homolog | 36 | 54699 | 6.49 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.08 |
| MEP50_MOUSE | Methylosome protein 50 | 36 | 36919 | 5.09 | 1 | 1 | 1 | 1 | 1 | 0.09 | 0.12 |
| SCFD2_MOUSE | Sec1 family domain-containing protein 2; OS = Mus musculus, GN = Scfd2, PE = 1, SV = 1 | 36 | 74703 | 6.35 | 4 | 1 | 4 | 1 | 4 | 0.11 | 0.06 |
| PTN9_MOUSE | Tyrosine-protein phosphatase nonreceptor type 9; OS = Mus musculus, GN = Ptpn9, PE = 1, SV = 2 | 36 | 67927 | 8.34 | 3 | 1 | 2 | 1 | 2 | 0.07 | 0.06 |
| MXRA8_MOUSE | Matrix remodeling-associated protein 8; OS = Mus musculus, GN = Mxra8, PE = 1, SV = 1 | 36 | 49719 | 6.66 | 2 | 1 | 2 | 1 | 2 | 0.05 | 0.09 |
| COASY_MOUSE | Bifunctional coenzyme A synthase; OS = Mus musculus, GN = Coasy, PE = 1, SV = 2 | 36 | 61985 | 6.61 | 4 | 2 | 3 | 2 | 3 | 0.13 | 0.14 |
| PFD1_MOUSE | Prefoldin subunit 1; OS = Mus musculus, GN = Pfdn1, PE = 1, SV = 1 | 35 | 14246 | 7.93 | 5 | 2 | 2 | 1 | 2 | 0.17 | 0.33 |
| COX7C_MOUSE | Cytochrome c oxidase subunit 7C, mitochondrial; OS = Mus musculus, GN = Cox7c, PE = 1, SV = 1 | 35 | 7328 | 11 | 2 | 1 | 1 | 1 | 1 | 0.33 | 0.72 |
| ZCCHV_MOUSE | Zinc finger CCCH-type antiviral protein 1; OS = Mus musculus, GN = Zc3hav1, PE = 1, SV = 1 | 35 | 106619 | 8.59 | 4 | 1 | 4 | 1 | 4 | 0.11 | 0.04 |
| ASC_MOUSE | Apoptosis-associated speck-like protein containing a CARD; OS = Mus musculus, GN = Pycard, PE = 1, SV = 1 | 35 | 21445 | 5.26 | 2 | 1 | 2 | 1 | 2 | 0.2 | 0.21 |
| KCY_MOUSE | UMP-CMP kinase; OS = Mus musculus, GN = Cmpk1, PE = 1, SV = 1 | 35 | 22151 | 5.68 | 4 | 2 | 4 | 2 | 4 | 0.3 | 0.45 |
| HINT1_MOUSE | Histidine triad nucleotide-binding protein 1; OS = Mus musculus, GN = Hint1, PE = 1, SV = 3 | 35 | 13768 | 6.36 | 6 | 1 | 1 | 1 | 1 | 0.11 | 0.35 |
| ARSA_MOUSE | Arylsulfatase A; OS = Mus musculus, GN = Arsa, PE = 1, SV = 2 | 35 | 53714 | 5.5 | 2 | 1 | 2 | 1 | 2 | 0.15 | 0.08 |
| MIC13_MOUSE | MICOS complex subunit MIC13; OS = Mus musculus, GN = Mic13, PE = 1, SV = 1 | 35 | 13365 | 8.68 | 2 | 1 | 2 | 1 | 2 | 0.27 | 0.36 |
| NUCB2_MOUSE | Nucleobindin-2 OS = Mus musculus, GN = Nucb2, PE = 1, SV = 2 | 35 | 50273 | 5.05 | 8 | 2 | 6 | 1 | 6 | 0.2 | 0.09 |
| CHMP5_MOUSE | Charged multivesicular body protein 5; OS = Mus musculus, GN = Chmp5, PE = 1, SV = 1 | 35 | 24560 | 4.65 | 5 | 1 | 2 | 1 | 2 | 0.2 | 0.18 |
| HOME3_MOUSE | Homer protein homolog 3 OS = Mus musculus GN = Homer3 PE = 1 SV = 2 | 35 | 39670 | 5.33 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.11 |
| NHLC3_MOUSE | NHL repeat-containing protein 3; OS = Mus musculus, GN = Nhlrc3, PE = 1, SV = 1 | 34 | 38171 | 5.81 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.11 |
| CTBP2_MOUSE | C-terminal-binding protein 2; OS = Mus musculus, GN = Ctbp2, PE = 1, SV = 2 | 34 | 48926 | 6.47 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.09 |
| NU160_MOUSE | Nuclear pore complex protein Nup160; OS = Mus musculus, GN = Nup160, PE = 1, SV = 2 | 34 | 158130 | 5.32 | 5 | 1 | 4 | 1 | 4 | 0.04 | 0.03 |
| RAB34_MOUSE | Ras-related protein Rab-34; OS = Mus musculus, GN = Rab34, PE = 1, SV = 2 | 34 | 29082 | 8.55 | 6 | 2 | 4 | 2 | 4 | 0.22 | 0.33 |
| COMD8_MOUSE | COMM domain-containing protein 8; OS = Mus musculus, GN = Commd8, PE = 1, SV = 1 | 34 | 20839 | 5.38 | 2 | 1 | 2 | 1 | 2 | 0.3 | 0.22 |
| HECD1_MOUSE | E3 ubiquitin-protein ligase HECTD1; OS = Mus musculus, GN = Hectd1, PE = 1, SV = 2 | 34 | 289905 | 5.26 | 12 | 1 | 11 | 1 | 11 | 0.08 | 0.01 |
| LYRIC_MOUSE | Protein LYRIC; OS = Mus musculus, GN = Mtdh, PE = 1, SV = 1 | 34 | 63808 | 9.34 | 5 | 3 | 4 | 3 | 4 | 0.2 | 0.22 |
| S35F6_MOUSE | Solute carrier family 35 member F6; OS = Mus musculus, GN = Slc35f6, PE = 1, SV = 1 | 34 | 40952 | 6.65 | 2 | 1 | 2 | 1 | 2 | 0.17 | 0.11 |
| SBDS_MOUSE | Ribosome maturation protein SBDS; OS = Mus musculus, GN = Sbds, PE = 1, SV = 4 | 34 | 28762 | 8.92 | 2 | 1 | 1 | 1 | 1 | 0.09 | 0.15 |
| RHG01_MOUSE | Rho GTPase-activating protein 1; OS = Mus musculus, GN = Arhgap1, PE = 1, SV = 1 | 34 | 50379 | 5.97 | 5 | 3 | 3 | 3 | 3 | 0.12 | 0.28 |
| FACR1_MOUSE | Fatty acyl-CoA reductase 1; OS = Mus musculus, GN = Far1, PE = 1, SV = 1 | 34 | 59397 | 9.26 | 4 | 1 | 2 | 1 | 2 | 0.09 | 0.07 |
| MPRD_MOUSE | Cation-dependent mannose-6-phosphate receptor; OS = Mus musculus, GN = M6pr, PE = 1, SV = 1 | 33 | 31152 | 5.24 | 3 | 2 | 2 | 1 | 2 | 0.13 | 0.14 |
| PNKP_MOUSE | Bifunctional polynucleotide phosphatase/kinase; OS = Mus musculus, GN = Pnkp, PE = 1, SV = 2 | 33 | 57188 | 8.03 | 9 | 1 | 5 | 1 | 5 | 0.15 | 0.08 |
| DCPS_MOUSE | m7GpppX diphosphatase; OS = Mus musculus, GN = Dcps, PE = 1, SV = 1 | 33 | 38964 | 6.02 | 2 | 1 | 2 | 1 | 2 | 0.1 | 0.11 |
| DHX29_MOUSE | ATP-dependent RNA helicase DHX29; OS = Mus musculus, GN = Dhx29, PE = 1, SV = 1 | 33 | 153879 | 8.13 | 11 | 2 | 9 | 1 | 9 | 0.1 | 0.03 |
| CN37_MOUSE | 2 ′,3 ′-Cyclic-nucleotide 3 ′-phosphodiesterase; OS = Mus musculus, GN = Cnp, PE = 1, SV = 3 | 33 | 47094 | 9.08 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.09 |
| TBCB_MOUSE | Tubulin-folding cofactor B; OS = Mus musculus, GN = Tbcb, PE = 1, SV = 2 | 33 | 27368 | 5.14 | 3 | 1 | 3 | 1 | 3 | 0.23 | 0.16 |
| SFPQ_MOUSE | Splicing factor, proline- and glutamine-rich; OS = Mus musculus, GN = Sfpq, PE = 1, SV = 1 | 33 | 75394 | 9.45 | 3 | 1 | 2 | 1 | 2 | 0.07 | 0.06 |
| GLYG_MOUSE | Glycogenin-1; OS = Mus musculus, GN = Gyg1, PE = 1, SV = 3 | 33 | 37378 | 5.06 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.12 |
| TSPO_MOUSE | Translocator protein; OS = Mus musculus, GN = Tspo, PE = 1, SV = 1 | 33 | 18829 | 9.52 | 2 | 1 | 2 | 1 | 2 | 0.3 | 0.24 |
| CND1_MOUSE | Condensin complex subunit 1; OS = Mus musculus, GN = Ncapd2, PE = 1, SV = 2 | 33 | 155567 | 5.98 | 4 | 1 | 3 | 1 | 3 | 0.04 | 0.03 |
| JIP4_MOUSE | C-Jun-amino-terminal kinase-interacting protein 4; OS = Mus musculus, GN = Spag9, PE = 1, SV = 2 | 33 | 146129 | 5.05 | 6 | 1 | 6 | 1 | 6 | 0.08 | 0.03 |
| NDUB9_MOUSE | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; OS = Mus musculus, GN = Ndufb9, PE = 1, SV = 3 | 33 | 21970 | 7.67 | 2 | 2 | 1 | 1 | 1 | 0.16 | 0.21 |
| RBGP1_MOUSE | Rab GTPase-activating protein 1; OS = Mus musculus, GN = Rabgap1, PE = 1, SV = 1 | 32 | 120722 | 5.14 | 5 | 1 | 3 | 1 | 3 | 0.03 | 0.04 |
| F13A_MOUSE | Coagulation factor XIII A chain; OS = Mus musculus, GN = F13a1, PE = 1, SV = 3 | 32 | 83155 | 5.63 | 4 | 3 | 2 | 2 | 2 | 0.06 | 0.11 |
| DOCK7_MOUSE | Dedicator of cytokinesis protein 7; OS = Mus musculus, GN = Dock7, PE = 1, SV = 3 | 32 | 241286 | 6.25 | 6 | 1 | 6 | 1 | 6 | 0.04 | 0.02 |
| IC1_MOUSE | Plasma protease C1 inhibitor; OS = Mus musculus, GN = Serping1, PE = 1, SV = 3 | 32 | 55549 | 5.87 | 2 | 1 | 2 | 1 | 2 | 0.13 | 0.08 |
| SRP54_MOUSE | Signal recognition particle 54 kDa protein; OS = Mus musculus, GN = Srp54, PE = 1, SV = 2 | 32 | 55684 | 8.87 | 15 | 3 | 10 | 3 | 10 | 0.38 | 0.25 |
| YIF1B_MOUSE | Protein YIF1B; OS = Mus musculus, GN = Yif1b, PE = 1, SV = 2 | 32 | 33961 | 9.19 | 5 | 1 | 2 | 1 | 2 | 0.14 | 0.13 |
| TPC13_MOUSE | Trafficking protein particle complex subunit 13; OS = Mus musculus, GN = Trappc13, PE = 2, SV = 1 | 32 | 46446 | 5.33 | 3 | 1 | 2 | 1 | 2 | 0.12 | 0.09 |
| MCL1_MOUSE | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog; OS = Mus musculus, GN = Mcl1, PE = 1, SV = 3 | 32 | 35195 | 5.88 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.13 |
| FPRP_MOUSE | Prostaglandin F2 receptor negative regulator; OS = Mus musculus, GN = Ptgfrn, PE = 1, SV = 2 | 32 | 98660 | 6.16 | 4 | 1 | 2 | 1 | 2 | 0.04 | 0.04 |
| SPT5H_MOUSE | Transcription elongation factor SPT5; OS = Mus musculus, GN = Supt5h, PE = 1, SV = 1 | 32 | 120589 | 4.93 | 4 | 1 | 4 | 1 | 4 | 0.06 | 0.04 |
| GPX8_MOUSE | Probable glutathione peroxidase 8; OS = Mus musculus, GN = Gpx8, PE = 1, SV = 1 | 32 | 24133 | 9.42 | 2 | 1 | 2 | 1 | 2 | 0.11 | 0.19 |
| COR1C_MOUSE | Coronin-1C; OS = Mus musculus, GN = Coro1c, PE = 1, SV = 2 | 32 | 53087 | 6.65 | 6 | 1 | 4 | 1 | 4 | 0.18 | 0.08 |
| T126A_MOUSE | Transmembrane protein 126A; OS = Mus musculus, GN = Tmem126a, PE = 1, SV = 1 | 32 | 21526 | 9.45 | 1 | 1 | 1 | 1 | 1 | 0.09 | 0.21 |
| TBD2B_MOUSE | TBC1 domain family member 2B; OS = Mus musculus, GN = Tbc1d2b, PE = 1, SV = 2 | 32 | 109881 | 5.74 | 3 | 1 | 3 | 1 | 3 | 0.05 | 0.04 |
| NDUA9_MOUSE | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial; OS = Mus musculus, GN = Ndufa9, PE = 1, SV = 2 | 31 | 42498 | 9.75 | 2 | 1 | 2 | 1 | 2 | 0.07 | 0.1 |
| GLMP_MOUSE | Glycosylated lysosomal membrane protein; OS = Mus musculus, GN = Glmp, PE = 1, SV = 1 | 31 | 43776 | 5.73 | 2 | 1 | 2 | 1 | 2 | 0.1 | 0.1 |
| PEF1_MOUSE | Peflin; OS = Mus musculus, GN = Pef1, PE = 1, SV = 1 | 31 | 29209 | 5.89 | 3 | 1 | 2 | 1 | 2 | 0.15 | 0.15 |
| TGM2_MOUSE | Protein-glutamine gamma-glutamyltransferase 2; OS = Mus musculus, GN = Tgm2, PE = 1, SV = 4 | 31 | 77012 | 4.98 | 3 | 1 | 3 | 1 | 3 | 0.05 | 0.06 |
| STXB3_MOUSE | Syntaxin-binding protein 3; OS = Mus musculus, GN = Stxbp3, PE = 1, SV = 1 | 31 | 67899 | 8.28 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.06 |
| APEH_MOUSE | Acylamino-acid-releasing enzyme; OS = Mus musculus, GN = Apeh, PE = 1, SV = 3 | 31 | 81529 | 5.36 | 5 | 2 | 5 | 2 | 5 | 0.14 | 0.11 |
| UFSP2_MOUSE | Ufm1-specific protease 2; OS = Mus musculus, GN = Ufsp2, PE = 1, SV = 1 | 31 | 52482 | 6.28 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.08 |
| CD166_MOUSE | CD166 antigen; OS = Mus musculus, GN = Alcam, PE = 1, SV = 3 | 31 | 65051 | 5.85 | 4 | 1 | 3 | 1 | 3 | 0.1 | 0.07 |
| HIG1A_MOUSE | HIG1 domain family member 1A, mitochondrial; OS = Mus musculus, GN = Higd1a, PE = 1, SV = 1 | 31 | 10418 | 9.79 | 1 | 1 | 1 | 1 | 1 | 0.23 | 0.47 |
| CYTB_MOUSE | Cystatin-B; OS = Mus musculus, GN = Cstb, PE = 1, SV = 1 | 31 | 11039 | 6.82 | 7 | 2 | 3 | 2 | 3 | 0.21 | 1.09 |
| TIMP1_MOUSE | Metalloproteinase inhibitor 1; OS = Mus musculus, GN = Timp1, PE = 1, SV = 2 | 31 | 22613 | 9.14 | 3 | 1 | 3 | 1 | 3 | 0.27 | 0.2 |
| SNP29_MOUSE | Synaptosomal-associated protein 29; OS = Mus musculus, GN = Snap29, PE = 1, SV = 1 | 31 | 29554 | 5.23 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.15 |
| CDV3_MOUSE | Protein CDV3; OS = Mus musculus, GN = Cdv3, PE = 1, SV = 2 | 30 | 29711 | 5.84 | 2 | 1 | 2 | 1 | 2 | 0.22 | 0.15 |
| FUBP1_MOUSE | Far upstream element-binding protein 1; OS = Mus musculus, GN = Fubp1, PE = 1, SV = 1 | 30 | 68497 | 7.74 | 5 | 1 | 4 | 1 | 4 | 0.1 | 0.06 |
| FEN1_MOUSE | Flap endonuclease 1; OS = Mus musculus, GN = Fen1, PE = 1, SV = 1 | 30 | 42288 | 8.54 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.1 |
| AAAT_MOUSE | Neutral amino acid transporter B(0); OS = Mus musculus, GN = Slc1a5, PE = 1, SV = 2 | 30 | 58445 | 8.14 | 5 | 1 | 4 | 1 | 4 | 0.08 | 0.07 |
| EMB_MOUSE | Embigin; OS = Mus musculus, GN = Emb, PE = 1, SV = 2 | 30 | 37041 | 5.7 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.12 |
| S2611_MOUSE | Sodium-independent sulfate anion transporter; OS = Mus musculus, GN = Slc26a11, PE = 2, SV = 2 | 30 | 64068 | 7.56 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.07 |
| BIEA_MOUSE | Biliverdin reductase A; OS = Mus musculus, GN = Blvra, PE = 1, SV = 1 | 30 | 33504 | 6.53 | 4 | 1 | 3 | 1 | 3 | 0.19 | 0.13 |
| TPD54_MOUSE | Tumor protein D54 OS = Mus musculus GN = Tpd52l2 PE = 1 SV = 1 | 30 | 24028 | 5.8 | 2 | 1 | 2 | 1 | 2 | 0.18 | 0.19 |
| SHIP2_MOUSE | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2; OS = Mus musculus, GN = Inppl1, PE = 1, SV = 1 | 30 | 138887 | 6.11 | 4 | 1 | 4 | 1 | 3 | 0.05 | 0.03 |
| LTOR5_MOUSE | Ragulator complex protein LAMTOR5; OS = Mus musculus, GN = Lamtor5, PE = 1, SV = 1 | 30 | 9636 | 4.69 | 2 | 1 | 2 | 1 | 2 | 0.44 | 0.52 |
| ZNT7_MOUSE | Zinc transporter 7; OS = Mus musculus, GN = Slc30a7, PE = 1, SV = 1 | 28 | 41763 | 6.33 | 5 | 2 | 4 | 2 | 4 | 0.19 | 0.22 |
| COX6C_MOUSE | Cytochrome c oxidase subunit 6C; OS = Mus musculus, GN = Cox6c, PE = 1, SV = 3 | 30 | 8464 | 10.13 | 2 | 1 | 2 | 1 | 2 | 0.33 | 0.61 |
| DRG2_MOUSE | Developmentally-regulated GTP-binding protein 2; OS = Mus musculus, GN = Drg2, PE = 1, SV = 1 | 29 | 40692 | 9.03 | 2 | 2 | 2 | 2 | 2 | 0.16 | 0.23 |
| DYST_MOUSE | Dystonin; OS = Mus musculus, GN = Dst, PE = 1, SV = 2 | 29 | 833701 | 5.2 | 22 | 2 | 22 | 2 | 22 | 0.05 | 0.01 |
| PSMG1_MOUSE | Proteasome assembly chaperone 1; OS = Mus musculus, GN = Psmg1, PE = 1, SV = 1 | 29 | 33083 | 6.05 | 4 | 2 | 2 | 2 | 2 | 0.15 | 0.29 |
| FBN1_MOUSE | Fibrillin-1; OS = Mus musculus, GN = Fbn1, PE = 1, SV = 2 | 29 | 312083 | 4.8 | 27 | 1 | 24 | 1 | 24 | 0.19 | 0.01 |
| DAG1_MOUSE | Dystroglycan; OS = Mus musculus, GN = Dag1, PE = 1, SV = 4 | 29 | 96844 | 8.59 | 4 | 1 | 3 | 1 | 3 | 0.04 | 0.04 |
| SNX5_MOUSE | Sorting nexin-5; OS = Mus musculus, GN = Snx5, PE = 1, SV = 1 | 29 | 46768 | 6.19 | 5 | 1 | 5 | 1 | 5 | 0.17 | 0.09 |
| TIM8B_MOUSE | Mitochondrial import inner membrane translocase subunit Tim8 B; OS = Mus musculus, GN = Timm8b, PE = 1, SV = 1 | 29 | 9281 | 5.02 | 1 | 1 | 1 | 1 | 1 | 0.13 | 0.54 |
| PLCB3_MOUSE | 1-Phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3; OS = Mus musculus, GN = Plcb3, PE = 1, SV = 2 | 29 | 139400 | 5.7 | 6 | 1 | 6 | 1 | 6 | 0.09 | 0.03 |
| RTCB_MOUSE | tRNA-splicing ligase RtcB homolog; OS = Mus musculus, GN = Rtcb, PE = 1, SV = 1 | 28 | 55214 | 6.77 | 5 | 2 | 4 | 2 | 4 | 0.19 | 0.16 |
| TM9S4_MOUSE | Transmembrane 9 superfamily member 4; OS = Mus musculus, GN = Tm9sf4, PE = 1, SV = 1 | 28 | 74644 | 6.86 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.06 |
| MBOA7_MOUSE | Lysophospholipid acyltransferase 7; OS = Mus musculus, GN = Mboat7, PE = 1, SV = 1 | 28 | 53400 | 8.96 | 2 | 1 | 1 | 1 | 1 | 0.05 | 0.08 |
| ARPC3_MOUSE | Actin-related protein 2/3 complex subunit 3; OS = Mus musculus, GN = Arpc3, PE = 1, SV = 3 | 28 | 20511 | 8.78 | 3 | 1 | 3 | 1 | 3 | 0.24 | 0.22 |
| RL27A_MOUSE | 60S ribosomal protein L27a; OS = Mus musculus, GN = Rpl27a, PE = 1, SV = 5 | 28 | 16595 | 11.12 | 11 | 2 | 4 | 1 | 4 | 0.29 | 0.28 |
| JAK1_MOUSE | Tyrosine-protein kinase JAK1; OS = Mus musculus, GN = Jak1, PE = 1, SV = 1 | 28 | 133282 | 7.63 | 10 | 2 | 6 | 2 | 6 | 0.08 | 0.06 |
| TBL1R_MOUSE | F-box-like/WD repeat-containing protein TBL1XR1; OS = Mus musculus, GN = Tbl1xr1, PE = 1, SV = 1 | 28 | 55626 | 5.33 | 4 | 1 | 2 | 1 | 2 | 0.1 | 0.08 |
| PIPNA_MOUSE | Phosphatidylinositol transfer protein alpha isoform; OS = Mus musculus, GN = Pitpna, PE = 1, SV = 2 | 28 | 31873 | 5.97 | 2 | 1 | 2 | 1 | 2 | 0.13 | 0.14 |
| GLGB_MOUSE | 1,4-Alpha-glucan-branching enzyme; OS = Mus musculus, GN = Gbe1, PE = 1, SV = 1 | 28 | 80313 | 5.97 | 2 | 2 | 1 | 1 | 1 | 0.02 | 0.05 |
| CY24B_MOUSE | Cytochrome b-245 heavy chain; OS = Mus musculus, GN = Cybb, PE = 1, SV = 1 | 27 | 65262 | 7.83 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.07 |
| SNX7_MOUSE | Sorting nexin-7; OS = Mus musculus, GN = Snx7, PE = 1, SV = 1 | 27 | 44971 | 4.99 | 3 | 2 | 2 | 1 | 2 | 0.14 | 0.1 |
| CSN8_MOUSE | COP9 signalosome complex subunit 8; OS = Mus musculus, GN = Cops8, PE = 1, SV = 1 | 27 | 23241 | 5.09 | 3 | 1 | 3 | 1 | 3 | 0.38 | 0.19 |
| MTNA_MOUSE | Methylthioribose-1-phosphate isomerase; OS = Mus musculus, GN = Mri1, PE = 1, SV = 1 | 27 | 39386 | 5.6 | 3 | 2 | 3 | 2 | 3 | 0.25 | 0.23 |
| SIR2_MOUSE | NAD-dependent protein deacetylase sirtuin-2; OS = Mus musculus, GN = Sirt2, PE = 1, SV = 2 | 27 | 43228 | 5.23 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.1 |
| QCR7_MOUSE | Cytochrome b-c1 complex subunit 7 OS = Mus musculus, GN = Uqcrb, PE = 1, SV = 3 | 27 | 13519 | 9.1 | 2 | 1 | 2 | 1 | 2 | 0.21 | 0.35 |
| ERBIN_MOUSE | Erbin; OS = Mus musculus, GN = Erbin, PE = 1, SV = 3 | 27 | 157150 | 5.46 | 4 | 1 | 4 | 1 | 4 | 0.05 | 0.03 |
| TTL12_MOUSE | Tubulin–tyrosine ligase-like protein 12; OS = Mus musculus, GN = Ttll12, PE = 1, SV = 1 | 27 | 73996 | 5.39 | 3 | 1 | 2 | 1 | 2 | 0.07 | 0.06 |
| FBLN3_MOUSE | EGF-containing fibulin-like extracellular matrix protein 1 | 27 | 54916 | 5.01 | 4 | 1 | 3 | 1 | 3 | 0.1 | 0.08 |
| PYRD_MOUSE | Dihydroorotate dehydrogenase (quinone), mitochondrial | 27 | 42674 | 9.56 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.1 |
| COMD2_MOUSE | COMM domain-containing protein 2 | 26 | 22834 | 6.14 | 3 | 1 | 3 | 1 | 3 | 0.28 | 0.2 |
| CBR1_MOUSE | Carbonyl reductase [NADPH] 1 | 26 | 30622 | 8.53 | 2 | 1 | 2 | 1 | 2 | 0.07 | 0.14 |
| CTL1_MOUSE | Choline transporter-like protein 1 | 26 | 73018 | 9.04 | 4 | 1 | 4 | 1 | 4 | 0.06 | 0.06 |
| CD47_MOUSE | Leukocyte surface antigen CD47 | 26 | 33076 | 8.93 | 2 | 1 | 2 | 1 | 2 | 0.14 | 0.13 |
| DJC24_MOUSE | DnaJ homolog subfamily C member 24 | 26 | 22066 | 5.63 | 2 | 1 | 2 | 1 | 2 | 0.17 | 0.21 |
| ACOD2_MOUSE | Acyl-CoA desaturase 2 | 26 | 40890 | 9.14 | 2 | 1 | 2 | 1 | 2 | 0.13 | 0.11 |
| PROS_MOUSE | Vitamin K-dependent protein S | 26 | 74886 | 5.61 | 8 | 1 | 5 | 1 | 5 | 0.11 | 0.06 |
| TTYH2_MOUSE | Protein tweety homolog 2 | 26 | 58969 | 5.67 | 2 | 1 | 2 | 1 | 2 | 0.05 | 0.07 |
| IF2P_MOUSE | Eukaryotic translation initiation factor 5B | 26 | 137532 | 5.47 | 10 | 2 | 9 | 2 | 9 | 0.14 | 0.06 |
| MCA3_MOUSE | Eukaryotic translation elongation factor 1 epsilon-1 | 26 | 19846 | 8.6 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.23 |
| TM214_MOUSE | Transmembrane protein 214 | 26 | 76381 | 9.41 | 5 | 1 | 5 | 1 | 5 | 0.16 | 0.06 |
| S10AA_MOUSE | Protein S100-A10 | 26 | 11179 | 6.27 | 4 | 1 | 1 | 1 | 1 | 0.27 | 0.44 |
| ARAP1_MOUSE | Arf-GAP with Rho-GAP domain, ANK repeat and pH domain-containing protein 1 | 25 | 162174 | 5.81 | 2 | 1 | 2 | 1 | 2 | 0.03 | 0.03 |
| LMF2_MOUSE | Lipase maturation factor 2 | 25 | 79947 | 9.99 | 1 | 1 | 1 | 1 | 1 | 0.02 | 0.05 |
| OLR1_MOUSE | Oxidized low-density lipoprotein receptor 1 | 25 | 41617 | 7.55 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.11 |
| UCRI_MOUSE | Cytochrome b-c1 complex subunit Rieske, mitochondrial | 25 | 29349 | 8.91 | 2 | 2 | 1 | 1 | 1 | 0.09 | 0.15 |
| MPPA_MOUSE | Mitochondrial-processing peptidase subunit alpha | 25 | 58242 | 6.36 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.07 |
| ARFG1_MOUSE | ADP-ribosylation factor GTPase-activating protein 1 | 25 | 45260 | 5.39 | 2 | 1 | 2 | 1 | 2 | 0.08 | 0.1 |
| UBP24_MOUSE | Ubiquitin carboxyl-terminal hydrolase 24 | 25 | 293814 | 5.82 | 4 | 1 | 4 | 1 | 4 | 0.03 | 0.01 |
| AIMP1_MOUSE | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1 | 25 | 33976 | 8.57 | 3 | 1 | 2 | 1 | 2 | 0.15 | 0.13 |
| STX17_MOUSE | Syntaxin-17 | 25 | 33201 | 6.28 | 2 | 1 | 2 | 1 | 2 | 0.15 | 0.13 |
| TM165_MOUSE | Transmembrane protein 165 | 25 | 34768 | 6.97 | 7 | 1 | 3 | 1 | 3 | 0.25 | 0.13 |
| ACOT2_MOUSE | Acyl-coenzyme A thioesterase 2, mitochondrial | 25 | 49626 | 6.88 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.09 |
| SPD2B_MOUSE | SH3 and PX domain-containing protein 2B | 25 | 101454 | 8.78 | 3 | 1 | 3 | 1 | 3 | 0.07 | 0.04 |
| VP13C_MOUSE | Vacuolar protein sorting-associated protein 13C | 25 | 419824 | 6.37 | 10 | 1 | 8 | 1 | 8 | 0.04 | 0.01 |
| LAP2B_MOUSE | Lamina-associated polypeptide 2, isoforms beta/delta/epsilon/gamma | 25 | 50342 | 9.45 | 7 | 1 | 7 | 1 | 7 | 0.24 | 0.09 |
| T106A_MOUSE | Transmembrane protein 106A | 25 | 29091 | 7.07 | 5 | 1 | 2 | 1 | 2 | 0.16 | 0.15 |
| NRP2_MOUSE | Neuropilin-2 | 24 | 104565 | 5.07 | 2 | 1 | 2 | 1 | 2 | 0.04 | 0.04 |
| VISTA_MOUSE | V-type immunoglobulin domain-containing suppressor of T cell activation | 24 | 33538 | 7.21 | 2 | 1 | 1 | 1 | 1 | 0.04 | 0.13 |
| LYPA1_MOUSE | Acyl-protein thioesterase 1 | 24 | 24671 | 6.14 | 2 | 1 | 1 | 1 | 1 | 0.17 | 0.18 |
| RIOK3_MOUSE | Serine/threonine-protein kinase RIO3 | 24 | 58667 | 5.47 | 4 | 1 | 3 | 1 | 3 | 0.09 | 0.07 |
| RFA1_MOUSE | Replication protein A 70 kDa DNA-binding subunit | 24 | 68994 | 8.13 | 5 | 1 | 4 | 1 | 4 | 0.11 | 0.06 |
| SRRM1_MOUSE | Serine/arginine repetitive matrix protein 1 | 24 | 106798 | 11.87 | 8 | 1 | 6 | 1 | 6 | 0.12 | 0.04 |
| GVIN1_MOUSE | Interferon-induced very large GTPase 1 | 24 | 280637 | 6.12 | 5 | 1 | 5 | 1 | 5 | 0.03 | 0.02 |
| INO1_MOUSE | Inositol-3-phosphate synthase 1 | 24 | 60893 | 5.99 | 3 | 1 | 3 | 1 | 3 | 0.1 | 0.07 |
| ADAS_MOUSE | Alkyldihydroxyacetonephosphate synthase, peroxisomal | 24 | 71638 | 7.25 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.06 |
| CASP3_MOUSE | Caspase-3 | 24 | 31454 | 6.45 | 3 | 1 | 3 | 1 | 3 | 0.18 | 0.14 |
| RIDA_MOUSE | 2-Iminobutanoate/2-iminopropanoate deaminase | 24 | 14247 | 8.74 | 4 | 2 | 2 | 1 | 2 | 0.19 | 0.33 |
| ARFG2_MOUSE | ADP-ribosylation factor GTPase-activating protein 2 | 24 | 56563 | 8.36 | 3 | 1 | 3 | 1 | 3 | 0.14 | 0.08 |
| DHC24_MOUSE | Delta(24)-sterol reductase | 24 | 60073 | 8.42 | 8 | 1 | 7 | 1 | 7 | 0.2 | 0.07 |
| RT16_MOUSE | 28S ribosomal protein S16, mitochondrial | 24 | 15182 | 9.74 | 1 | 1 | 1 | 1 | 1 | 0.15 | 0.31 |
| PTH_MOUSE | Probable peptidyl-tRNA hydrolase | 24 | 22147 | 9.92 | 1 | 1 | 1 | 1 | 1 | 0.1 | 0.2 |
| SWP70_MOUSE | Switch-associated protein 70 | 23 | 68953 | 5.78 | 2 | 1 | 2 | 1 | 2 | 0.07 | 0.06 |
| VWA8_MOUSE | von Willebrand factor A domain-containing protein 8 | 23 | 213287 | 6.14 | 8 | 1 | 7 | 1 | 7 | 0.08 | 0.02 |
| UBXN4_MOUSE | UBX domain-containing protein 4 | 23 | 56437 | 6.23 | 5 | 1 | 4 | 1 | 4 | 0.23 | 0.08 |
| AEBP1_MOUSE | Adipocyte enhancer-binding protein 1 | 23 | 128284 | 5.02 | 6 | 1 | 5 | 1 | 5 | 0.09 | 0.03 |
| GCR_MOUSE | Glucocorticoid receptor | 23 | 85998 | 5.99 | 8 | 1 | 7 | 1 | 7 | 0.1 | 0.05 |
| IAH1_MOUSE | Isoamyl acetate-hydrolyzing esterase 1 homolog | 23 | 27956 | 5.34 | 3 | 1 | 2 | 1 | 2 | 0.12 | 0.16 |
| VPS16_MOUSE | Vacuolar protein sorting-associated protein 16 homolog | 22 | 94868 | 6.56 | 6 | 2 | 5 | 1 | 5 | 0.1 | 0.05 |
| LY6C1_MOUSE | Lymphocyte antigen 6C1 | 22 | 14183 | 5.76 | 2 | 1 | 2 | 1 | 2 | 0.29 | 0.33 |
| BCAT1_MOUSE | Branched-chain-amino-acid aminotransferase, cytosolic | 22 | 42764 | 5.25 | 3 | 1 | 3 | 1 | 3 | 0.12 | 0.1 |
| E2AK2_MOUSE | Interferon-induced, double-stranded RNA-activated protein kinase | 22 | 58243 | 8.76 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.07 |
| FLII_MOUSE | Protein flightless-1 homolog | 21 | 144712 | 5.75 | 9 | 1 | 9 | 1 | 9 | 0.1 | 0.03 |
| CHCH2_MOUSE | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2 | 21 | 15651 | 9.78 | 2 | 1 | 2 | 1 | 2 | 0.24 | 0.3 |
| TNPO2_MOUSE | Transportin-2 | 21 | 100391 | 4.85 | 7 | 1 | 6 | 1 | 4 | 0.14 | 0.04 |
| UBFD1_MOUSE | Ubiquitin domain-containing protein UBFD1 | 21 | 40118 | 8.98 | 7 | 1 | 4 | 1 | 4 | 0.2 | 0.11 |
| WWOX_MOUSE | WW domain-containing oxidoreductase | 21 | 46483 | 6.54 | 2 | 1 | 2 | 1 | 2 | 0.11 | 0.09 |
| DYR_MOUSE | Dihydrofolate reductase | 21 | 21592 | 8.56 | 3 | 1 | 2 | 1 | 2 | 0.23 | 0.21 |
| ARHG1_MOUSE | Rho guanine nucleotide exchange factor 1 | 21 | 102741 | 5.43 | 3 | 1 | 3 | 1 | 3 | 0.06 | 0.04 |
| ATX2L_MOUSE | Ataxin-2-like protein | 20 | 110580 | 8.94 | 2 | 1 | 2 | 1 | 2 | 0.05 | 0.04 |
| PSD10_MOUSE | 26S proteasome non-ATPase regulatory subunit 10 | 20 | 25068 | 5.68 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.18 |
| TMF1_MOUSE | TATA element modulatory factor | 20 | 121729 | 4.83 | 5 | 2 | 3 | 1 | 3 | 0.06 | 0.03 |
| ANFY1_MOUSE | Rabankyrin-5 | 20 | 128571 | 5.58 | 5 | 2 | 4 | 1 | 4 | 0.07 | 0.03 |
| MAGD1_MOUSE | Melanoma-associated antigen D1 | 20 | 85617 | 7.01 | 5 | 1 | 4 | 1 | 4 | 0.1 | 0.05 |
| YIF1A_MOUSE | Protein YIF1A | 19 | 32114 | 9.11 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.14 |
| TR10B_MOUSE | Tumor necrosis factor receptor superfamily member 10B | 19 | 42138 | 6.75 | 4 | 1 | 3 | 1 | 3 | 0.13 | 0.1 |
| QCR8_MOUSE | Cytochrome b-c1 complex subunit 8 | 19 | 9762 | 10.26 | 1 | 1 | 1 | 1 | 1 | 0.28 | 0.51 |
| ATP5J_MOUSE | ATP synthase-coupling factor 6, mitochondrial | 18 | 12489 | 9.36 | 6 | 1 | 4 | 1 | 4 | 0.4 | 0.38 |
| YAP1_MOUSE | Transcriptional coactivator YAP1 | 18 | 52351 | 4.96 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.08 |
| PARP9_MOUSE | Poly [ADP-ribose] polymerase 9 | 17 | 96597 | 6.68 | 3 | 1 | 3 | 1 | 3 | 0.09 | 0.04 |
| MTCH2_MOUSE | Mitochondrial carrier homolog 2 | 17 | 33477 | 8.59 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.13 |
| NXN_MOUSE | Nucleoredoxin | 17 | 48314 | 4.84 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.09 |
- aProtein score is calculated from the score of the peptide attributed to the protein. bpI is (predicted) isoelectric point. cNumber of matches is spectrum number matched to protein #1. dNumber of significant matches is spectrum number that matches protein and exceeds the identification criteria. eNumber of sequences is number of peptides matched to protein #2. fNumber of significant sequences is number of peptides exceeding the identification criteria matched to proteins. gNumber of unique sequences is a unique #3 number of peptides matched to proteins. hSequence coverage is the ratio of the total number of matched peptide residues to the total length of the protein. iExponentially modified protein abundance index (http://www.matrixscience.com/help/quant_empai_help.html). #1When multiple spectra are matched to the same peptide, the way of counting into one is called "peptide number". #2When multiple spectra are matched to the same peptide, the method of counting is called "spectrum number". #3"Unique" is a peptide that is not matched to other proteins and has been assigned only to the relevant protein.
| UniProt/Swiss-Prot ID | Description | Protein scorea | Protein mass (kDa) | pIb | Num. of matchesc | Num. of significant matchesd | Num. of sequencese | Num. of significant sequencesf | Num. of unique sequencesg | Sequence coverageh | emPAIi |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MYH9_MOUSE | Myosin-9 | 3652 | 226232 | 5.54 | 238 | 130 | 81 | 52 | 74 | 0.52 | 1.93 |
| MYH10_MOUSE | Myosin-10 | 793 | 228855 | 5.43 | 66 | 31 | 25 | 13 | 18 | 0.19 | 0.32 |
| SERPH_MOUSE | Serpin H1 | 3042 | 46504 | 8.88 | 217 | 141 | 22 | 21 | 22 | 0.59 | 7.57 |
| G3P_MOUSE | Glyceraldehyde-3-phosphate dehydrogenase | 3019 | 35787 | 8.44 | 240 | 131 | 15 | 12 | 9 | 0.67 | 8.1 |
| S10AB_MOUSE | Protein S100-A11 | 2948 | 11075 | 5.28 | 127 | 109 | 5 | 4 | 3 | 0.66 | 37.69 |
| CLH1_MOUSE | Clathrin heavy chain 1 | 2893 | 191435 | 5.48 | 163 | 110 | 62 | 45 | 62 | 0.57 | 1.92 |
| ACTN1_MOUSE | Alpha-actinin-1 | 2445 | 103004 | 5.23 | 136 | 82 | 42 | 31 | 27 | 0.65 | 2.98 |
| ACTN4_MOUSE | Alpha-actinin-4 | 1377 | 104911 | 5.25 | 111 | 60 | 36 | 24 | 21 | 0.56 | 1.82 |
| KPYM_MOUSE | Pyruvate kinase PKM | 2269 | 57808 | 7.18 | 136 | 93 | 33 | 26 | 14 | 0.69 | 7.7 |
| HS90A_MOUSE | Heat shock protein HSP 90-alpha | 1754 | 84735 | 4.93 | 181 | 93 | 28 | 22 | 21 | 0.52 | 2.79 |
| ENPL_MOUSE | Endoplasmin | 1610 | 92418 | 4.74 | 101 | 64 | 34 | 24 | 16 | 0.48 | 2.39 |
| TRAP1_MOUSE | Heat shock protein 75 kDa, mitochondrial | 123 | 80159 | 6.25 | 6 | 6 | 4 | 4 | 4 | 0.11 | 0.23 |
| ENOA_MOUSE | Alpha-enolase | 2147 | 47111 | 6.37 | 163 | 78 | 20 | 14 | 7 | 0.69 | 4.37 |
| FLNA_MOUSE | Filamin-A | 2029 | 281046 | 5.68 | 241 | 136 | 64 | 42 | 61 | 0.43 | 0.93 |
| FLNB_MOUSE | Filamin-B | 1355 | 277651 | 5.46 | 119 | 68 | 56 | 40 | 52 | 0.43 | 0.86 |
| FLNC_MOUSE | Filamin-C | 351 | 290937 | 5.63 | 66 | 26 | 36 | 16 | 31 | 0.24 | 0.3 |
| GRP75_MOUSE | Stress-70 protein, mitochondrial | 961 | 73416 | 5.81 | 123 | 47 | 16 | 11 | 16 | 0.36 | 0.98 |
| UBA1_MOUSE | Ubiquitin-like modifier-activating enzyme 1 | 1729 | 117734 | 5.43 | 87 | 64 | 29 | 22 | 29 | 0.58 | 1.52 |
| TLN1_MOUSE | Talin-1 | 1723 | 269653 | 5.84 | 122 | 71 | 43 | 30 | 32 | 0.35 | 0.64 |
| FAS_MOUSE | Fatty acid synthase | 1679 | 272257 | 6.13 | 109 | 68 | 54 | 36 | 54 | 0.42 | 0.77 |
| EF2_MOUSE | Elongation factor 2 | 1674 | 95253 | 6.41 | 114 | 76 | 31 | 25 | 31 | 0.61 | 2.27 |
| VIME_MOUSE | Vimentin | 1667 | 53655 | 5.06 | 152 | 77 | 37 | 26 | 37 | 0.67 | 8.53 |
| ASSY_MOUSE | Argininosuccinate synthase | 1574 | 46555 | 8.36 | 96 | 68 | 22 | 15 | 18 | 0.66 | 6.84 |
| ANXA2_MOUSE | Annexin A2 | 1565 | 38652 | 7.55 | 126 | 98 | 14 | 11 | 14 | 0.54 | 2.27 |
| VDAC1_MOUSE | Voltage-dependent anion-selective channel protein 1 | 1559 | 32331 | 8.55 | 161 | 108 | 13 | 8 | 13 | 0.65 | 2.18 |
| VDAC2_MOUSE | Voltage-dependent anion-selective channel protein 2 | 312 | 31713 | 7.44 | 76 | 23 | 8 | 6 | 8 | 0.42 | 1.19 |
| DYHC1_MOUSE | Cytoplasmic dynein 1 heavy chain 1 | 1401 | 531710 | 6.03 | 112 | 64 | 65 | 36 | 65 | 0.26 | 0.34 |
| CH60_MOUSE | 60 kDa heat shock protein, mitochondrial | 1376 | 60917 | 5.91 | 76 | 42 | 22 | 18 | 22 | 0.54 | 2.67 |
| PDIA1_MOUSE | Protein disulfide-isomerase | 1325 | 57023 | 4.77 | 92 | 45 | 19 | 15 | 19 | 0.41 | 1.99 |
| PLEC_MOUSE | Plectin | 1320 | 533861 | 5.74 | 113 | 55 | 74 | 36 | 74 | 0.26 | 0.33 |
| TKT_MOUSE | Transketolase | 1319 | 67588 | 7.23 | 83 | 61 | 22 | 20 | 22 | 0.62 | 2.66 |
| TBB6_MOUSE | Tubulin beta-6 chain | 846 | 50058 | 4.8 | 83 | 31 | 19 | 13 | 7 | 0.61 | 2.48 |
| ANXA5_MOUSE | Annexin A5 | 1219 | 35730 | 4.83 | 83 | 50 | 17 | 14 | 15 | 0.67 | 6.21 |
| CATB_MOUSE | Cathepsin B | 1206 | 37256 | 5.57 | 37 | 31 | 7 | 5 | 7 | 0.45 | 1.44 |
| GELS_MOUSE | Gelsolin | 1190 | 85888 | 5.83 | 68 | 48 | 19 | 15 | 13 | 0.56 | 1.29 |
| GELS_HORSE | Gelsolin | 546 | 80777 | 5.58 | 33 | 24 | 9 | 6 | 3 | 0.28 | 0.51 |
| IMB1_MOUSE | Importin subunit beta-1 | 1155 | 97122 | 4.68 | 71 | 53 | 29 | 22 | 29 | 0.61 | 2.2 |
| FPPS_MOUSE | Farnesyl pyrophosphate synthase | 1095 | 40556 | 5.49 | 44 | 31 | 14 | 11 | 14 | 0.68 | 2.43 |
| PSMD2_MOUSE | 26S proteasome non-ATPase regulatory subunit 2 | 1057 | 100139 | 5.06 | 51 | 39 | 19 | 15 | 19 | 0.44 | 0.95 |
| TCPA_MOUSE | T-complex protein 1 subunit alpha | 1055 | 60411 | 5.82 | 44 | 31 | 16 | 11 | 16 | 0.54 | 1.29 |
| ESTD_MOUSE | S-Formylglutathione hydrolase | 1044 | 31299 | 6.7 | 53 | 36 | 15 | 11 | 15 | 0.79 | 4.6 |
| ATPA_MOUSE | ATP synthase subunit alpha, mitochondrial | 1041 | 59716 | 9.22 | 59 | 42 | 16 | 14 | 4 | 0.49 | 2.06 |
| IQGA1_MOUSE | Ras GTPase-activating-like protein IQGAP1 | 1034 | 188624 | 6.07 | 67 | 37 | 35 | 19 | 35 | 0.36 | 0.56 |
| WDR1_MOUSE | WD repeat-containing protein 1 | 1014 | 66365 | 6.11 | 64 | 42 | 19 | 15 | 19 | 0.65 | 1.57 |
| TCPE_MOUSE | T-complex protein 1 subunit epsilon | 1002 | 59586 | 5.72 | 51 | 33 | 15 | 9 | 15 | 0.55 | 1.16 |
| SPB6_MOUSE | Serpin B6 | 988 | 42571 | 5.53 | 46 | 35 | 13 | 11 | 13 | 0.56 | 2.92 |
| ACLY_MOUSE | ATP-citrate synthase | 961 | 119651 | 7.13 | 64 | 44 | 26 | 20 | 26 | 0.42 | 1.08 |
| LRP1_MOUSE | Prolow-density lipoprotein receptor-related protein 1 | 939 | 504411 | 5.14 | 76 | 46 | 37 | 25 | 37 | 0.16 | 0.27 |
| CAN2_MOUSE | Calpain-2 catalytic subunit | 937 | 79822 | 4.86 | 55 | 37 | 18 | 15 | 17 | 0.53 | 1.31 |
| ALDOA_MOUSE | Fructose-bisphosphate aldolase A | 923 | 39331 | 8.31 | 75 | 49 | 15 | 13 | 15 | 0.62 | 3.89 |
| SPTB2_MOUSE | Spectrin beta chain, nonerythrocytic 1 | 900 | 274052 | 5.4 | 74 | 42 | 33 | 21 | 33 | 0.24 | 0.38 |
| LDHA_MOUSE | L-Lactate dehydrogenase A chain | 888 | 36475 | 7.62 | 64 | 43 | 17 | 12 | 13 | 0.68 | 3.92 |
| LDHB_MOUSE | L-Lactate dehydrogenase B chain | 144 | 36549 | 5.7 | 9 | 6 | 3 | 3 | 3 | 0.19 | 0.41 |
| 1433B_MOUSE | 14-3-3 protein beta/alpha | 483 | 28069 | 4.77 | 30 | 19 | 8 | 5 | 4 | 0.5 | 1.81 |
| TCPZ_MOUSE | T-complex protein 1 subunit zeta | 854 | 57968 | 6.63 | 51 | 37 | 13 | 10 | 13 | 0.41 | 1.05 |
| COF1_MOUSE | Cofilin-1 | 795 | 18548 | 8.22 | 57 | 34 | 12 | 9 | 11 | 0.68 | 6.37 |
| COF2_MOUSE | Cofilin-2 | 363 | 18698 | 7.66 | 17 | 10 | 5 | 5 | 4 | 0.46 | 2.01 |
| SYAC_MOUSE | Alanine–tRNA ligase, cytoplasmic | 785 | 106841 | 5.45 | 46 | 33 | 19 | 13 | 19 | 0.35 | 0.66 |
| PLST_MOUSE | Plastin-3 | 784 | 70697 | 5.42 | 56 | 35 | 21 | 15 | 21 | 0.54 | 1.42 |
| ANXA1_MOUSE | Annexin A1 | 774 | 38710 | 6.97 | 49 | 30 | 11 | 9 | 11 | 0.46 | 1.92 |
| THIO_MOUSE | Thioredoxin | 767 | 11668 | 4.8 | 33 | 19 | 7 | 3 | 7 | 0.78 | 1.85 |
| PRDX6_MOUSE | Peroxiredoxin-6 | 751 | 24855 | 5.71 | 40 | 28 | 11 | 8 | 11 | 0.61 | 2.78 |
| SPTN1_MOUSE | Spectrin alpha chain, nonerythrocytic 1 | 749 | 284422 | 5.2 | 77 | 34 | 42 | 24 | 42 | 0.27 | 0.42 |
| LEG1_MOUSE | Galectin-1 | 735 | 14856 | 5.32 | 56 | 36 | 11 | 9 | 11 | 0.82 | 26.15 |
| TCPH_MOUSE | T-complex protein 1 subunit eta | 734 | 59614 | 7.95 | 38 | 26 | 16 | 12 | 16 | 0.5 | 1.48 |
| HNRPF_MOUSE | Heterogeneous nuclear ribonucleoprotein F | 710 | 45701 | 5.31 | 33 | 25 | 9 | 7 | 3 | 0.46 | 1.27 |
| EF1G_MOUSE | Elongation factor 1-gamma | 689 | 50029 | 6.31 | 44 | 28 | 15 | 13 | 15 | 0.57 | 1.95 |
| PDIA3_MOUSE | Protein disulfide-isomerase A3 | 682 | 56643 | 5.88 | 64 | 37 | 23 | 16 | 14 | 0.54 | 2.5 |
| TENA_MOUSE | Tenascin | 679 | 231659 | 4.77 | 33 | 23 | 17 | 11 | 17 | 0.16 | 0.22 |
| ATPB_MOUSE | ATP synthase subunit beta, mitochondrial | 676 | 56265 | 5.19 | 69 | 33 | 23 | 15 | 7 | 0.74 | 2.27 |
| RCN2_MOUSE | Reticulocalbin-2 | 663 | 37248 | 4.28 | 23 | 20 | 9 | 8 | 9 | 0.46 | 1.44 |
| TCPB_MOUSE | T-complex protein 1 subunit beta | 654 | 57441 | 5.97 | 32 | 19 | 17 | 12 | 17 | 0.49 | 1.39 |
| LYAG_MOUSE | Lysosomal alpha-glucosidase | 630 | 106180 | 5.53 | 45 | 34 | 16 | 13 | 16 | 0.34 | 0.81 |
| CKAP4_MOUSE | Cytoskeleton-associated protein 4 | 626 | 63654 | 5.46 | 41 | 23 | 14 | 7 | 14 | 0.38 | 0.58 |
| DPYL2_MOUSE | Dihydropyrimidinase-related protein 2 | 626 | 62239 | 5.95 | 47 | 26 | 24 | 13 | 21 | 0.63 | 1.39 |
| DPYL3_MOUSE | Dihydropyrimidinase-related protein 3 | 602 | 61897 | 6.04 | 26 | 19 | 15 | 8 | 12 | 0.57 | 0.96 |
| ESYT1_MOUSE | Extended synaptotagmin-1 | 621 | 121478 | 5.63 | 25 | 21 | 9 | 7 | 9 | 0.19 | 0.27 |
| NEDD4_MOUSE | E3 ubiquitin-protein ligase NEDD4 | 611 | 102642 | 5.12 | 40 | 26 | 17 | 13 | 17 | 0.31 | 0.77 |
| SYVC_MOUSE | Valine–tRNA ligase | 604 | 140127 | 7.9 | 37 | 26 | 14 | 9 | 14 | 0.24 | 0.35 |
| GSTO1_MOUSE | Glutathione S-transferase omega-1 | 600 | 27480 | 6.92 | 50 | 30 | 13 | 7 | 13 | 0.65 | 2.87 |
| SEPT2_MOUSE | Septin-2 | 599 | 41499 | 6.1 | 24 | 19 | 9 | 8 | 8 | 0.38 | 1.46 |
| 6PGD_MOUSE | 6-Phosphogluconate dehydrogenase, decarboxylating | 599 | 53213 | 6.81 | 50 | 32 | 18 | 12 | 18 | 0.63 | 2 |
| RPN1_MOUSE | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 | 597 | 68486 | 6.02 | 35 | 22 | 15 | 11 | 15 | 0.4 | 0.96 |
| NUCL_MOUSE | Nucleolin | 588 | 76677 | 4.69 | 37 | 22 | 15 | 9 | 15 | 0.27 | 0.63 |
| GDIB_MOUSE | Rab GDP dissociation inhibitor beta | 583 | 50505 | 5.93 | 40 | 26 | 13 | 10 | 11 | 0.45 | 1.48 |
| TPM4_MOUSE | Tropomyosin alpha-4 chain | 581 | 28450 | 4.65 | 51 | 29 | 10 | 9 | 10 | 0.44 | 3.3 |
| PPCE_MOUSE | Prolyl endopeptidase | 572 | 80700 | 5.44 | 30 | 21 | 12 | 10 | 12 | 0.29 | 0.68 |
| NCPR_MOUSE | NADPH–cytochrome P450 reductase | 570 | 76995 | 5.34 | 31 | 19 | 13 | 8 | 13 | 0.34 | 0.54 |
| CISY_MOUSE | Citrate synthase, mitochondrial | 563 | 51703 | 8.72 | 28 | 24 | 12 | 11 | 12 | 0.44 | 2.09 |
| DDX3X_MOUSE | ATP-dependent RNA helicase DDX3X | 555 | 73056 | 6.73 | 28 | 21 | 12 | 8 | 12 | 0.32 | 0.58 |
| P5CS_MOUSE | Delta-1-pyrroline-5-carboxylate synthase | 553 | 87212 | 7.18 | 26 | 18 | 10 | 5 | 10 | 0.2 | 0.27 |
| MBB1A_MOUSE | Myb-binding protein 1A | 547 | 151942 | 9.08 | 35 | 21 | 17 | 10 | 17 | 0.23 | 0.32 |
| EF1B_MOUSE | Elongation factor 1-beta | 546 | 24678 | 4.53 | 29 | 24 | 9 | 9 | 9 | 0.57 | 3.53 |
| UGGG1_MOUSE | UDP-glucose:glycoprotein glucosyltransferase 1 | 545 | 176323 | 5.4 | 49 | 29 | 23 | 14 | 23 | 0.29 | 0.46 |
| RPN2_MOUSE | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2 | 542 | 69020 | 5.54 | 27 | 19 | 12 | 11 | 12 | 0.37 | 0.94 |
| HS105_MOUSE | Heat shock protein 105 kDa | 541 | 96346 | 5.39 | 27 | 17 | 16 | 11 | 15 | 0.29 | 0.61 |
| HSP74_MOUSE | Heat shock 70 kDa protein 4 | 440 | 94073 | 5.15 | 26 | 20 | 17 | 16 | 2 | 0.3 | 1.04 |
| PROF1_MOUSE | Profilin-1 | 536 | 14948 | 8.46 | 79 | 30 | 8 | 7 | 8 | 0.61 | 19.18 |
| GANAB_MOUSE | Neutral alpha-glucosidase AB | 523 | 106844 | 5.67 | 49 | 27 | 20 | 15 | 20 | 0.46 | 0.8 |
| THIC_MOUSE | Acetyl-CoA acetyltransferase, cytosolic | 523 | 41271 | 7.16 | 30 | 20 | 8 | 6 | 8 | 0.5 | 0.83 |
| ANXA6_MOUSE | Annexin A6 | 518 | 75837 | 5.34 | 39 | 20 | 19 | 10 | 19 | 0.4 | 0.73 |
| P4HA1_MOUSE | Prolyl 4-hydroxylase subunit alpha-1 | 510 | 60872 | 5.62 | 32 | 26 | 13 | 10 | 13 | 0.44 | 1.13 |
| NDKB_MOUSE | Nucleoside diphosphate kinase B | 508 | 17352 | 6.97 | 31 | 19 | 7 | 4 | 4 | 0.63 | 2.27 |
| KAD1_MOUSE | Adenylate kinase isoenzyme 1 | 499 | 21526 | 5.67 | 26 | 19 | 8 | 5 | 8 | 0.57 | 1.61 |
| SAP_MOUSE | Prosaposin | 497 | 61381 | 5.07 | 48 | 30 | 11 | 5 | 11 | 0.31 | 0.72 |
| PGH1_MOUSE | Prostaglandin G/H synthase 1 | 491 | 68998 | 6.36 | 29 | 21 | 10 | 8 | 10 | 0.21 | 0.72 |
| HMCS1_MOUSE | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 487 | 57532 | 5.65 | 31 | 22 | 15 | 13 | 15 | 0.49 | 1.76 |
| CAP1_MOUSE | Adenylyl cyclase-associated protein 1 | 478 | 51532 | 7.16 | 39 | 22 | 11 | 8 | 11 | 0.36 | 0.91 |
| MK01_MOUSE | Mitogen-activated protein kinase 1 | 474 | 41249 | 6.5 | 21 | 16 | 7 | 5 | 6 | 0.44 | 0.65 |
| ARF4_MOUSE | ADP-ribosylation factor 4 | 470 | 20384 | 6.59 | 35 | 20 | 12 | 8 | 6 | 0.8 | 6.52 |
| 2AAA_MOUSE | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | 470 | 65281 | 5 | 22 | 15 | 14 | 11 | 14 | 0.42 | 1.02 |
| CPNS1_MOUSE | Calpain small subunit 1 | 469 | 28445 | 5.41 | 24 | 18 | 6 | 5 | 6 | 0.39 | 1.4 |
| FSCN1_MOUSE | Fascin | 463 | 54474 | 6.44 | 37 | 22 | 14 | 10 | 14 | 0.51 | 1.32 |
| PDIA4_MOUSE | Protein disulfide-isomerase A4 | 461 | 71938 | 5.16 | 32 | 20 | 15 | 10 | 15 | 0.39 | 0.79 |
| TSP1_MOUSE | Thrombospondin-1 | 460 | 129564 | 4.72 | 33 | 15 | 16 | 8 | 15 | 0.19 | 0.29 |
| G6PI_MOUSE | Glucose-6-phosphate isomerase | 459 | 62727 | 8.14 | 35 | 17 | 15 | 9 | 15 | 0.47 | 1.08 |
| NP1L1_MOUSE | Nucleosome assembly protein 1-like 1 | 458 | 45317 | 4.36 | 30 | 19 | 11 | 9 | 10 | 0.47 | 1.51 |
| ADT1_MOUSE | ADP/ATP translocase 1 | 413 | 32883 | 9.73 | 25 | 19 | 11 | 9 | 5 | 0.49 | 3.01 |
| IPO5_MOUSE | Importin-5 | 449 | 123511 | 4.82 | 38 | 24 | 22 | 13 | 22 | 0.37 | 0.61 |
| RINI_MOUSE | Ribonuclease inhibitor | 440 | 49784 | 4.69 | 21 | 16 | 12 | 9 | 12 | 0.55 | 1.12 |
| AT1A1_MOUSE | Sodium/potassium-transporting ATPase subunit alpha-1 | 436 | 112910 | 5.3 | 19 | 15 | 12 | 10 | 12 | 0.2 | 0.45 |
| GFPT1_MOUSE | Glutamine–fructose-6-phosphate aminotransferase [isomerizing] 1 | 433 | 78489 | 6.39 | 19 | 14 | 11 | 7 | 11 | 0.27 | 0.45 |
| PSA_MOUSE | Puromycin-sensitive aminopeptidase | 432 | 103260 | 5.61 | 19 | 12 | 7 | 3 | 7 | 0.17 | 0.13 |
| PDIA6_MOUSE | Protein disulfide-isomerase A6 | 424 | 48070 | 5 | 36 | 26 | 14 | 11 | 14 | 0.48 | 1.59 |
| DEST_MOUSE | Destrin | 421 | 18509 | 8.14 | 26 | 14 | 9 | 7 | 9 | 0.54 | 4.9 |
| MYOF_MOUSE | Myoferlin | 421 | 233177 | 5.83 | 29 | 19 | 19 | 13 | 19 | 0.15 | 0.31 |
| ALDH2_MOUSE | Aldehyde dehydrogenase, mitochondrial | 419 | 56502 | 7.53 | 28 | 16 | 10 | 7 | 10 | 0.4 | 0.8 |
| HNRPU_MOUSE | Heterogeneous nuclear ribonucleoprotein U | 417 | 87863 | 5.92 | 32 | 20 | 13 | 9 | 13 | 0.27 | 0.53 |
| ANXA4_MOUSE | Annexin A4 | 415 | 35893 | 5.43 | 14 | 10 | 8 | 6 | 8 | 0.36 | 1 |
| PDC6I_MOUSE | Programmed cell death 6-interacting protein | 414 | 95964 | 6.15 | 25 | 17 | 14 | 8 | 14 | 0.28 | 0.48 |
| MARCS_MOUSE | Myristoylated alanine-rich C-kinase substrate | 413 | 29644 | 4.34 | 65 | 43 | 2 | 1 | 2 | 0.15 | 0.15 |
| CALU_MOUSE | Calumenin | 308 | 37041 | 4.49 | 29 | 16 | 9 | 8 | 3 | 0.44 | 1.74 |
| VIGLN_MOUSE | Vigilin | 410 | 141655 | 6.43 | 32 | 18 | 19 | 10 | 19 | 0.24 | 0.34 |
| FBLN2_MOUSE | Fibulin-2 | 409 | 131746 | 4.58 | 20 | 15 | 12 | 9 | 12 | 0.18 | 0.33 |
| OAT_MOUSE | Ornithine aminotransferase, mitochondrial | 408 | 48324 | 6.19 | 18 | 12 | 9 | 6 | 9 | 0.37 | 0.68 |
| TPP1_MOUSE | Tripeptidyl-peptidase 1 | 404 | 61304 | 6.1 | 9 | 8 | 2 | 2 | 2 | 0.1 | 0.15 |
| RL9_MOUSE | 60S ribosomal protein L9 | 398 | 21868 | 9.96 | 19 | 15 | 7 | 5 | 7 | 0.49 | 2.1 |
| FLII_MOUSE | Protein flightless-1 homolog | 397 | 144712 | 5.75 | 27 | 15 | 15 | 8 | 15 | 0.28 | 0.3 |
| UGDH_MOUSE | UDP-glucose 6-dehydrogenase | 397 | 54797 | 7.49 | 35 | 21 | 19 | 13 | 19 | 0.6 | 1.69 |
| ECHA_MOUSE | Trifunctional enzyme subunit alpha, mitochondrial | 396 | 82617 | 9.24 | 19 | 13 | 9 | 6 | 9 | 0.25 | 0.35 |
| PUR2_MOUSE | Trifunctional purine biosynthetic protein adenosine-3 | 395 | 107436 | 6.25 | 24 | 14 | 10 | 5 | 10 | 0.22 | 0.31 |
| PPIA_MOUSE | Peptidyl-prolyl cis-trans isomerase A | 393 | 17960 | 7.74 | 31 | 21 | 9 | 8 | 9 | 0.65 | 8.83 |
| 6PGL_MOUSE | 6-Phosphogluconolactonase | 389 | 27237 | 5.55 | 11 | 8 | 5 | 4 | 5 | 0.39 | 0.84 |
| CAND1_MOUSE | Cullin-associated NEDD8-dissociated protein 1 | 385 | 136245 | 5.52 | 46 | 27 | 16 | 10 | 16 | 0.3 | 0.4 |
| GARS_MOUSE | Glycine–tRNA ligase | 385 | 81826 | 6.24 | 29 | 17 | 15 | 10 | 15 | 0.42 | 0.67 |
| SND1_MOUSE | Staphylococcal nuclease domain-containing protein 1 | 381 | 102025 | 7.08 | 39 | 20 | 20 | 13 | 20 | 0.4 | 0.78 |
| ESYT2_MOUSE | Extended synaptotagmin-2 | 380 | 94081 | 7.63 | 22 | 14 | 9 | 6 | 9 | 0.17 | 0.31 |
| IDHC_MOUSE | Isocitrate dehydrogenase [NADP] cytoplasmic | 374 | 46644 | 6.73 | 30 | 16 | 17 | 9 | 16 | 0.54 | 1.23 |
| COPB_MOUSE | Coatomer subunit beta | 371 | 106998 | 5.69 | 35 | 19 | 16 | 12 | 16 | 0.31 | 0.6 |
| CNPY2_MOUSE | Protein canopy homolog 2 | 371 | 20754 | 4.95 | 5 | 5 | 2 | 2 | 2 | 0.19 | 0.49 |
| MYADM_MOUSE | Myeloid-associated differentiation marker | 371 | 35261 | 8.69 | 32 | 7 | 3 | 2 | 3 | 0.14 | 0.27 |
| SYIC_MOUSE | Isoleucine–tRNA ligase, cytoplasmic | 368 | 144179 | 6.14 | 33 | 17 | 13 | 8 | 13 | 0.19 | 0.26 |
| DHE3_MOUSE | Glutamate dehydrogenase 1, mitochondrial | 366 | 61298 | 8.05 | 26 | 14 | 12 | 8 | 12 | 0.28 | 0.85 |
| CLIC1_MOUSE | Chloride intracellular channel protein 1 | 363 | 26996 | 5.09 | 29 | 19 | 10 | 8 | 10 | 0.58 | 2.98 |
| RCN3_MOUSE | Reticulocalbin-3 | 362 | 37978 | 4.74 | 13 | 7 | 4 | 3 | 4 | 0.31 | 0.39 |
| FINC_MOUSE | Fibronectin | 361 | 272368 | 5.39 | 31 | 15 | 14 | 8 | 14 | 0.11 | 0.13 |
| ADK_MOUSE | Adenosine kinase | 360 | 40123 | 5.84 | 18 | 15 | 7 | 6 | 5 | 0.4 | 0.86 |
| DHB4_MOUSE | Peroxisomal multifunctional enzyme type 2 | 359 | 79432 | 8.76 | 12 | 8 | 5 | 2 | 4 | 0.11 | 0.11 |
| CALX_MOUSE | Calnexin | 357 | 67236 | 4.5 | 32 | 19 | 14 | 10 | 14 | 0.38 | 0.86 |
| RLA0_MOUSE | 60S acidic ribosomal protein P0 | 355 | 34195 | 5.91 | 26 | 19 | 11 | 7 | 11 | 0.51 | 1.64 |
| PYRG2_MOUSE | CTP synthase 2 | 190 | 65473 | 6.05 | 12 | 7 | 7 | 3 | 6 | 0.27 | 0.21 |
| CATZ_MOUSE | Cathepsin Z | 353 | 33974 | 6.13 | 16 | 11 | 3 | 1 | 3 | 0.23 | 0.28 |
| GNAI2_MOUSE | Guanine nucleotide-binding protein G(i) subunit alpha-2 | 352 | 40463 | 5.28 | 17 | 11 | 9 | 6 | 9 | 0.36 | 0.85 |
| CH10_MOUSE | 10 kDa heat shock protein, mitochondrial | 352 | 10956 | 7.93 | 30 | 18 | 5 | 2 | 5 | 0.39 | 1.09 |
| VATB2_MOUSE | V-type proton ATPase subunit B, brain isoform | 350 | 56515 | 5.57 | 20 | 12 | 10 | 7 | 10 | 0.41 | 0.68 |
| MDHM_MOUSE | Malate dehydrogenase, mitochondrial | 346 | 35589 | 8.93 | 37 | 17 | 10 | 8 | 10 | 0.43 | 1.54 |
| MYL9_MOUSE | Myosin regulatory light polypeptide 9 | 192 | 19841 | 4.8 | 15 | 7 | 7 | 5 | 4 | 0.62 | 1.82 |
| ADHX_MOUSE | Alcohol dehydrogenase class-3 | 341 | 39522 | 6.97 | 11 | 9 | 6 | 5 | 6 | 0.4 | 0.69 |
| APT_MOUSE | Adenine phosphoribosyltransferase | 338 | 19712 | 6.31 | 20 | 10 | 7 | 5 | 7 | 0.56 | 1.84 |
| IPO4_MOUSE | Importin-4 | 338 | 119198 | 4.92 | 24 | 17 | 12 | 8 | 12 | 0.21 | 0.32 |
| AATM_MOUSE | Aspartate aminotransferase, mitochondrial | 336 | 47381 | 9.13 | 26 | 14 | 11 | 6 | 11 | 0.41 | 0.69 |
| SC31A_MOUSE | Protein transport protein Sec31A | 334 | 133486 | 6.3 | 24 | 18 | 13 | 10 | 13 | 0.21 | 0.37 |
| MAOX_MOUSE | NADP-dependent malic enzyme | 329 | 63913 | 7.16 | 35 | 18 | 13 | 10 | 13 | 0.47 | 0.92 |
| NDKA_MOUSE | Nucleoside diphosphate kinase A | 324 | 17197 | 6.84 | 23 | 16 | 5 | 2 | 2 | 0.47 | 1.05 |
| TAGL2_MOUSE | Transgelin-2 | 323 | 22381 | 8.39 | 18 | 11 | 7 | 6 | 7 | 0.49 | 2.63 |
| PPAC_MOUSE | Low molecular weight phosphotyrosine protein phosphatase | 323 | 18180 | 6.3 | 9 | 7 | 5 | 3 | 5 | 0.44 | 0.97 |
| P4HA2_MOUSE | Prolyl 4-hydroxylase subunit alpha-2 | 322 | 60964 | 5.55 | 26 | 19 | 9 | 8 | 9 | 0.27 | 0.73 |
| IF4G1_MOUSE | Eukaryotic translation initiation factor 4 gamma 1 | 322 | 175967 | 5.3 | 24 | 16 | 11 | 9 | 11 | 0.11 | 0.27 |
| MRC2_MOUSE | C-type mannose receptor 2 | 321 | 166968 | 5.65 | 32 | 16 | 17 | 9 | 17 | 0.21 | 0.29 |
| DCTN1_MOUSE | Dynactin subunit 1 | 319 | 141588 | 5.66 | 14 | 10 | 6 | 3 | 6 | 0.1 | 0.13 |
| DPYL1_MOUSE | Dihydropyrimidinase-related protein 1 | 317 | 62129 | 6.63 | 9 | 5 | 6 | 2 | 3 | 0.17 | 0.14 |
| MVD1_MOUSE | Diphosphomevalonate decarboxylase | 315 | 44044 | 5.89 | 11 | 8 | 6 | 5 | 6 | 0.34 | 0.6 |
| PUR9_MOUSE | Bifunctional purine biosynthesis protein PURH | 315 | 64177 | 6.3 | 19 | 11 | 11 | 7 | 8 | 0.31 | 0.58 |
| SNX9_MOUSE | Sorting nexin-9 | 313 | 66504 | 5.35 | 16 | 12 | 8 | 6 | 8 | 0.26 | 0.46 |
| AR6P1_MOUSE | ADP-ribosylation factor-like protein 6-interacting protein 1 | 312 | 23421 | 9.38 | 6 | 6 | 1 | 1 | 1 | 0.14 | 0.42 |
| XPO2_MOUSE | Exportin-2 | 312 | 110384 | 5.52 | 20 | 10 | 12 | 6 | 12 | 0.26 | 0.26 |
| ANXA3_MOUSE | Annexin A3 | 312 | 36362 | 5.5 | 17 | 10 | 6 | 4 | 6 | 0.34 | 0.58 |
| OST48_MOUSE | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase 48 kDa subunit | 310 | 48997 | 5.52 | 21 | 15 | 9 | 8 | 9 | 0.41 | 1.34 |
| TCPD_MOUSE | T-complex protein 1 subunit delta | 310 | 58030 | 8.24 | 15 | 9 | 9 | 6 | 9 | 0.29 | 0.54 |
| PLOD3_MOUSE | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 | 309 | 84869 | 5.81 | 29 | 18 | 17 | 10 | 17 | 0.41 | 0.64 |
| RL5_MOUSE | 60S ribosomal protein L5 | 308 | 34379 | 9.78 | 14 | 8 | 5 | 4 | 5 | 0.33 | 0.83 |
| CATD_MOUSE | Cathepsin D | 308 | 44925 | 6.71 | 39 | 18 | 10 | 8 | 10 | 0.58 | 2.04 |
| SC22B_MOUSE | Vesicle-trafficking protein SEC22b | 307 | 24725 | 8.67 | 18 | 12 | 7 | 4 | 7 | 0.6 | 1.3 |
| PLOD1_MOUSE | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | 306 | 83542 | 6.08 | 17 | 10 | 10 | 6 | 10 | 0.24 | 0.42 |
| TCTP_MOUSE | Translationally-controlled tumor protein | 306 | 19450 | 4.76 | 28 | 14 | 6 | 4 | 6 | 0.48 | 1.89 |
| TCPG_MOUSE | T-complex protein 1 subunit gamma | 304 | 60591 | 6.28 | 17 | 12 | 11 | 8 | 11 | 0.37 | 0.86 |
| SC23A_MOUSE | Protein transport protein Sec23A | 303 | 86106 | 6.64 | 10 | 7 | 6 | 3 | 6 | 0.16 | 0.16 |
| SAHH_MOUSE | Adenosylhomocysteinase | 302 | 47657 | 6.08 | 28 | 15 | 11 | 8 | 11 | 0.42 | 1.01 |
| TCPQ_MOUSE | T-complex protein 1 subunit theta | 298 | 59518 | 5.44 | 17 | 8 | 14 | 6 | 14 | 0.39 | 0.52 |
| PSMD1_MOUSE | 26S proteasome non-ATPase regulatory subunit 1 | 298 | 105663 | 5.25 | 37 | 15 | 15 | 8 | 15 | 0.32 | 0.37 |
| COCA1_MOUSE | Collagen alpha-1(XII) chain | 296 | 340004 | 5.47 | 24 | 7 | 14 | 6 | 14 | 0.09 | 0.08 |
| GCN1_MOUSE | eIF-2-alpha kinase activator GCN1 | 295 | 292834 | 7.14 | 22 | 7 | 15 | 4 | 15 | 0.1 | 0.06 |
| USO1_MOUSE | General vesicular transport factor p115 | 295 | 106917 | 4.85 | 20 | 15 | 10 | 5 | 10 | 0.22 | 0.22 |
| UAP1L_MOUSE | UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 | 295 | 56578 | 5.27 | 20 | 15 | 7 | 6 | 6 | 0.21 | 0.56 |
| UAP1_MOUSE | UDP-N-acetylhexosamine pyrophosphorylase | 93 | 58572 | 6.04 | 10 | 4 | 5 | 2 | 4 | 0.12 | 0.15 |
| PTBP1_MOUSE | Polypyrimidine tract-binding protein 1 | 289 | 56443 | 8.47 | 19 | 11 | 9 | 8 | 9 | 0.39 | 0.81 |
| EIF3A_MOUSE | Eukaryotic translation initiation factor 3 subunit A | 288 | 161838 | 6.38 | 18 | 11 | 10 | 5 | 10 | 0.12 | 0.14 |
| GSTP1_MOUSE | Glutathione S-transferase P 1 | 286 | 23594 | 7.68 | 31 | 12 | 8 | 5 | 8 | 0.74 | 2.4 |
| SERA_MOUSE | D-3-Phosphoglycerate dehydrogenase | 279 | 56549 | 6.12 | 15 | 10 | 9 | 7 | 9 | 0.32 | 0.8 |
| GDIR1_MOUSE | Rho GDP-dissociation inhibitor 1 | 279 | 23393 | 5.12 | 29 | 18 | 9 | 6 | 9 | 0.42 | 1.88 |
| VINC_MOUSE | Vinculin | 278 | 116644 | 5.77 | 27 | 14 | 15 | 10 | 15 | 0.26 | 0.43 |
| PNPH_MOUSE | Purine nucleoside phosphorylase | 277 | 32256 | 5.78 | 12 | 8 | 5 | 4 | 5 | 0.37 | 0.67 |
| PSMD7_MOUSE | 26S proteasome non-ATPase regulatory subunit 7 | 275 | 36517 | 6.29 | 17 | 12 | 8 | 6 | 8 | 0.54 | 0.98 |
| LRC59_MOUSE | Leucine-rich repeat-containing protein 59 | 274 | 34856 | 9.57 | 14 | 9 | 6 | 2 | 6 | 0.32 | 0.27 |
| UBP14_MOUSE | Ubiquitin carboxyl-terminal hydrolase 14 | 273 | 55966 | 5.15 | 14 | 11 | 6 | 4 | 6 | 0.25 | 0.35 |
| CNN3_MOUSE | Calponin-3 | 272 | 36406 | 5.46 | 20 | 12 | 8 | 6 | 8 | 0.45 | 1.22 |
| SDHA_MOUSE | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 270 | 72539 | 7.06 | 16 | 11 | 7 | 4 | 7 | 0.27 | 0.26 |
| VAT1_MOUSE | Synaptic vesicle membrane protein VAT-1 homolog | 269 | 43069 | 5.95 | 29 | 12 | 10 | 7 | 10 | 0.38 | 0.97 |
| RLA2_MOUSE | 60S acidic ribosomal protein P2 | 267 | 11644 | 4.42 | 17 | 10 | 5 | 3 | 5 | 0.7 | 1.85 |
| ARLY_MOUSE | Argininosuccinate lyase | 264 | 51707 | 6.48 | 10 | 8 | 5 | 3 | 5 | 0.22 | 0.38 |
| DDX1_MOUSE | ATP-dependent RNA helicase DDX1 | 263 | 82448 | 6.8 | 18 | 9 | 12 | 6 | 12 | 0.29 | 0.36 |
| PYR1_MOUSE | CAD protein | 262 | 243084 | 6 | 22 | 8 | 14 | 6 | 14 | 0.13 | 0.11 |
| AATC_MOUSE | Aspartate aminotransferase, cytoplasmic | 260 | 46219 | 6.68 | 16 | 7 | 7 | 4 | 7 | 0.3 | 0.43 |
| STIP1_MOUSE | Stress-induced-phosphoprotein 1 | 259 | 62542 | 6.4 | 18 | 8 | 9 | 6 | 9 | 0.31 | 0.49 |
| ZYX_MOUSE | Zyxin | 257 | 60507 | 5.99 | 15 | 11 | 5 | 3 | 5 | 0.17 | 0.32 |
| RL3_MOUSE | 60S ribosomal protein L3 | 257 | 46081 | 10.22 | 18 | 10 | 7 | 5 | 7 | 0.25 | 0.57 |
| CO1A1_MOUSE | Collagen alpha-1(I) chain | 253 | 137948 | 5.65 | 19 | 14 | 9 | 6 | 8 | 0.12 | 0.24 |
| PCOC1_MOUSE | Procollagen C-endopeptidase enhancer 1 | 253 | 50136 | 8.73 | 11 | 8 | 7 | 4 | 7 | 0.33 | 0.51 |
| RL6_MOUSE | 60S ribosomal protein L6 | 250 | 33489 | 10.69 | 13 | 9 | 8 | 5 | 8 | 0.25 | 0.86 |
| PP1A_MOUSE | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 249 | 37516 | 5.94 | 19 | 13 | 8 | 7 | 4 | 0.38 | 1.17 |
| FUMH_MOUSE | Fumarate hydratase, mitochondrial | 246 | 54322 | 9.12 | 20 | 11 | 8 | 5 | 8 | 0.35 | 0.59 |
| PCBP2_MOUSE | Poly(rC)-binding protein 2 | 154 | 38197 | 6.33 | 25 | 11 | 7 | 4 | 5 | 0.33 | 0.55 |
| LKHA4_MOUSE | Leukotriene A-4 hydrolase | 246 | 69007 | 5.98 | 15 | 10 | 9 | 4 | 9 | 0.22 | 0.27 |
| TOM40_MOUSE | Mitochondrial import receptor subunit TOM40 homolog | 244 | 37871 | 7.64 | 18 | 10 | 9 | 6 | 9 | 0.48 | 0.93 |
| SH3L3_MOUSE | SH3 domain-binding glutamic acid-rich-like protein 3 | 243 | 10470 | 5.02 | 15 | 10 | 3 | 1 | 3 | 0.65 | 1.17 |
| SYNC_MOUSE | Asparagine–tRNA ligase, cytoplasmic | 242 | 64238 | 5.62 | 22 | 12 | 15 | 6 | 15 | 0.38 | 0.48 |
| PUR6_MOUSE | Multifunctional protein ADE2 | 242 | 46976 | 6.94 | 23 | 10 | 10 | 5 | 10 | 0.44 | 0.7 |
| AL7A1_MOUSE | Alpha-aminoadipic semialdehyde dehydrogenase | 242 | 58824 | 7.16 | 13 | 9 | 6 | 5 | 6 | 0.27 | 0.43 |
| CP51A_MOUSE | Lanosterol 14-alpha demethylase | 242 | 56739 | 8.6 | 16 | 8 | 9 | 3 | 9 | 0.34 | 0.25 |
| SYSC_MOUSE | Serine–tRNA ligase, cytoplasmic | 241 | 58352 | 5.95 | 19 | 9 | 10 | 4 | 10 | 0.35 | 0.33 |
| NDUS1_MOUSE | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | 239 | 79726 | 5.51 | 15 | 10 | 9 | 4 | 9 | 0.22 | 0.3 |
| EIF3H_MOUSE | Eukaryotic translation initiation factor 3 subunit H | 238 | 39807 | 6.2 | 22 | 14 | 11 | 7 | 11 | 0.5 | 1.3 |
| PSB6_MOUSE | Proteasome subunit beta type-6 | 237 | 25362 | 4.97 | 7 | 7 | 4 | 4 | 4 | 0.47 | 0.92 |
| PSA6_MOUSE | Proteasome subunit alpha type-6 | 235 | 27355 | 6.34 | 9 | 8 | 3 | 3 | 3 | 0.29 | 0.58 |
| AMPL_MOUSE | Cytosol aminopeptidase | 234 | 56106 | 7.62 | 17 | 9 | 11 | 6 | 11 | 0.34 | 0.68 |
| TXND5_MOUSE | Thioredoxin domain-containing protein 5 | 232 | 46386 | 5.51 | 13 | 7 | 7 | 4 | 7 | 0.29 | 0.43 |
| TAGL_MOUSE | Transgelin | 231 | 22561 | 8.85 | 19 | 13 | 8 | 6 | 8 | 0.49 | 2 |
| AIMP2_MOUSE | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | 229 | 35355 | 7.7 | 15 | 9 | 6 | 3 | 6 | 0.42 | 0.42 |
| RET1_MOUSE | Retinol-binding protein 1 | 229 | 15836 | 5.1 | 9 | 6 | 4 | 3 | 4 | 0.44 | 1.17 |
| IPYR_MOUSE | Inorganic pyrophosphatase | 229 | 32646 | 5.37 | 14 | 7 | 7 | 3 | 7 | 0.31 | 0.46 |
| GSTM2_MOUSE | Glutathione S-transferase Mu 2 | 228 | 25700 | 6.9 | 14 | 9 | 7 | 6 | 6 | 0.39 | 1.62 |
| GSTM1_MOUSE | Glutathione S-transferase Mu 1 | 197 | 25953 | 7.71 | 14 | 8 | 5 | 4 | 4 | 0.33 | 0.89 |
| SYRC_MOUSE | Arginine–tRNA ligase, cytoplasmic | 228 | 75625 | 7.48 | 23 | 13 | 12 | 7 | 12 | 0.24 | 0.47 |
| WDR61_MOUSE | WD repeat-containing protein 61 | 228 | 33752 | 5.1 | 6 | 4 | 3 | 2 | 3 | 0.18 | 0.28 |
| IMA4_MOUSE | Importin subunit alpha-4 | 178 | 57737 | 4.8 | 10 | 7 | 6 | 4 | 4 | 0.32 | 0.34 |
| RL7_MOUSE | 60S ribosomal protein L7 | 227 | 31400 | 10.89 | 13 | 7 | 6 | 3 | 5 | 0.24 | 0.49 |
| PSME2_MOUSE | Proteasome activator complex subunit 2 | 227 | 27040 | 5.54 | 14 | 7 | 5 | 4 | 5 | 0.34 | 0.84 |
| SCRB2_MOUSE | Lysosome membrane protein 2 | 225 | 54009 | 4.99 | 13 | 8 | 6 | 4 | 6 | 0.2 | 0.36 |
| AT2A2_MOUSE | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 224 | 114784 | 5.23 | 26 | 12 | 13 | 7 | 13 | 0.24 | 0.29 |
| SODM_MOUSE | Superoxide dismutase [Mn], mitochondrial | 222 | 24588 | 8.8 | 14 | 8 | 5 | 3 | 5 | 0.5 | 0.96 |
| SYLC_MOUSE | Leucine–tRNA ligase, cytoplasmic | 222 | 134106 | 6.64 | 22 | 7 | 13 | 4 | 13 | 0.19 | 0.13 |
| PTGIS_MOUSE | Prostacyclin synthase | 222 | 57011 | 6.26 | 28 | 17 | 13 | 9 | 6 | 0.52 | 0.93 |
| PYGB_MOUSE | Glycogen phosphorylase, brain form | 219 | 96668 | 6.28 | 16 | 10 | 9 | 6 | 9 | 0.16 | 0.3 |
| MPCP_MOUSE | Phosphate carrier protein, mitochondrial | 218 | 39606 | 9.36 | 15 | 11 | 8 | 5 | 8 | 0.32 | 0.88 |
| LAMP1_MOUSE | Lysosome-associated membrane glycoprotein 1 | 217 | 43837 | 8.66 | 8 | 7 | 4 | 3 | 4 | 0.16 | 0.46 |
| COPA_MOUSE | Coatomer subunit alpha | 217 | 138344 | 7.69 | 23 | 11 | 16 | 9 | 16 | 0.23 | 0.31 |
| KINH_MOUSE | Kinesin-1 heavy chain | 216 | 109484 | 6.06 | 13 | 6 | 10 | 4 | 10 | 0.19 | 0.17 |
| LG3BP_MOUSE | Galectin-3-binding protein | 216 | 64450 | 5 | 12 | 5 | 8 | 5 | 8 | 0.29 | 0.38 |
| PSB4_MOUSE | Proteasome subunit beta type-4 | 215 | 29097 | 5.47 | 5 | 3 | 3 | 2 | 3 | 0.23 | 0.33 |
| IDI1_MOUSE | Isopentenyl-diphosphate delta-isomerase 1 | 215 | 26272 | 5.79 | 19 | 12 | 8 | 7 | 8 | 0.54 | 2.01 |
| CNN2_MOUSE | Calponin-2 | 215 | 33134 | 7.53 | 17 | 9 | 9 | 4 | 9 | 0.59 | 1.12 |
| NAA15_MOUSE | N-Alpha-acetyltransferase 15, NatA auxiliary subunit | 212 | 100897 | 7.68 | 19 | 9 | 9 | 5 | 9 | 0.18 | 0.23 |
| IF2P_MOUSE | Eukaryotic translation initiation factor 5B | 212 | 137532 | 5.47 | 16 | 9 | 7 | 4 | 7 | 0.14 | 0.16 |
| LGMN_MOUSE | Legumain | 211 | 49341 | 5.92 | 10 | 9 | 3 | 3 | 3 | 0.17 | 0.4 |
| DJC10_MOUSE | DnaJ homolog subfamily C member 10 | 210 | 90525 | 6.53 | 15 | 10 | 6 | 3 | 6 | 0.13 | 0.2 |
| PFKAL_MOUSE | ATP-dependent 6-phosphofructokinase, liver type | 210 | 85305 | 6.74 | 11 | 8 | 6 | 6 | 6 | 0.12 | 0.34 |
| UGPA_MOUSE | UTP–glucose-1-phosphate uridylyltransferase | 210 | 56944 | 7.18 | 8 | 6 | 6 | 5 | 6 | 0.26 | 0.44 |
| RTN4_MOUSE | Reticulon-4 | 210 | 126535 | 4.47 | 11 | 9 | 6 | 4 | 6 | 0.12 | 0.14 |
| CD34_MOUSE | Hematopoietic progenitor cell antigen CD34 | 208 | 40957 | 5.2 | 3 | 3 | 1 | 1 | 1 | 0.05 | 0.11 |
| TPP2_MOUSE | Tripeptidyl-peptidase 2 | 208 | 139790 | 6.13 | 15 | 8 | 9 | 4 | 9 | 0.16 | 0.13 |
| PCNA_MOUSE | Proliferating cell nuclear antigen | 208 | 28766 | 4.66 | 15 | 8 | 8 | 6 | 8 | 0.63 | 1.37 |
| TSPO_MOUSE | Translocator protein | 205 | 18829 | 9.52 | 12 | 8 | 4 | 2 | 4 | 0.46 | 0.92 |
| SNAA_MOUSE | Alpha-soluble NSF attachment protein | 202 | 33168 | 5.3 | 13 | 7 | 7 | 4 | 7 | 0.43 | 0.65 |
| PDXD1_MOUSE | Pyridoxal-dependent decarboxylase domain-containing protein 1 | 202 | 87281 | 5.31 | 6 | 4 | 4 | 3 | 4 | 0.1 | 0.15 |
| PRDX1_MOUSE | Peroxiredoxin-1 | 202 | 22162 | 8.26 | 43 | 18 | 16 | 7 | 14 | 0.69 | 2.67 |
| DHX9_MOUSE | ATP-dependent RNA helicase A | 201 | 149381 | 6.39 | 19 | 12 | 11 | 6 | 11 | 0.16 | 0.18 |
| MIC60_MOUSE | MICOS complex subunit Mic60 | 200 | 83848 | 6.18 | 14 | 8 | 4 | 3 | 4 | 0.1 | 0.16 |
| AACS_MOUSE | Acetoacetyl-CoA synthetase | 199 | 75152 | 6.25 | 24 | 9 | 11 | 7 | 11 | 0.36 | 0.56 |
| SERC_MOUSE | Phosphoserine aminotransferase | 199 | 40447 | 8.15 | 23 | 12 | 11 | 5 | 11 | 0.52 | 0.85 |
| HYOU1_MOUSE | Hypoxia-upregulated protein 1 | 198 | 111112 | 5.12 | 14 | 7 | 9 | 4 | 9 | 0.18 | 0.21 |
| INF2_MOUSE | Inverted formin-2 | 198 | 138474 | 5.09 | 18 | 8 | 10 | 4 | 10 | 0.14 | 0.13 |
| PSMD6_MOUSE | 26S proteasome non-ATPase regulatory subunit 6 | 197 | 45507 | 5.38 | 4 | 4 | 2 | 2 | 2 | 0.1 | 0.2 |
| AL9A1_MOUSE | 4-Trimethylaminobutyraldehyde dehydrogenase | 196 | 53480 | 6.63 | 9 | 6 | 3 | 2 | 3 | 0.17 | 0.26 |
| RN213_MOUSE | E3 ubiquitin-protein ligase RNF213 | 196 | 584411 | 6.35 | 23 | 8 | 21 | 7 | 21 | 0.07 | 0.05 |
| CX7A2_MOUSE | Cytochrome c oxidase subunit 7A2, mitochondrial | 196 | 9285 | 10.28 | 7 | 6 | 3 | 2 | 3 | 0.59 | 1.37 |
| PSB5_MOUSE | Proteasome subunit beta type-5 | 196 | 28514 | 6.52 | 10 | 8 | 5 | 4 | 5 | 0.32 | 0.79 |
| TMX3_MOUSE | Protein disulfide-isomerase TMX3 | 196 | 51815 | 5.02 | 4 | 4 | 2 | 2 | 2 | 0.11 | 0.17 |
| DLDH_MOUSE | Dihydrolipoyl dehydrogenase, mitochondrial | 195 | 54238 | 7.99 | 16 | 12 | 5 | 4 | 5 | 0.21 | 0.47 |
| AEBP1_MOUSE | Adipocyte enhancer-binding protein 1 | 194 | 128284 | 5.02 | 4 | 3 | 2 | 1 | 2 | 0.02 | 0.03 |
| MP2K1_MOUSE | Dual specificity mitogen-activated protein kinase 1 | 194 | 43446 | 6.24 | 9 | 5 | 6 | 2 | 5 | 0.32 | 0.21 |
| MP2K2_MOUSE | Dual specificity mitogen-activated protein kinase 2 | 191 | 44374 | 6.58 | 5 | 5 | 2 | 2 | 1 | 0.12 | 0.21 |
| STRAP_MOUSE | Serine-threonine kinase receptor-associated protein | 193 | 38418 | 4.99 | 12 | 10 | 6 | 5 | 6 | 0.35 | 0.72 |
| XPO1_MOUSE | Exportin-1 | 192 | 123013 | 5.72 | 18 | 10 | 12 | 8 | 12 | 0.2 | 0.31 |
| EIF3I_MOUSE | Eukaryotic translation initiation factor 3 subunit I | 191 | 36438 | 5.38 | 13 | 8 | 6 | 3 | 6 | 0.36 | 0.41 |
| CPNE1_MOUSE | Copine-1 | 190 | 58849 | 5.4 | 11 | 7 | 7 | 4 | 7 | 0.3 | 0.33 |
| EIF3K_MOUSE | Eukaryotic translation initiation factor 3 subunit K | 189 | 25070 | 4.81 | 7 | 4 | 4 | 3 | 4 | 0.33 | 0.64 |
| MIF_MOUSE | Macrophage migration inhibitory factor | 189 | 12496 | 6.79 | 13 | 7 | 5 | 3 | 5 | 0.63 | 2.67 |
| CYFP1_MOUSE | Cytoplasmic FMR1-interacting protein 1 | 189 | 145148 | 6.46 | 14 | 6 | 10 | 3 | 10 | 0.16 | 0.09 |
| MYO1C_MOUSE | Unconventional myosin-Ic | 188 | 121868 | 9.41 | 30 | 7 | 18 | 6 | 18 | 0.29 | 0.23 |
| JAK1_MOUSE | Tyrosine-protein kinase JAK1 | 188 | 133282 | 7.63 | 11 | 5 | 6 | 1 | 6 | 0.1 | 0.03 |
| ARPC2_MOUSE | Actin-related protein 2/3 complex subunit 2 | 188 | 34336 | 6.84 | 14 | 10 | 7 | 4 | 7 | 0.41 | 0.62 |
| CSAD_MOUSE | Cysteine sulfinic acid decarboxylase | 187 | 55109 | 6.17 | 5 | 3 | 3 | 1 | 3 | 0.11 | 0.08 |
| PA2G4_MOUSE | Proliferation-associated protein 2G4 | 185 | 43671 | 6.41 | 4 | 4 | 3 | 3 | 3 | 0.15 | 0.33 |
| DJB11_MOUSE | DnaJ homolog subfamily B member 11 | 185 | 40530 | 5.92 | 13 | 7 | 5 | 3 | 5 | 0.2 | 0.36 |
| NSDHL_MOUSE | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | 184 | 40660 | 7.71 | 20 | 8 | 7 | 6 | 7 | 0.39 | 0.85 |
| P3H1_MOUSE | Prolyl 3-hydroxylase 1 | 184 | 83598 | 5.03 | 21 | 11 | 7 | 3 | 7 | 0.2 | 0.16 |
| S10AA_MOUSE | Protein S100-A10 | 184 | 11179 | 6.27 | 5 | 5 | 2 | 2 | 2 | 0.44 | 1.06 |
| DHB12_MOUSE | Very-long-chain 3-oxoacyl-CoA reductase | 182 | 34719 | 9.55 | 15 | 10 | 7 | 5 | 7 | 0.45 | 0.82 |
| SPRC_MOUSE | SPARC | 182 | 34428 | 4.77 | 11 | 6 | 5 | 3 | 5 | 0.25 | 0.44 |
| DHB7_MOUSE | 3-Keto-steroid reductase | 179 | 37293 | 6.25 | 5 | 5 | 4 | 4 | 4 | 0.21 | 0.56 |
| MPRD_MOUSE | Cation-dependent mannose-6-phosphate receptor | 179 | 31152 | 5.24 | 5 | 4 | 3 | 2 | 3 | 0.18 | 0.3 |
| FKB10_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP10 | 179 | 64656 | 5.38 | 17 | 7 | 9 | 4 | 9 | 0.32 | 0.29 |
| FRIL1_MOUSE | Ferritin light chain 1 | 178 | 20790 | 5.66 | 13 | 8 | 4 | 3 | 4 | 0.43 | 0.81 |
| PGP_MOUSE | Glycerol-3-phosphate phosphatase | 178 | 34519 | 5.21 | 3 | 3 | 1 | 1 | 1 | 0.06 | 0.13 |
| IMPA1_MOUSE | Inositol monophosphatase 1 | 178 | 30416 | 5.08 | 7 | 6 | 3 | 2 | 3 | 0.12 | 0.31 |
| SYCC_MOUSE | Cysteine–tRNA ligase, cytoplasmic | 178 | 94800 | 6.32 | 12 | 5 | 7 | 5 | 7 | 0.18 | 0.25 |
| FKBP4_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP4 | 177 | 51540 | 5.54 | 9 | 8 | 3 | 3 | 3 | 0.12 | 0.27 |
| PPIB_MOUSE | Peptidyl-prolyl cis-trans isomerase B | 177 | 23699 | 9.56 | 10 | 5 | 6 | 2 | 6 | 0.33 | 0.42 |
| NASP_MOUSE | Nuclear autoantigenic sperm protein | 177 | 83903 | 4.35 | 10 | 10 | 4 | 4 | 4 | 0.14 | 0.22 |
| MPRI_MOUSE | Cation-independent mannose-6-phosphate receptor | 177 | 273639 | 5.47 | 14 | 6 | 12 | 4 | 12 | 0.1 | 0.06 |
| NQO1_MOUSE | NAD(P)H dehydrogenase [quinone] 1 | 175 | 30940 | 8.74 | 19 | 7 | 5 | 4 | 5 | 0.49 | 0.71 |
| GCSH_MOUSE | Glycine cleavage system H protein, mitochondrial | 173 | 18625 | 4.78 | 5 | 5 | 1 | 1 | 1 | 0.22 | 0.55 |
| ITB1_MOUSE | Integrin beta-1 | 173 | 88173 | 5.68 | 22 | 10 | 12 | 6 | 12 | 0.29 | 0.33 |
| CO1A2_MOUSE | Collagen alpha-2(I) chain | 173 | 129478 | 9.27 | 20 | 12 | 10 | 4 | 10 | 0.12 | 0.14 |
| HNRPM_MOUSE | Heterogeneous nuclear ribonucleoprotein M | 172 | 77597 | 8.8 | 18 | 6 | 11 | 4 | 11 | 0.24 | 0.24 |
| GHITM_MOUSE | Growth hormone-inducible transmembrane protein | 172 | 37250 | 9.82 | 5 | 3 | 2 | 1 | 2 | 0.09 | 0.25 |
| TTL12_MOUSE | Tubulin–tyrosine ligase-like protein 12 | 172 | 73996 | 5.39 | 15 | 7 | 8 | 5 | 8 | 0.27 | 0.33 |
| RAB1B_MOUSE | Ras-related protein Rab-1B | 118 | 22173 | 5.55 | 14 | 5 | 7 | 5 | 2 | 0.53 | 1.53 |
| ACO13_MOUSE | Acyl-coenzyme A thioesterase 13 | 171 | 15173 | 8.95 | 8 | 7 | 3 | 3 | 3 | 0.41 | 1.24 |
| MMP14_MOUSE | Matrix metalloproteinase-14 | 171 | 65877 | 8.07 | 6 | 5 | 4 | 3 | 4 | 0.13 | 0.21 |
| PSA7_MOUSE | Proteasome subunit alpha type-7 | 171 | 27838 | 8.59 | 11 | 8 | 6 | 4 | 6 | 0.38 | 0.81 |
| LEG3_MOUSE | Galectin-3 | 170 | 27498 | 8.46 | 24 | 11 | 7 | 5 | 7 | 0.35 | 1.12 |
| PMVK_MOUSE | Phosphomevalonate kinase | 169 | 21902 | 5.7 | 2 | 2 | 1 | 1 | 1 | 0.1 | 0.21 |
| TPIS_MOUSE | Triosephosphate isomerase | 169 | 32171 | 5.56 | 17 | 7 | 8 | 4 | 8 | 0.39 | 0.67 |
| SEPT9_MOUSE | Septin-9 | 168 | 65534 | 9.01 | 11 | 4 | 7 | 3 | 7 | 0.22 | 0.21 |
| UBR4_MOUSE | E3 ubiquitin-protein ligase UBR4 | 167 | 571927 | 5.72 | 24 | 10 | 16 | 6 | 16 | 0.07 | 0.05 |
| GPDM_MOUSE | Glycerol-3-phosphate dehydrogenase, mitochondrial | 167 | 80902 | 6.17 | 27 | 10 | 16 | 8 | 16 | 0.35 | 0.51 |
| GLCM_MOUSE | Glucosylceramidase | 166 | 57585 | 7.64 | 28 | 11 | 9 | 6 | 9 | 0.39 | 0.66 |
| LA_MOUSE | Lupus La protein homolog | 166 | 47727 | 9.77 | 9 | 7 | 7 | 5 | 7 | 0.23 | 0.55 |
| VCAM1_MOUSE | Vascular cell adhesion protein 1 | 166 | 81265 | 5.21 | 7 | 6 | 3 | 3 | 3 | 0.07 | 0.17 |
| AP2A1_MOUSE | AP-2 complex subunit alpha-1 | 165 | 107596 | 6.63 | 18 | 6 | 10 | 5 | 7 | 0.19 | 0.21 |
| HMOX2_MOUSE | Heme oxygenase 2 | 164 | 35716 | 5.61 | 5 | 2 | 3 | 1 | 3 | 0.23 | 0.12 |
| ITAV_MOUSE | Integrin alpha-V | 164 | 115287 | 5.41 | 15 | 9 | 11 | 7 | 11 | 0.16 | 0.29 |
| ECI1_MOUSE | Enoyl-CoA delta isomerase 1, mitochondrial | 163 | 32230 | 9.12 | 4 | 4 | 2 | 2 | 2 | 0.15 | 0.29 |
| SYEP_MOUSE | Bifunctional glutamate/proline–tRNA ligase | 162 | 169972 | 7.75 | 33 | 10 | 18 | 7 | 18 | 0.22 | 0.19 |
| KAD2_MOUSE | Adenylate kinase 2, mitochondrial | 162 | 26452 | 6.96 | 5 | 5 | 2 | 2 | 2 | 0.16 | 0.37 |
| NOMO1_MOUSE | Nodal modulator 1 | 161 | 133336 | 5.75 | 19 | 9 | 11 | 4 | 11 | 0.19 | 0.13 |
| C1TM_MOUSE | Monofunctional C1-tetrahydrofolate synthase, mitochondrial | 161 | 105662 | 6.58 | 9 | 7 | 4 | 3 | 4 | 0.09 | 0.13 |
| AKAP2_MOUSE | A-kinase anchor protein 2 | 160 | 98519 | 5.13 | 4 | 3 | 3 | 2 | 3 | 0.05 | 0.09 |
| SFPQ_MOUSE | Splicing factor, proline- and glutamine-rich | 160 | 75394 | 9.45 | 5 | 3 | 2 | 1 | 2 | 0.05 | 0.06 |
| BACH_MOUSE | Cytosolic acyl coenzyme A thioester hydrolase | 159 | 42510 | 8.9 | 15 | 7 | 9 | 5 | 9 | 0.37 | 0.63 |
| GLRX1_MOUSE | Glutaredoxin-1 | 159 | 11863 | 8.67 | 7 | 6 | 2 | 1 | 2 | 0.47 | 0.98 |
| DHC24_MOUSE | Delta(24)-sterol reductase | 159 | 60073 | 8.42 | 16 | 8 | 6 | 4 | 6 | 0.23 | 0.32 |
| NMT1_MOUSE | Glycylpeptide N-tetradecanoyltransferase 1 | 159 | 56852 | 8.04 | 6 | 5 | 3 | 2 | 3 | 0.15 | 0.25 |
| PRDX3_MOUSE | Thioredoxin-dependent peroxide reductase, mitochondrial | 158 | 28109 | 7.15 | 7 | 6 | 4 | 3 | 4 | 0.35 | 0.56 |
| CAVN1_MOUSE | Caveolae-associated protein 1 | 157 | 43927 | 5.43 | 8 | 5 | 4 | 2 | 4 | 0.18 | 0.21 |
| PALLD_MOUSE | Palladin | 156 | 152037 | 5.87 | 11 | 8 | 7 | 5 | 7 | 0.1 | 0.15 |
| NPC1_MOUSE | Niemann-Pick C1 protein | 156 | 142791 | 5.44 | 4 | 3 | 2 | 1 | 2 | 0.04 | 0.03 |
| SRPRB_MOUSE | Signal recognition particle receptor subunit beta | 156 | 29561 | 9.34 | 4 | 3 | 3 | 2 | 3 | 0.16 | 0.32 |
| PCYOX_MOUSE | Prenylcysteine oxidase | 156 | 56459 | 6.44 | 10 | 4 | 7 | 3 | 7 | 0.27 | 0.34 |
| TRBM_MOUSE | Thrombomodulin | 155 | 61827 | 4.5 | 9 | 8 | 2 | 2 | 2 | 0.13 | 0.22 |
| PSB2_MOUSE | Proteasome subunit beta type-2 | 154 | 22892 | 6.52 | 25 | 11 | 6 | 6 | 6 | 0.54 | 1.95 |
| AB1IP_MOUSE | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | 154 | 74272 | 5.23 | 13 | 5 | 5 | 2 | 5 | 0.14 | 0.12 |
| AGM1_MOUSE | Phosphoacetylglucosamine mutase | 154 | 59415 | 5.8 | 8 | 6 | 3 | 2 | 3 | 0.13 | 0.23 |
| APEH_MOUSE | Acylamino-acid-releasing enzyme | 153 | 81529 | 5.36 | 7 | 6 | 5 | 4 | 5 | 0.16 | 0.23 |
| CATL1_MOUSE | Cathepsin L1 | 153 | 37523 | 6.37 | 12 | 10 | 6 | 4 | 6 | 0.44 | 0.56 |
| ATOX1_MOUSE | Copper transport protein ATOX1 | 151 | 7334 | 6.04 | 8 | 7 | 2 | 1 | 2 | 0.53 | 0.72 |
| ALD2_MOUSE | Aldose reductase-related protein 2 | 151 | 36098 | 5.97 | 11 | 8 | 7 | 5 | 7 | 0.36 | 0.78 |
| PPIC_MOUSE | Peptidyl-prolyl cis-trans isomerase C | 151 | 22780 | 6.96 | 7 | 6 | 4 | 3 | 4 | 0.46 | 1.07 |
| NRDC_MOUSE | Nardilysin | 150 | 132808 | 4.77 | 14 | 6 | 8 | 3 | 8 | 0.15 | 0.1 |
| MD2L1_MOUSE | Mitotic spindle assembly checkpoint protein MAD2A | 150 | 23583 | 5.17 | 6 | 3 | 2 | 1 | 2 | 0.17 | 0.19 |
| BROX_MOUSE | BRO1 domain-containing protein BROX | 149 | 46172 | 7.59 | 6 | 4 | 4 | 2 | 4 | 0.22 | 0.2 |
| SYTC_MOUSE | Threonine–tRNA ligase, cytoplasmic | 148 | 83303 | 7.03 | 21 | 9 | 13 | 5 | 13 | 0.25 | 0.29 |
| DNJA1_MOUSE | DnaJ homolog subfamily A member 1 | 148 | 44839 | 6.65 | 14 | 9 | 6 | 4 | 6 | 0.31 | 0.59 |
| NNRE_MOUSE | NAD(P)H-hydrate epimerase | 147 | 30953 | 7.59 | 11 | 9 | 4 | 3 | 4 | 0.3 | 0.49 |
| CPNE3_MOUSE | Copine-3 | 147 | 59547 | 5.52 | 6 | 5 | 3 | 3 | 3 | 0.1 | 0.23 |
| PTGR1_MOUSE | Prostaglandin reductase 1 | 147 | 35537 | 8.09 | 9 | 5 | 4 | 2 | 4 | 0.26 | 0.42 |
| C1QBP_MOUSE | Complement component 1 Q subcomponent-binding protein, mitochondrial | 146 | 30994 | 4.82 | 18 | 8 | 6 | 5 | 6 | 0.45 | 0.95 |
| ERG7_MOUSE | Lanosterol synthase | 145 | 83088 | 5.96 | 13 | 5 | 9 | 3 | 9 | 0.23 | 0.22 |
| PAPS1_MOUSE | Bifunctional 3 ′-phosphoadenosine 5 ′-phosphosulfate synthase 1 | 144 | 70749 | 6.31 | 11 | 4 | 8 | 4 | 8 | 0.21 | 0.27 |
| MTPN_MOUSE | Myotrophin | 143 | 12853 | 5.27 | 11 | 7 | 6 | 3 | 6 | 0.62 | 1.59 |
| VATA_MOUSE | V-type proton ATPase catalytic subunit A | 143 | 68283 | 5.42 | 16 | 6 | 7 | 3 | 7 | 0.25 | 0.2 |
| ATPG_MOUSE | ATP synthase subunit gamma, mitochondrial | 143 | 32865 | 9.06 | 7 | 5 | 4 | 2 | 4 | 0.26 | 0.29 |
| DDB1_MOUSE | DNA damage-binding protein 1 | 143 | 126772 | 5.14 | 20 | 10 | 11 | 5 | 11 | 0.21 | 0.18 |
| RL4_MOUSE | 60S ribosomal protein L4 | 143 | 47124 | 11.01 | 16 | 6 | 12 | 5 | 12 | 0.37 | 0.55 |
| NDUB4_MOUSE | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 | 143 | 15072 | 9.89 | 4 | 4 | 1 | 1 | 1 | 0.16 | 0.31 |
| CRTAP_MOUSE | Cartilage-associated protein | 143 | 46140 | 5.46 | 12 | 7 | 5 | 4 | 5 | 0.2 | 0.43 |
| TFR1_MOUSE | Transferrin receptor protein 1 | 143 | 85677 | 6.13 | 11 | 7 | 3 | 3 | 3 | 0.08 | 0.16 |
| EI3JA_MOUSE | Eukaryotic translation initiation factor 3 subunit J-A | 142 | 29326 | 4.69 | 5 | 3 | 2 | 2 | 2 | 0.1 | 0.33 |
| ATLA3_MOUSE | Atlastin-3 | 142 | 60537 | 5.73 | 15 | 7 | 10 | 5 | 10 | 0.36 | 0.41 |
| PUR4_MOUSE | Phosphoribosylformylglycinamidine synthase | 142 | 144538 | 5.43 | 10 | 7 | 8 | 5 | 8 | 0.14 | 0.16 |
| IPO9_MOUSE | Importin-9 | 142 | 115978 | 4.71 | 16 | 9 | 8 | 5 | 8 | 0.2 | 0.2 |
| PSMD3_MOUSE | 26S proteasome non-ATPase regulatory subunit 3 | 141 | 60680 | 8.48 | 10 | 8 | 6 | 5 | 6 | 0.14 | 0.41 |
| AK1A1_MOUSE | Alcohol dehydrogenase [NADP(+)] | 141 | 36564 | 6.9 | 5 | 3 | 4 | 2 | 4 | 0.27 | 0.26 |
| DCTN3_MOUSE | Dynactin subunit 3 | 140 | 20965 | 5.84 | 4 | 3 | 3 | 2 | 3 | 0.31 | 0.8 |
| SWP70_MOUSE | Switch-associated protein 70 | 138 | 68953 | 5.78 | 13 | 7 | 6 | 5 | 6 | 0.17 | 0.35 |
| ATPO_MOUSE | ATP synthase subunit O, mitochondrial | 137 | 23349 | 10 | 7 | 4 | 2 | 2 | 2 | 0.17 | 0.43 |
| UN45A_MOUSE | Protein unc-45 homolog A | 137 | 103382 | 6.01 | 9 | 3 | 7 | 2 | 7 | 0.13 | 0.08 |
| SCOT1_MOUSE | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial | 137 | 55953 | 8.73 | 20 | 9 | 10 | 7 | 10 | 0.36 | 0.68 |
| BCAT1_MOUSE | Branched-chain-amino-acid aminotransferase, cytosolic | 137 | 42764 | 5.25 | 8 | 7 | 3 | 3 | 3 | 0.21 | 0.48 |
| ASNS_MOUSE | Asparagine synthetase [glutamine-hydrolyzing] | 137 | 64241 | 6.12 | 16 | 8 | 7 | 4 | 7 | 0.19 | 0.3 |
| PPGB_MOUSE | Lysosomal protective protein | 136 | 53809 | 5.56 | 7 | 4 | 3 | 2 | 3 | 0.1 | 0.17 |
| PGBM_MOUSE | Basement membrane-specific heparan sulfate proteoglycan core protein | 136 | 398039 | 5.88 | 25 | 6 | 13 | 2 | 13 | 0.07 | 0.02 |
| CSN7A_MOUSE | COP9 signalosome complex subunit 7a | 136 | 30206 | 7.68 | 14 | 7 | 6 | 3 | 6 | 0.51 | 0.51 |
| HCD2_MOUSE | 3-Hydroxyacyl-CoA dehydrogenase type-2 | 136 | 27402 | 8.53 | 6 | 3 | 4 | 2 | 4 | 0.2 | 0.35 |
| BZW2_MOUSE | Basic leucine zipper and W2 domain-containing protein 2 | 135 | 48033 | 6.26 | 6 | 4 | 3 | 1 | 3 | 0.22 | 0.19 |
| TOM70_MOUSE | Mitochondrial import receptor subunit TOM70 | 135 | 67547 | 7.46 | 10 | 7 | 6 | 4 | 4 | 0.24 | 0.28 |
| XPP1_MOUSE | Xaa-Pro aminopeptidase 1 | 135 | 69547 | 5.33 | 14 | 7 | 7 | 4 | 4 | 0.18 | 0.27 |
| NXP20_MOUSE | Protein Noxp20 | 135 | 60975 | 4.49 | 6 | 6 | 1 | 1 | 1 | 0.07 | 0.15 |
| PAI1_MOUSE | Plasminogen activator inhibitor 1 | 134 | 45141 | 6.17 | 9 | 3 | 7 | 2 | 7 | 0.32 | 0.2 |
| ROA0_MOUSE | Heterogeneous nuclear ribonucleoprotein A0 | 134 | 30512 | 9.35 | 9 | 3 | 4 | 2 | 4 | 0.24 | 0.31 |
| GORS2_MOUSE | Golgi reassembly-stacking protein 2 | 134 | 47009 | 4.68 | 4 | 3 | 3 | 2 | 3 | 0.14 | 0.19 |
| SMC2_MOUSE | Structural maintenance of chromosomes protein 2 | 131 | 134156 | 8.54 | 8 | 5 | 5 | 2 | 5 | 0.06 | 0.06 |
| RAB21_MOUSE | Ras-related protein Rab-21 | 132 | 24091 | 8.11 | 7 | 5 | 5 | 4 | 5 | 0.21 | 0.99 |
| SAC1_MOUSE | Phosphatidylinositide phosphatase SAC1 | 132 | 66901 | 6.85 | 8 | 3 | 5 | 3 | 5 | 0.16 | 0.21 |
| HSPB1_MOUSE | Heat shock protein beta-1 | 131 | 23000 | 6.12 | 14 | 9 | 7 | 4 | 7 | 0.66 | 1.05 |
| GLU2B_MOUSE | Glucosidase 2 subunit beta | 131 | 58756 | 4.41 | 9 | 6 | 3 | 2 | 3 | 0.12 | 0.24 |
| IMA1_MOUSE | Importin subunit alpha-1 | 130 | 57892 | 5.49 | 10 | 5 | 6 | 4 | 6 | 0.27 | 0.33 |
| SEC13_MOUSE | Protein SEC13 homolog | 130 | 35543 | 5.15 | 19 | 5 | 4 | 3 | 4 | 0.19 | 0.42 |
| 4F2_MOUSE | 4F2 cell-surface antigen heavy chain | 130 | 58300 | 5.62 | 5 | 5 | 2 | 2 | 2 | 0.1 | 0.15 |
| CSN4_MOUSE | COP9 signalosome complex subunit 4 | 130 | 46256 | 5.57 | 9 | 4 | 6 | 2 | 6 | 0.25 | 0.2 |
| ROAA_MOUSE | Heterogeneous nuclear ribonucleoprotein A/B | 129 | 30812 | 7.68 | 11 | 9 | 6 | 6 | 6 | 0.2 | 1.24 |
| ERF3A_MOUSE | Eukaryotic peptide chain release factor GTP-binding subunit ERF3A | 129 | 68582 | 5.12 | 19 | 10 | 9 | 6 | 9 | 0.18 | 0.44 |
| CUL4A_MOUSE | Cullin-4A | 128 | 87697 | 8.53 | 4 | 2 | 3 | 2 | 1 | 0.07 | 0.1 |
| CUL4B_MOUSE | Cullin-4B | 102 | 110630 | 8.56 | 7 | 3 | 6 | 3 | 4 | 0.12 | 0.12 |
| AHSA1_MOUSE | Activator of 90 kDa heat shock protein ATPase homolog 1 | 128 | 38093 | 5.41 | 6 | 3 | 4 | 2 | 4 | 0.23 | 0.24 |
| PDLI1_MOUSE | PDZ and LIM domain protein 1 | 127 | 35752 | 6.38 | 9 | 5 | 5 | 4 | 5 | 0.25 | 0.59 |
| ECHM_MOUSE | Enoyl-CoA hydratase, mitochondrial | 127 | 31454 | 8.76 | 8 | 5 | 5 | 3 | 5 | 0.33 | 0.49 |
| ITA5_MOUSE | Integrin alpha-5 | 126 | 114971 | 5.65 | 13 | 6 | 6 | 2 | 6 | 0.09 | 0.08 |
| NDUBA_MOUSE | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | 126 | 21010 | 8.19 | 6 | 4 | 4 | 3 | 4 | 0.31 | 0.8 |
| RENBP_MOUSE | N-Acylglucosamine 2-epimerase | 126 | 49739 | 5.69 | 8 | 5 | 5 | 3 | 5 | 0.21 | 0.29 |
| NU155_MOUSE | Nuclear pore complex protein Nup155 | 125 | 155019 | 5.77 | 9 | 6 | 5 | 3 | 5 | 0.06 | 0.08 |
| DDAH1_MOUSE | N(G),N(G)-Dimethylarginine dimethylaminohydrolase 1 | 125 | 31361 | 5.64 | 6 | 4 | 5 | 3 | 5 | 0.4 | 0.49 |
| G6PE_MOUSE | GDH/6PGL endoplasmic bifunctional protein | 124 | 88872 | 6.44 | 16 | 3 | 9 | 3 | 9 | 0.21 | 0.15 |
| CO5A1_MOUSE | Collagen alpha-1(V) chain | 124 | 183564 | 4.86 | 7 | 3 | 6 | 2 | 6 | 0.09 | 0.05 |
| PLAP_MOUSE | Phospholipase A-2-activating protein | 124 | 87166 | 5.77 | 6 | 2 | 5 | 1 | 5 | 0.15 | 0.05 |
| PDLI5_MOUSE | PDZ and LIM domain protein 5 | 123 | 63259 | 8.61 | 15 | 6 | 7 | 3 | 7 | 0.21 | 0.22 |
| NAGAB_MOUSE | Alpha-N-acetylgalactosaminidase | 122 | 47204 | 6.02 | 7 | 5 | 6 | 4 | 6 | 0.23 | 0.42 |
| PDIA5_MOUSE | Protein disulfide-isomerase A5 | 121 | 59229 | 7.25 | 5 | 5 | 4 | 4 | 4 | 0.19 | 0.33 |
| PSMD5_MOUSE | 26S proteasome non-ATPase regulatory subunit 5 | 121 | 55937 | 5.13 | 12 | 6 | 7 | 4 | 7 | 0.26 | 0.35 |
| FRIH_MOUSE | Ferritin heavy chain | 120 | 21053 | 5.53 | 9 | 6 | 5 | 3 | 5 | 0.48 | 0.8 |
| OSTF1_MOUSE | Osteoclast-stimulating factor 1 | 119 | 23768 | 5.46 | 6 | 2 | 4 | 1 | 4 | 0.19 | 0.19 |
| PFD5_MOUSE | Prefoldin subunit 5 | 119 | 17345 | 5.93 | 7 | 4 | 4 | 3 | 4 | 0.47 | 1.04 |
| PCKGM_MOUSE | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | 119 | 70482 | 6.92 | 16 | 7 | 10 | 7 | 10 | 0.26 | 0.51 |
| SAE2_MOUSE | SUMO-activating enzyme subunit 2 | 119 | 70525 | 5.09 | 8 | 6 | 5 | 4 | 5 | 0.21 | 0.27 |
| NSF_MOUSE | Vesicle-fusing ATPase | 119 | 82561 | 6.52 | 7 | 3 | 7 | 3 | 7 | 0.17 | 0.16 |
| THY1_MOUSE | Thy-1 membrane glycoprotein | 118 | 18069 | 9.16 | 9 | 2 | 3 | 1 | 3 | 0.25 | 0.26 |
| RRBP1_MOUSE | Ribosome-binding protein 1 | 117 | 172776 | 9.35 | 12 | 3 | 9 | 3 | 9 | 0.1 | 0.08 |
| CYB5_MOUSE | Cytochrome b5 | 117 | 15232 | 4.96 | 9 | 4 | 3 | 1 | 3 | 0.43 | 1.94 |
| ECI2_MOUSE | Enoyl-CoA delta isomerase 2, mitochondrial | 117 | 43240 | 9.08 | 4 | 3 | 3 | 2 | 3 | 0.13 | 0.21 |
| TCAF2_MOUSE | TRPM8 channel-associated factor 2 | 117 | 101532 | 6.07 | 5 | 5 | 3 | 3 | 3 | 0.06 | 0.13 |
| RCN1_MOUSE | Reticulocalbin-1 | 117 | 38090 | 4.7 | 9 | 6 | 5 | 3 | 5 | 0.27 | 0.39 |
| ELP1_MOUSE | Elongator complex protein 1 | 117 | 149489 | 5.67 | 9 | 2 | 6 | 1 | 6 | 0.09 | 0.03 |
| NHLC2_MOUSE | NHL repeat-containing protein 2 | 117 | 78381 | 5.33 | 10 | 6 | 3 | 2 | 3 | 0.12 | 0.17 |
| NQO2_MOUSE | Ribosyldihydronicotinamide dehydrogenase [quinone] | 117 | 26231 | 6.54 | 6 | 4 | 4 | 3 | 4 | 0.32 | 0.61 |
| C1TC_MOUSE | C-1-tetrahydrofolate synthase, cytoplasmic | 116 | 101136 | 6.7 | 12 | 7 | 6 | 4 | 6 | 0.16 | 0.18 |
| TWF1_MOUSE | Twinfilin-1 | 115 | 40054 | 6.21 | 7 | 3 | 7 | 3 | 6 | 0.36 | 0.37 |
| HPCL1_MOUSE | Hippocalcin-like protein 1 | 115 | 22324 | 5.32 | 5 | 2 | 5 | 2 | 5 | 0.43 | 0.45 |
| PEBP1_MOUSE | Phosphatidylethanolamine-binding protein 1 | 114 | 20817 | 5.19 | 16 | 6 | 5 | 3 | 5 | 0.57 | 0.81 |
| FSTL1_MOUSE | Follistatin-related protein 1 | 114 | 34532 | 5.58 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.13 |
| TIGAR_MOUSE | Fructose-2,6-bisphosphatase TIGAR | 114 | 29172 | 8.45 | 3 | 2 | 2 | 1 | 2 | 0.16 | 0.15 |
| HMOX1_MOUSE | Heme oxygenase 1 | 113 | 32908 | 6.08 | 14 | 6 | 4 | 3 | 4 | 0.3 | 0.46 |
| FKB1A_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP1A | 112 | 11915 | 7.88 | 10 | 5 | 4 | 3 | 4 | 0.66 | 1.79 |
| GT251_MOUSE | Procollagen galactosyltransferase 1 | 112 | 71015 | 6.83 | 10 | 5 | 8 | 4 | 8 | 0.18 | 0.27 |
| CD81_MOUSE | CD81 antigen | 111 | 25797 | 5.54 | 5 | 4 | 2 | 2 | 2 | 0.18 | 0.38 |
| IDH3A_MOUSE | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | 111 | 39613 | 6.27 | 4 | 3 | 3 | 2 | 3 | 0.16 | 0.23 |
| VMA5A_MOUSE | von Willebrand factor A domain-containing protein 5A | 111 | 87087 | 6.15 | 6 | 5 | 4 | 3 | 4 | 0.09 | 0.15 |
| LONM_MOUSE | Lon protease homolog, mitochondrial | 111 | 105776 | 6.15 | 9 | 4 | 6 | 2 | 6 | 0.16 | 0.08 |
| ECHB_MOUSE | Trifunctional enzyme subunit beta, mitochondrial | 111 | 51353 | 9.43 | 10 | 2 | 6 | 2 | 6 | 0.23 | 0.18 |
| SRPRA_MOUSE | Signal recognition particle receptor subunit alpha | 110 | 69579 | 9.07 | 6 | 3 | 5 | 2 | 5 | 0.12 | 0.13 |
| EIF3D_MOUSE | Eukaryotic translation initiation factor 3 subunit D | 109 | 63948 | 5.79 | 8 | 4 | 5 | 2 | 5 | 0.2 | 0.14 |
| CX6A1_MOUSE | Cytochrome c oxidase subunit 6A1, mitochondrial | 109 | 12344 | 9.97 | 5 | 4 | 2 | 1 | 2 | 0.45 | 0.39 |
| RISC_MOUSE | Retinoid-inducible serine carboxypeptidase | 108 | 50932 | 5.48 | 16 | 8 | 10 | 6 | 10 | 0.3 | 0.77 |
| PRDX2_MOUSE | Peroxiredoxin-2 | 108 | 21765 | 5.2 | 12 | 6 | 6 | 4 | 6 | 0.46 | 1.13 |
| RL18_MOUSE | 60S ribosomal protein L18 | 108 | 21631 | 11.79 | 2 | 2 | 2 | 2 | 2 | 0.14 | 0.46 |
| PTGR3_MOUSE | Prostaglandin reductase-3 | 108 | 40503 | 7.01 | 6 | 5 | 3 | 3 | 3 | 0.23 | 0.51 |
| PPID_MOUSE | Peptidyl-prolyl cis-trans isomerase D | 108 | 40717 | 7.08 | 4 | 2 | 3 | 1 | 3 | 0.15 | 0.11 |
| SGTA_MOUSE | Small glutamine-rich tetratricopeptide repeat-containing protein alpha | 106 | 34301 | 4.99 | 7 | 6 | 2 | 2 | 2 | 0.05 | 0.27 |
| HYEP_MOUSE | Epoxide hydrolase 1 | 106 | 52543 | 8.43 | 28 | 6 | 15 | 3 | 15 | 0.43 | 0.37 |
| RAGP1_MOUSE | Ran GTPase-activating protein 1 | 105 | 63491 | 4.59 | 10 | 5 | 8 | 5 | 8 | 0.21 | 0.39 |
| NCBP1_MOUSE | Nuclear cap-binding protein subunit 1 | 105 | 91868 | 6.17 | 9 | 3 | 8 | 2 | 8 | 0.15 | 0.1 |
| FKBP9_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP9 | 104 | 62956 | 5.03 | 10 | 5 | 6 | 4 | 6 | 0.17 | 0.3 |
| NB5R3_MOUSE | NADH-cytochrome b5 reductase 3 | 104 | 34106 | 8.55 | 13 | 5 | 7 | 5 | 7 | 0.55 | 0.84 |
| CX6B1_MOUSE | Cytochrome c oxidase subunit 6B1 | 104 | 10065 | 8.96 | 4 | 3 | 2 | 2 | 2 | 0.29 | 1.23 |
| COMT_MOUSE | Catechol O-methyltransferase | 104 | 29467 | 5.52 | 10 | 7 | 4 | 3 | 4 | 0.24 | 0.53 |
| BIN1_MOUSE | Myc box-dependent-interacting protein 1 | 103 | 64430 | 4.95 | 10 | 3 | 7 | 2 | 7 | 0.22 | 0.14 |
| AN32B_MOUSE | Acidic leucine-rich nuclear phosphoprotein 32 family member B | 102 | 31060 | 3.89 | 6 | 3 | 3 | 2 | 3 | 0.16 | 0.31 |
| TIF1B_MOUSE | Transcription intermediary factor 1-beta | 101 | 88791 | 5.52 | 6 | 4 | 3 | 3 | 3 | 0.06 | 0.15 |
| SH3L2_MOUSE | SH3 domain-binding glutamic acid-rich-like protein 2 | 101 | 12247 | 5.48 | 3 | 3 | 2 | 2 | 2 | 0.26 | 0.95 |
| ERO1A_MOUSE | ERO1-like protein alpha | 101 | 54050 | 6.12 | 8 | 5 | 5 | 4 | 5 | 0.15 | 0.36 |
| LMNA_MOUSE | Prelamin-A/C | 101 | 74193 | 6.54 | 16 | 4 | 11 | 3 | 11 | 0.26 | 0.18 |
| ODPB_MOUSE | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | 101 | 38912 | 6.41 | 6 | 4 | 5 | 4 | 5 | 0.29 | 0.53 |
| MPPA_MOUSE | Mitochondrial-processing peptidase subunit alpha | 100 | 58242 | 6.36 | 2 | 1 | 1 | 1 | 1 | 0.03 | 0.07 |
| EIF2A_MOUSE | Eukaryotic translation initiation factor 2A | 100 | 64363 | 9.04 | 8 | 4 | 6 | 3 | 6 | 0.2 | 0.21 |
| GSHR_MOUSE | Glutathione reductase, mitochondrial | 100 | 53629 | 8.19 | 5 | 3 | 3 | 1 | 3 | 0.15 | 0.08 |
| ADPRH_MOUSE | [Protein ADP-ribosylarginine] hydrolase | 99 | 40042 | 5.46 | 8 | 5 | 3 | 3 | 3 | 0.22 | 0.37 |
| DDX58_MOUSE | Probable ATP-dependent RNA helicase DDX58 | 99 | 105908 | 6.23 | 9 | 4 | 4 | 2 | 4 | 0.09 | 0.08 |
| AP1M1_MOUSE | AP-1 complex subunit mu-1 | 99 | 48512 | 6.82 | 4 | 3 | 2 | 2 | 2 | 0.09 | 0.19 |
| MVP_MOUSE | Major vault protein | 98 | 95865 | 5.43 | 11 | 4 | 7 | 3 | 7 | 0.18 | 0.14 |
| COPE_MOUSE | Coatomer subunit epsilon | 98 | 34545 | 4.94 | 24 | 6 | 6 | 2 | 6 | 0.4 | 0.27 |
| DPP2_MOUSE | Dipeptidyl peptidase 2 | 98 | 56218 | 5.17 | 3 | 2 | 2 | 1 | 2 | 0.07 | 0.08 |
| PURA2_MOUSE | Adenylosuccinate synthetase isozyme 2 | 98 | 49990 | 5.98 | 12 | 3 | 7 | 3 | 7 | 0.25 | 0.28 |
| PGS2_MOUSE | Decorin | 98 | 39784 | 8.87 | 5 | 2 | 3 | 1 | 3 | 0.15 | 0.11 |
| COPB2_MOUSE | Coatomer subunit beta ′ | 98 | 102384 | 5.17 | 19 | 8 | 12 | 6 | 12 | 0.27 | 0.28 |
| HCDH_MOUSE | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 97 | 34442 | 8.76 | 5 | 3 | 4 | 2 | 4 | 0.23 | 0.27 |
| CTBP1_MOUSE | C-terminal-binding protein 1 | 97 | 47715 | 6.28 | 6 | 3 | 3 | 1 | 3 | 0.12 | 0.09 |
| BAG6_MOUSE | Large proline-rich protein BAG6 | 97 | 120962 | 5.46 | 6 | 2 | 4 | 2 | 4 | 0.08 | 0.07 |
| MAP1B_MOUSE | Microtubule-associated protein 1B | 97 | 270089 | 4.76 | 16 | 5 | 13 | 3 | 13 | 0.11 | 0.05 |
| ATX10_MOUSE | Ataxin-10 | 97 | 53673 | 5.12 | 11 | 5 | 5 | 3 | 5 | 0.19 | 0.26 |
| PRRC1_MOUSE | Protein PRRC1 | 97 | 46268 | 5.62 | 9 | 4 | 6 | 3 | 6 | 0.28 | 0.31 |
| RRAS_MOUSE | Ras-related protein R-Ras | 97 | 23749 | 6.32 | 4 | 3 | 3 | 2 | 3 | 0.22 | 0.42 |
| CPT1A_MOUSE | Carnitine O-palmitoyltransferase 1, liver isoform | 97 | 88195 | 8.83 | 4 | 2 | 2 | 2 | 2 | 0.09 | 0.1 |
| SPRE_MOUSE | Sepiapterin reductase | 97 | 27865 | 5.58 | 3 | 3 | 2 | 2 | 2 | 0.22 | 0.35 |
| NEK7_MOUSE | Serine/threonine-protein kinase Nek7 | 96 | 34514 | 8.49 | 3 | 2 | 2 | 1 | 2 | 0.16 | 0.13 |
| FACE1_MOUSE | CAAX prenyl protease 1 homolog | 95 | 54699 | 6.49 | 6 | 2 | 4 | 1 | 4 | 0.17 | 0.08 |
| STAM2_MOUSE | Signal transducing adapter molecule 2 | 95 | 57419 | 4.94 | 7 | 6 | 3 | 2 | 3 | 0.15 | 0.24 |
| TMED5_MOUSE | Transmembrane emp24 domain-containing protein 5 | 94 | 26155 | 4.81 | 3 | 3 | 1 | 1 | 1 | 0.12 | 0.17 |
| SYDC_MOUSE | Aspartate–tRNA ligase, cytoplasmic | 94 | 57111 | 6.07 | 13 | 3 | 9 | 3 | 9 | 0.28 | 0.24 |
| ACADM_MOUSE | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 94 | 46452 | 8.6 | 3 | 3 | 2 | 2 | 2 | 0.11 | 0.2 |
| PDCL3_MOUSE | Phosducin-like protein 3 | 94 | 27564 | 4.66 | 7 | 5 | 4 | 3 | 4 | 0.36 | 0.57 |
| SYWC_MOUSE | Tryptophan–tRNA ligase, cytoplasmic | 94 | 54323 | 6.44 | 10 | 4 | 5 | 1 | 5 | 0.2 | 0.08 |
| NIBL1_MOUSE | Niban-like protein 1 | 94 | 84765 | 5.65 | 14 | 8 | 7 | 5 | 7 | 0.15 | 0.34 |
| CYB5B_MOUSE | Cytochrome b5 type B | 94 | 16308 | 4.79 | 3 | 2 | 1 | 1 | 1 | 0.23 | 0.29 |
| TNPO1_MOUSE | Transportin-1 | 93 | 102291 | 4.84 | 17 | 9 | 12 | 6 | 9 | 0.28 | 0.28 |
| SCFD1_MOUSE | Sec1 family domain-containing protein 1 | 93 | 72277 | 5.98 | 8 | 3 | 5 | 3 | 5 | 0.19 | 0.19 |
| PRDX5_MOUSE | Peroxiredoxin-5, mitochondrial | 93 | 21884 | 9.1 | 13 | 3 | 7 | 3 | 7 | 0.48 | 0.76 |
| PUR8_MOUSE | Adenylosuccinate lyase | 92 | 54831 | 6.9 | 7 | 5 | 6 | 4 | 6 | 0.23 | 0.36 |
| IAH1_MOUSE | Isoamyl acetate-hydrolyzing esterase 1 homolog | 92 | 27956 | 5.34 | 3 | 2 | 1 | 1 | 1 | 0.07 | 0.16 |
| VA0D1_MOUSE | V-type proton ATPase subunit d 1 | 91 | 40275 | 4.89 | 5 | 3 | 5 | 3 | 5 | 0.33 | 0.36 |
| CMTD1_MOUSE | Catechol O-methyltransferase domain-containing protein 1 | 91 | 28943 | 8.61 | 2 | 2 | 1 | 1 | 1 | 0.1 | 0.15 |
| AIFM1_MOUSE | Apoptosis-inducing factor 1, mitochondrial | 90 | 66724 | 9.23 | 4 | 2 | 3 | 2 | 3 | 0.1 | 0.13 |
| MSMO1_MOUSE | Methylsterol monooxygenase 1 | 90 | 34750 | 7.29 | 8 | 3 | 5 | 2 | 5 | 0.31 | 0.43 |
| PON2_MOUSE | Serum paraoxonase/arylesterase 2 | 90 | 39592 | 5.49 | 2 | 1 | 2 | 1 | 2 | 0.1 | 0.11 |
| LTOR3_MOUSE | Ragulator complex protein LAMTOR3 | 90 | 13544 | 6.73 | 4 | 3 | 3 | 2 | 3 | 0.4 | 0.83 |
| ACACA_MOUSE | Acetyl-CoA carboxylase 1 | 90 | 265088 | 5.97 | 12 | 4 | 10 | 3 | 10 | 0.09 | 0.07 |
| PGPI_MOUSE | Pyroglutamyl-peptidase 1 | 90 | 22919 | 5.21 | 4 | 4 | 2 | 2 | 2 | 0.21 | 0.43 |
| CTND1_MOUSE | Catenin delta-1 | 89 | 104860 | 6.41 | 9 | 6 | 5 | 2 | 5 | 0.12 | 0.08 |
| NDUA9_MOUSE | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | 89 | 42498 | 9.75 | 6 | 4 | 4 | 2 | 4 | 0.16 | 0.22 |
| PMM2_MOUSE | Phosphomannomutase 2 | 89 | 27639 | 6.01 | 2 | 2 | 1 | 1 | 1 | 0.11 | 0.16 |
| FAAA_MOUSE | Fumarylacetoacetase | 89 | 46146 | 6.7 | 4 | 4 | 3 | 3 | 3 | 0.16 | 0.31 |
| TCP4_MOUSE | Activated RNA polymerase II transcriptional coactivator p15 | 89 | 14418 | 9.6 | 2 | 2 | 1 | 1 | 1 | 0.19 | 0.33 |
| IDHP_MOUSE | Isocitrate dehydrogenase [NADP], mitochondrial | 89 | 50874 | 8.88 | 12 | 4 | 7 | 3 | 6 | 0.27 | 0.39 |
| DUS3_MOUSE | Dual specificity protein phosphatase 3 | 89 | 20459 | 6.07 | 2 | 1 | 2 | 1 | 2 | 0.19 | 0.22 |
| LIMA1_MOUSE | LIM domain and actin-binding protein 1 | 88 | 84008 | 6.18 | 6 | 2 | 4 | 1 | 4 | 0.09 | 0.05 |
| MESD_MOUSE | LRP chaperone MESD | 88 | 25191 | 6.06 | 4 | 3 | 3 | 2 | 3 | 0.23 | 0.39 |
| CYGB_MOUSE | Cytoglobin | 88 | 21452 | 6.32 | 2 | 2 | 1 | 1 | 1 | 0.13 | 0.21 |
| MA2A1_MOUSE | Alpha-mannosidase 2 | 88 | 131548 | 8.17 | 9 | 4 | 8 | 4 | 8 | 0.14 | 0.14 |
| DPP3_MOUSE | Dipeptidyl peptidase 3 | 88 | 82846 | 5.26 | 11 | 8 | 7 | 5 | 7 | 0.22 | 0.29 |
| NDUC2_MOUSE | NADH dehydrogenase [ubiquinone] 1 subunit C2 | 87 | 14154 | 9.24 | 2 | 2 | 1 | 1 | 1 | 0.16 | 0.33 |
| CNPY4_MOUSE | Protein canopy homolog 4 | 86 | 28076 | 4.72 | 5 | 3 | 3 | 2 | 3 | 0.22 | 0.34 |
| CATH_MOUSE | Pro-cathepsin H | 86 | 37146 | 8.68 | 3 | 2 | 2 | 1 | 2 | 0.14 | 0.12 |
| PSMD8_MOUSE | 26S proteasome non-ATPase regulatory subunit 8 | 86 | 39905 | 9.61 | 9 | 3 | 6 | 2 | 6 | 0.4 | 0.23 |
| RCC1_MOUSE | Regulator of chromosome condensation | 86 | 44903 | 8.34 | 5 | 3 | 4 | 2 | 4 | 0.19 | 0.2 |
| ORN_MOUSE | Oligoribonuclease, mitochondrial | 86 | 26722 | 6.67 | 2 | 1 | 2 | 1 | 2 | 0.18 | 0.17 |
| G6PC3_MOUSE | Glucose-6-phosphatase 3 | 86 | 38756 | 8.42 | 2 | 2 | 2 | 2 | 2 | 0.1 | 0.24 |
| IMA5_MOUSE | Importin subunit alpha-5 | 86 | 60144 | 4.93 | 5 | 4 | 2 | 2 | 2 | 0.11 | 0.15 |
| PSMD4_MOUSE | 26S proteasome non-ATPase regulatory subunit 4 | 86 | 40678 | 4.67 | 7 | 4 | 4 | 2 | 4 | 0.17 | 0.23 |
| GSTT3_MOUSE | Glutathione S-transferase theta-3 | 85 | 27385 | 7.63 | 3 | 3 | 1 | 1 | 1 | 0.14 | 0.16 |
| PSA1_MOUSE | Proteasome subunit alpha type-1 | 85 | 29528 | 6 | 9 | 6 | 6 | 3 | 6 | 0.41 | 0.75 |
| GPNMB_MOUSE | Transmembrane glycoprotein NMB | 85 | 63635 | 7.55 | 6 | 4 | 3 | 3 | 3 | 0.06 | 0.22 |
| FUBP2_MOUSE | Far upstream element-binding protein 2 | 84 | 76728 | 6.9 | 9 | 3 | 6 | 2 | 6 | 0.12 | 0.12 |
| RS21_MOUSE | 40S ribosomal protein S21 | 84 | 9136 | 8.71 | 6 | 3 | 1 | 1 | 1 | 0.17 | 0.56 |
| ABRAL_MOUSE | Costars family protein ABRACL | 84 | 9025 | 5.52 | 5 | 1 | 2 | 1 | 2 | 0.46 | 0.56 |
| UMPS_MOUSE | Uridine 5 ′-monophosphate synthase | 84 | 52259 | 6.17 | 7 | 3 | 3 | 2 | 3 | 0.15 | 0.17 |
| PSA3_MOUSE | Proteasome subunit alpha type-3 | 84 | 28387 | 5.29 | 3 | 2 | 2 | 1 | 2 | 0.11 | 0.16 |
| HGS_MOUSE | Hepatocyte growth factor-regulated tyrosine kinase substrate | 83 | 85961 | 5.84 | 10 | 2 | 7 | 2 | 7 | 0.17 | 0.1 |
| RIR1_MOUSE | Ribonucleoside-diphosphate reductase large subunit | 83 | 90153 | 6.27 | 8 | 2 | 6 | 2 | 6 | 0.16 | 0.1 |
| PGH2_MOUSE | Prostaglandin G/H synthase 2 | 83 | 68969 | 7 | 9 | 3 | 6 | 2 | 6 | 0.21 | 0.13 |
| CSDE1_MOUSE | Cold shock domain-containing protein E1 | 83 | 88735 | 5.97 | 5 | 1 | 5 | 1 | 5 | 0.12 | 0.05 |
| SAE1_MOUSE | SUMO-activating enzyme subunit 1 | 82 | 38596 | 5.24 | 9 | 6 | 3 | 3 | 3 | 0.17 | 0.38 |
| NPS3B_MOUSE | Protein NipSnap homolog 3B | 82 | 28290 | 9.51 | 3 | 3 | 1 | 1 | 1 | 0.11 | 0.16 |
| TBCD_MOUSE | Tubulin-specific chaperone D | 82 | 133236 | 6.08 | 5 | 4 | 4 | 3 | 4 | 0.08 | 0.1 |
| COR1C_MOUSE | Coronin-1C | 82 | 53087 | 6.65 | 7 | 3 | 5 | 3 | 5 | 0.17 | 0.27 |
| SCMC1_MOUSE | Calcium-binding mitochondrial carrier protein SCaMC-1 | 82 | 52868 | 7.02 | 9 | 4 | 4 | 1 | 4 | 0.15 | 0.08 |
| ECM29_MOUSE | Proteasome-associated protein ECM29 homolog | 82 | 203573 | 6.68 | 21 | 7 | 16 | 6 | 16 | 0.18 | 0.13 |
| LICH_MOUSE | Lysosomal acid lipase/cholesteryl ester hydrolase | 81 | 45296 | 8.16 | 4 | 3 | 3 | 2 | 3 | 0.15 | 0.2 |
| IMA7_MOUSE | Importin subunit alpha-7 | 81 | 59926 | 4.86 | 9 | 4 | 6 | 4 | 6 | 0.29 | 0.32 |
| CLPT1_MOUSE | Cleft lip and palate transmembrane protein 1 homolog | 81 | 75243 | 5.88 | 4 | 1 | 3 | 1 | 3 | 0.11 | 0.06 |
| RWDD1_MOUSE | RWD domain-containing protein 1 | 81 | 27768 | 4.18 | 6 | 4 | 3 | 2 | 3 | 0.3 | 0.35 |
| DDX21_MOUSE | Nucleolar RNA helicase 2 | 80 | 93493 | 9.19 | 7 | 5 | 3 | 2 | 3 | 0.08 | 0.09 |
| XPO5_MOUSE | Exportin-5 | 80 | 136883 | 5.57 | 16 | 8 | 7 | 3 | 7 | 0.11 | 0.13 |
| SDCB1_MOUSE | Syntenin-1 | 80 | 32359 | 6.66 | 9 | 5 | 4 | 2 | 4 | 0.25 | 0.29 |
| GLOD4_MOUSE | Glyoxalase domain-containing protein 4 | 80 | 33296 | 5.28 | 8 | 5 | 3 | 2 | 3 | 0.19 | 0.28 |
| PLD3_MOUSE | Phospholipase D3 | 80 | 54354 | 6.07 | 10 | 4 | 7 | 3 | 7 | 0.34 | 0.36 |
| LARP1_MOUSE | La-related protein 1 | 80 | 121050 | 8.87 | 6 | 3 | 4 | 1 | 4 | 0.05 | 0.04 |
| NDUAA_MOUSE | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | 80 | 40578 | 7.63 | 2 | 2 | 2 | 2 | 2 | 0.09 | 0.23 |
| EIF3C_MOUSE | Eukaryotic translation initiation factor 3 subunit C | 80 | 105465 | 5.55 | 14 | 4 | 10 | 3 | 10 | 0.18 | 0.13 |
| GNPI1_MOUSE | Glucosamine-6-phosphate isomerase 1 | 79 | 32528 | 6.13 | 5 | 3 | 3 | 3 | 3 | 0.21 | 0.47 |
| ADDA_MOUSE | Alpha-adducin | 79 | 80596 | 5.62 | 6 | 2 | 4 | 1 | 4 | 0.12 | 0.05 |
| SEPT5_MOUSE | Septin-5 | 79 | 42721 | 6.21 | 16 | 5 | 9 | 4 | 9 | 0.42 | 0.63 |
| TADBP_MOUSE | TAR DNA-binding protein 43 | 79 | 44519 | 6.26 | 3 | 3 | 1 | 1 | 1 | 0.07 | 0.1 |
| MAP4_MOUSE | Microtubule-associated protein 4 | 79 | 117357 | 4.9 | 7 | 2 | 5 | 1 | 5 | 0.09 | 0.04 |
| RAB9A_MOUSE | Ras-related protein Rab-9A | 79 | 22895 | 5.43 | 4 | 4 | 2 | 2 | 2 | 0.19 | 0.43 |
| AGAL_MOUSE | Alpha-galactosidase A | 79 | 47611 | 5.44 | 6 | 4 | 5 | 3 | 5 | 0.21 | 0.3 |
| ULA1_MOUSE | NEDD8-activating enzyme E1 regulatory subunit | 78 | 60236 | 5.34 | 4 | 3 | 3 | 2 | 3 | 0.15 | 0.15 |
| EI2BA_MOUSE | Translation initiation factor eIF-2B subunit alpha | 78 | 33795 | 8.48 | 4 | 2 | 3 | 2 | 3 | 0.17 | 0.28 |
| LPPRC_MOUSE | Leucine-rich PPR motif-containing protein, mitochondrial | 78 | 156516 | 6.42 | 10 | 6 | 7 | 4 | 7 | 0.12 | 0.11 |
| SYK_MOUSE | Lysine–tRNA ligase | 78 | 67796 | 5.65 | 10 | 5 | 6 | 4 | 6 | 0.17 | 0.28 |
| SWI5_MOUSE | DNA repair protein SWI5 homolog | 78 | 10254 | 4.67 | 1 | 1 | 1 | 1 | 1 | 0.19 | 0.49 |
| FAM3C_MOUSE | Protein FAM3C | 77 | 24737 | 8.52 | 5 | 2 | 2 | 1 | 2 | 0.13 | 0.18 |
| BLMH_MOUSE | Bleomycin hydrolase | 77 | 52477 | 6.04 | 8 | 3 | 6 | 2 | 6 | 0.26 | 0.17 |
| TOIP1_MOUSE | Torsin-1A-interacting protein 1 | 77 | 66740 | 6.58 | 2 | 2 | 2 | 2 | 2 | 0.04 | 0.13 |
| NUDC_MOUSE | Nuclear migration protein nudC | 77 | 38334 | 5.17 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.11 |
| GSHB_MOUSE | Glutathione synthetase | 77 | 52214 | 5.56 | 4 | 3 | 3 | 2 | 3 | 0.14 | 0.17 |
| SPTC2_MOUSE | Serine palmitoyltransferase 2 | 77 | 62941 | 8.43 | 8 | 4 | 4 | 1 | 4 | 0.14 | 0.07 |
| CP20A_MOUSE | Cytochrome P450 20A1 | 77 | 52116 | 6.48 | 4 | 2 | 3 | 1 | 3 | 0.13 | 0.08 |
| USP9X_MOUSE | Probable ubiquitin carboxyl-terminal hydrolase FAF-X | 76 | 290526 | 5.57 | 19 | 6 | 10 | 3 | 10 | 0.08 | 0.04 |
| ASPH_MOUSE | Aspartyl/asparaginyl beta-hydroxylase | 75 | 82991 | 4.97 | 9 | 4 | 8 | 4 | 8 | 0.17 | 0.22 |
| ALDR_MOUSE | Aldose reductase | 76 | 35709 | 6.71 | 4 | 2 | 3 | 1 | 3 | 0.19 | 0.12 |
| MGAT1_MOUSE | Alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase | 76 | 51658 | 9.02 | 6 | 5 | 2 | 2 | 2 | 0.12 | 0.18 |
| ATP5H_MOUSE | ATP synthase subunit d, mitochondrial | 76 | 18738 | 5.52 | 5 | 3 | 4 | 3 | 4 | 0.42 | 0.93 |
| AT5F1_MOUSE | ATP synthase F(0) complex subunit B1, mitochondrial | 76 | 28930 | 9.11 | 5 | 1 | 5 | 1 | 5 | 0.34 | 0.15 |
| IF2B_MOUSE | Eukaryotic translation initiation factor 2 subunit 2 | 76 | 38068 | 5.61 | 6 | 3 | 4 | 2 | 4 | 0.19 | 0.24 |
| HEAT3_MOUSE | HEAT repeat-containing protein 3 | 75 | 74258 | 4.91 | 5 | 3 | 3 | 2 | 3 | 0.08 | 0.12 |
| LCLT1_MOUSE | Lysocardiolipin acyltransferase 1 | 75 | 44371 | 8.77 | 2 | 2 | 2 | 2 | 2 | 0.14 | 0.21 |
| MCM6_MOUSE | DNA replication licensing factor MCM6 | 75 | 92809 | 5.32 | 4 | 3 | 4 | 3 | 4 | 0.09 | 0.14 |
| NDUA8_MOUSE | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | 75 | 19979 | 8.76 | 3 | 2 | 2 | 1 | 2 | 0.23 | 0.23 |
| POSTN_MOUSE | Periostin | 75 | 93085 | 7.27 | 11 | 4 | 6 | 2 | 6 | 0.18 | 0.09 |
| ACDSB_MOUSE | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial | 74 | 47843 | 8 | 4 | 1 | 3 | 1 | 3 | 0.13 | 0.09 |
| EIF1_MOUSE | Eukaryotic translation initiation factor 1 | 74 | 12739 | 6.89 | 2 | 1 | 2 | 1 | 2 | 0.33 | 0.38 |
| IF1A_MOUSE | Eukaryotic translation initiation factor 1A | 73 | 16492 | 5.07 | 4 | 1 | 4 | 1 | 1 | 0.26 | 0.28 |
| CHMP5_MOUSE | Charged multivesicular body protein 5 | 73 | 24560 | 4.65 | 2 | 2 | 1 | 1 | 1 | 0.12 | 0.18 |
| S10A4_MOUSE | Protein S100-A4 | 73 | 11714 | 5.23 | 6 | 5 | 3 | 3 | 3 | 0.25 | 1.82 |
| ITM2C_MOUSE | Integral membrane protein 2C | 73 | 30463 | 8.83 | 2 | 2 | 1 | 1 | 1 | 0.08 | 0.15 |
| MEP50_MOUSE | Methylosome protein 50 | 73 | 36919 | 5.09 | 2 | 2 | 1 | 1 | 1 | 0.09 | 0.12 |
| NDUS8_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial | 73 | 24023 | 5.89 | 3 | 3 | 2 | 2 | 2 | 0.09 | 0.41 |
| ZFPL1_MOUSE | Zinc finger protein-like 1 | 73 | 34133 | 8.57 | 7 | 2 | 3 | 2 | 3 | 0.2 | 0.27 |
| USMG5_MOUSE | Upregulated during skeletal muscle growth protein 5 | 72 | 6377 | 9.84 | 2 | 2 | 1 | 1 | 1 | 0.26 | 0.87 |
| VATL_MOUSE | V-type proton ATPase 16 kDa proteolipid subunit | 72 | 15798 | 9.1 | 1 | 1 | 1 | 1 | 1 | 0.2 | 0.3 |
| SFRP1_MOUSE | Secreted frizzled-related protein 1 | 72 | 35389 | 9.17 | 3 | 2 | 2 | 1 | 2 | 0.12 | 0.12 |
| AAAS_MOUSE | Aladin | 72 | 59393 | 6.39 | 3 | 3 | 1 | 1 | 1 | 0.03 | 0.07 |
| ABD12_MOUSE | Monoacylglycerol lipase ABHD12 | 72 | 45241 | 8.9 | 3 | 1 | 2 | 1 | 2 | 0.07 | 0.1 |
| SYIM_MOUSE | Isoleucine–tRNA ligase, mitochondrial | 72 | 112732 | 6.37 | 7 | 5 | 5 | 3 | 5 | 0.11 | 0.12 |
| MAPK2_MOUSE | MAP kinase-activated protein kinase 2 | 71 | 44022 | 8.77 | 7 | 2 | 5 | 2 | 5 | 0.19 | 0.21 |
| BIEA_MOUSE | Biliverdin reductase A | 71 | 33504 | 6.53 | 6 | 5 | 4 | 4 | 4 | 0.25 | 0.64 |
| AP3D1_MOUSE | AP-3 complex subunit delta-1 | 71 | 134996 | 7.04 | 4 | 3 | 3 | 2 | 3 | 0.04 | 0.06 |
| ALG5_MOUSE | Dolichyl-phosphate beta-glucosyltransferase | 71 | 36767 | 8.83 | 4 | 2 | 2 | 2 | 2 | 0.12 | 0.25 |
| CD99_MOUSE | CD99 antigen | 71 | 16772 | 5.14 | 2 | 2 | 1 | 1 | 1 | 0.27 | 0.28 |
| SYQ_MOUSE | Glutamine–tRNA ligase | 70 | 87621 | 6.93 | 8 | 2 | 5 | 2 | 5 | 0.12 | 0.1 |
| TCEA1_MOUSE | Transcription elongation factor A protein 1 | 70 | 33859 | 8.64 | 5 | 5 | 1 | 1 | 1 | 0.09 | 0.13 |
| XDH_MOUSE | Xanthine dehydrogenase/oxidase | 70 | 146468 | 7.62 | 14 | 6 | 6 | 3 | 6 | 0.1 | 0.12 |
| ANXA7_MOUSE | Annexin A7 | 70 | 49893 | 5.91 | 5 | 2 | 5 | 2 | 5 | 0.16 | 0.18 |
| IPYR2_MOUSE | Inorganic pyrophosphatase 2, mitochondrial | 70 | 38090 | 6.51 | 8 | 3 | 5 | 2 | 5 | 0.16 | 0.24 |
| CY1_MOUSE | Cytochrome c1, heme protein, mitochondrial | 70 | 35305 | 9.24 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.12 |
| NP1L4_MOUSE | Nucleosome assembly protein 1-like 4 | 70 | 42653 | 4.56 | 6 | 3 | 5 | 3 | 4 | 0.24 | 0.34 |
| NU133_MOUSE | Nuclear pore complex protein Nup133 | 70 | 128539 | 5.06 | 5 | 2 | 5 | 2 | 5 | 0.09 | 0.07 |
| SIAS_MOUSE | Sialic acid synthase | 70 | 39998 | 6.61 | 2 | 1 | 1 | 1 | 1 | 0.1 | 0.11 |
| ETFA_MOUSE | Electron transfer flavoprotein subunit alpha, mitochondrial | 69 | 34988 | 8.62 | 10 | 5 | 5 | 3 | 5 | 0.33 | 0.43 |
| GALM_MOUSE | Aldose 1-epimerase | 69 | 37775 | 6.26 | 3 | 1 | 2 | 1 | 2 | 0.12 | 0.12 |
| ABCF1_MOUSE | ATP-binding cassette sub-family F member 1 | 69 | 94887 | 6.15 | 3 | 2 | 2 | 1 | 2 | 0.06 | 0.05 |
| T176B_MOUSE | Transmembrane protein 176B | 68 | 28349 | 8.03 | 2 | 1 | 2 | 1 | 2 | 0.22 | 0.16 |
| ICAL_MOUSE | Calpastatin | 68 | 84871 | 5.37 | 6 | 3 | 4 | 2 | 4 | 0.1 | 0.1 |
| THIM_MOUSE | 3-Ketoacyl-CoA thiolase, mitochondrial | 68 | 41803 | 8.33 | 3 | 1 | 3 | 1 | 3 | 0.13 | 0.1 |
| CAPR1_MOUSE | Caprin-1 | 67 | 78121 | 5.14 | 7 | 2 | 5 | 2 | 5 | 0.11 | 0.11 |
| PTPA_MOUSE | Serine/threonine-protein phosphatase 2A activator | 67 | 36687 | 5.95 | 2 | 2 | 2 | 2 | 2 | 0.11 | 0.25 |
| ATP5L_MOUSE | ATP synthase subunit g, mitochondrial | 67 | 11417 | 9.74 | 3 | 2 | 2 | 1 | 2 | 0.32 | 0.43 |
| UBA6_MOUSE | Ubiquitin-like modifier-activating enzyme 6 | 67 | 117891 | 5.75 | 7 | 1 | 6 | 1 | 6 | 0.11 | 0.04 |
| VASP_MOUSE | Vasodilator-stimulated phosphoprotein | 67 | 39642 | 8.69 | 5 | 2 | 3 | 2 | 3 | 0.08 | 0.23 |
| E41L2_MOUSE | Band 4.1-like protein 2 | 67 | 109873 | 5.31 | 8 | 4 | 5 | 3 | 5 | 0.07 | 0.12 |
| GLRX3_MOUSE | Glutaredoxin-3 | 67 | 37754 | 5.42 | 16 | 7 | 8 | 6 | 8 | 0.46 | 0.94 |
| TRXR1_MOUSE | Thioredoxin reductase 1, cytoplasmic | 66 | 67042 | 7.42 | 12 | 4 | 6 | 2 | 6 | 0.19 | 0.13 |
| IASPP_MOUSE | RelA-associated inhibitor | 66 | 88921 | 6.39 | 3 | 1 | 3 | 1 | 3 | 0.1 | 0.05 |
| NUP85_MOUSE | Nuclear pore complex protein Nup85 | 66 | 74728 | 5.37 | 6 | 3 | 4 | 1 | 4 | 0.09 | 0.06 |
| DDX18_MOUSE | ATP-dependent RNA helicase DDX18 | 66 | 74134 | 9.54 | 6 | 4 | 3 | 2 | 3 | 0.07 | 0.12 |
| NEUL_MOUSE | Neurolysin, mitochondrial | 66 | 80378 | 6.01 | 10 | 4 | 7 | 2 | 7 | 0.18 | 0.11 |
| MLEC_MOUSE | Malectin | 66 | 32322 | 5.73 | 8 | 3 | 6 | 2 | 6 | 0.37 | 0.47 |
| GPC4_MOUSE | Glypican-4 | 65 | 62546 | 5.96 | 3 | 1 | 3 | 1 | 3 | 0.13 | 0.07 |
| KIME_MOUSE | Mevalonate kinase | 65 | 41851 | 6.22 | 4 | 3 | 2 | 1 | 2 | 0.14 | 0.1 |
| EIF3L_MOUSE | Eukaryotic translation initiation factor 3 subunit L | 65 | 66570 | 6.01 | 5 | 3 | 4 | 2 | 4 | 0.13 | 0.13 |
| G3BP1_MOUSE | Ras GTPase-activating protein-binding protein 1 | 65 | 51797 | 5.41 | 10 | 3 | 9 | 3 | 9 | 0.31 | 0.27 |
| CTNB1_MOUSE | Catenin beta-1 | 64 | 85416 | 5.53 | 10 | 3 | 5 | 2 | 5 | 0.13 | 0.1 |
| ACADL_MOUSE | Long-chain specific acyl-CoA dehydrogenase, mitochondrial | 64 | 47877 | 8.53 | 10 | 3 | 5 | 3 | 3 | 0.17 | 0.3 |
| HXK1_MOUSE | Hexokinase-1 | 64 | 108234 | 6.37 | 7 | 3 | 6 | 3 | 6 | 0.09 | 0.12 |
| TF_MOUSE | Tissue factor | 64 | 32914 | 9.4 | 2 | 2 | 1 | 1 | 1 | 0.09 | 0.13 |
| MYEF2_MOUSE | Myelin expression factor 2 | 64 | 63254 | 8.96 | 3 | 1 | 3 | 1 | 3 | 0.05 | 0.07 |
| ANM5_MOUSE | Protein arginine N-methyltransferase 5 | 64 | 72634 | 5.99 | 16 | 3 | 7 | 1 | 7 | 0.25 | 0.12 |
| MED11_MOUSE | Mediator of RNA polymerase II transcription subunit 11 | 63 | 13123 | 5.71 | 3 | 3 | 1 | 1 | 1 | 0.21 | 0.86 |
| HEAT6_MOUSE | HEAT repeat-containing protein 6 | 63 | 128840 | 6.88 | 5 | 3 | 3 | 1 | 3 | 0.06 | 0.03 |
| NAMPT_MOUSE | Nicotinamide phosphoribosyltransferase | 63 | 55413 | 6.69 | 3 | 2 | 2 | 2 | 2 | 0.09 | 0.16 |
| MYO9B_MOUSE | Unconventional myosin-Ixb | 63 | 238685 | 8.82 | 3 | 1 | 3 | 1 | 3 | 0.03 | 0.02 |
| AL1L2_MOUSE | Mitochondrial 10-formyltetrahydrofolate dehydrogenase | 63 | 101526 | 5.93 | 5 | 1 | 4 | 1 | 4 | 0.08 | 0.04 |
| HSBP1_MOUSE | Heat shock factor-binding protein 1 | 63 | 8605 | 4.11 | 2 | 1 | 1 | 1 | 1 | 0.25 | 0.59 |
| MYG1_MOUSE | UPF0160 protein MYG1, mitochondrial | 63 | 42696 | 6.54 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.1 |
| RBM3_MOUSE | RNA-binding protein 3 | 62 | 16595 | 6.84 | 11 | 4 | 4 | 2 | 4 | 0.46 | 0.64 |
| SLIT3_MOUSE | Slit homolog 3 protein | 62 | 167617 | 7.95 | 13 | 2 | 7 | 2 | 7 | 0.11 | 0.05 |
| PPM1B_MOUSE | Protein phosphatase 1B | 62 | 42768 | 5.04 | 3 | 1 | 2 | 1 | 2 | 0.11 | 0.1 |
| DPEP1_MOUSE | Dipeptidase 1 | 62 | 45693 | 5.9 | 4 | 1 | 3 | 1 | 3 | 0.23 | 0.1 |
| F10A1_MOUSE | Hsc70-interacting protein | 62 | 41630 | 5.19 | 6 | 3 | 3 | 2 | 3 | 0.12 | 0.22 |
| MCFD2_MOUSE | Multiple coagulation factor deficiency protein 2 homolog | 62 | 16158 | 4.55 | 1 | 1 | 1 | 1 | 1 | 0.17 | 0.29 |
| RM12_MOUSE | 39S ribosomal protein L12, mitochondrial | 62 | 21695 | 9.34 | 8 | 2 | 3 | 1 | 3 | 0.2 | 0.46 |
| FDFT_MOUSE | Squalene synthase | 61 | 48123 | 5.91 | 5 | 2 | 2 | 2 | 2 | 0.06 | 0.19 |
| LGUL_MOUSE | Lactoylglutathione lyase | 61 | 20796 | 5.24 | 8 | 3 | 4 | 1 | 4 | 0.33 | 0.22 |
| CDK4_MOUSE | Cyclin-dependent kinase 4 | 54 | 33729 | 6.16 | 6 | 2 | 5 | 2 | 5 | 0.31 | 0.28 |
| CDC37_MOUSE | Hsp90 co-chaperone Cdc37 | 61 | 44565 | 5.24 | 4 | 2 | 3 | 2 | 2 | 0.13 | 0.21 |
| CDK1_MOUSE | Cyclin-dependent kinase 1 | 61 | 34085 | 8.39 | 10 | 6 | 5 | 3 | 5 | 0.27 | 0.44 |
| LETM1_MOUSE | Mitochondrial proton/calcium exchanger protein | 61 | 82937 | 6.16 | 7 | 1 | 6 | 1 | 6 | 0.15 | 0.05 |
| PGM1_MOUSE | Phosphoglucomutase-1 | 60 | 61380 | 6.14 | 6 | 3 | 6 | 3 | 6 | 0.29 | 0.23 |
| LAMP2_MOUSE | Lysosome-associated membrane glycoprotein 2 | 60 | 45652 | 7.05 | 12 | 4 | 8 | 3 | 8 | 0.25 | 0.31 |
| PKD2_MOUSE | Polycystin-2 | 60 | 108914 | 5.47 | 4 | 1 | 4 | 1 | 4 | 0.08 | 0.04 |
| UCHL1_MOUSE | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 60 | 24822 | 5.14 | 7 | 4 | 5 | 3 | 5 | 0.38 | 0.65 |
| COPD_MOUSE | Coatomer subunit delta | 60 | 57193 | 5.89 | 18 | 4 | 12 | 3 | 12 | 0.34 | 0.24 |
| IRGM1_MOUSE | Immunity-related GTPase family M protein 1 | 60 | 46522 | 8.56 | 4 | 3 | 2 | 1 | 2 | 0.13 | 0.09 |
| AKA12_MOUSE | A-kinase anchor protein 12 | 60 | 180586 | 4.39 | 11 | 2 | 5 | 2 | 5 | 0.05 | 0.05 |
| MP2K4_MOUSE | Dual specificity mitogen-activated protein kinase 4 | 60 | 44085 | 8.28 | 5 | 2 | 5 | 2 | 5 | 0.27 | 0.21 |
| NCLN_MOUSE | Nicalin | 59 | 62868 | 6.03 | 3 | 2 | 2 | 2 | 2 | 0.06 | 0.14 |
| LMF2_MOUSE | Lipase maturation factor 2 | 58 | 79947 | 9.99 | 4 | 3 | 2 | 2 | 2 | 0.06 | 0.11 |
| FHL1_MOUSE | Four and a half LIM domains protein 1 | 59 | 31867 | 8.76 | 6 | 3 | 3 | 2 | 3 | 0.17 | 0.3 |
| ARHG7_MOUSE | Rho guanine nucleotide exchange factor 7 | 59 | 96995 | 6.35 | 2 | 1 | 2 | 1 | 2 | 0.04 | 0.04 |
| PP14B_MOUSE | Protein phosphatase 1 regulatory subunit 14B | 59 | 15947 | 4.74 | 10 | 6 | 2 | 2 | 2 | 0.29 | 0.67 |
| MANF_MOUSE | Mesencephalic astrocyte-derived neurotrophic factor | 59 | 20361 | 8.34 | 12 | 4 | 3 | 3 | 3 | 0.21 | 0.84 |
| ACOT2_MOUSE | Acyl-coenzyme A thioesterase 2, mitochondrial | 59 | 49626 | 6.88 | 4 | 3 | 2 | 2 | 2 | 0.08 | 0.18 |
| S23IP_MOUSE | SEC23-interacting protein | 59 | 110711 | 5.63 | 11 | 6 | 6 | 3 | 6 | 0.14 | 0.12 |
| ERP29_MOUSE | Endoplasmic reticulum resident protein 29 | 58 | 28805 | 5.9 | 7 | 3 | 2 | 2 | 2 | 0.14 | 0.33 |
| CO6A1_MOUSE | Collagen alpha-1(VI) chain | 58 | 108422 | 5.2 | 5 | 2 | 4 | 1 | 4 | 0.1 | 0.04 |
| DBNL_MOUSE | Drebrin-like protein | 58 | 48670 | 4.9 | 4 | 2 | 2 | 1 | 2 | 0.1 | 0.09 |
| SCPDL_MOUSE | Saccharopine dehydrogenase-like oxidoreductase | 58 | 47099 | 8.86 | 6 | 3 | 3 | 1 | 3 | 0.2 | 0.09 |
| PP1R7_MOUSE | Protein phosphatase 1 regulatory subunit 7 | 58 | 41266 | 4.85 | 5 | 3 | 2 | 1 | 2 | 0.14 | 0.22 |
| MGST1_MOUSE | Microsomal glutathione S-transferase 1 | 58 | 17540 | 9.67 | 2 | 1 | 1 | 1 | 1 | 0.21 | 0.26 |
| MON2_MOUSE | Protein MON2 homolog | 58 | 188958 | 5.77 | 6 | 2 | 5 | 2 | 5 | 0.08 | 0.05 |
| PARK7_MOUSE | Protein/nucleic acid deglycase DJ-1 | 57 | 20008 | 6.32 | 3 | 2 | 3 | 2 | 3 | 0.27 | 0.51 |
| WASC4_MOUSE | WASH complex subunit 4 | 57 | 136283 | 6.99 | 5 | 2 | 5 | 2 | 5 | 0.09 | 0.06 |
| AKAP1_MOUSE | A-kinase anchor protein 1, mitochondrial | 57 | 92137 | 4.91 | 2 | 1 | 2 | 1 | 2 | 0.03 | 0.05 |
| MALD1_MOUSE | MARVEL domain-containing protein 1 | 57 | 19074 | 9.77 | 5 | 3 | 2 | 1 | 2 | 0.21 | 0.24 |
| SPEE_MOUSE | Spermidine synthase | 57 | 33973 | 5.31 | 7 | 3 | 5 | 3 | 5 | 0.3 | 0.44 |
| RFIP5_MOUSE | Rab11 family-interacting protein 5 | 57 | 69510 | 9.17 | 5 | 3 | 2 | 1 | 2 | 0.04 | 0.06 |
| EXOC1_MOUSE | Exocyst complex component 1 | 57 | 101825 | 6.09 | 6 | 3 | 5 | 2 | 5 | 0.13 | 0.09 |
| DOCK7_MOUSE | Dedicator of cytokinesis protein 7 | 56 | 241286 | 6.25 | 15 | 2 | 8 | 2 | 8 | 0.08 | 0.04 |
| NDUA4_MOUSE | Cytochrome c oxidase subunit NDUFA4 | 56 | 9321 | 9.52 | 4 | 2 | 2 | 1 | 2 | 0.46 | 0.54 |
| TGFI1_MOUSE | Transforming growth factor beta-1-induced transcript 1 protein | 56 | 50068 | 6.28 | 8 | 4 | 3 | 2 | 3 | 0.18 | 0.18 |
| HUWE1_MOUSE | E3 ubiquitin-protein ligase HUWE1 | 56 | 482332 | 5.1 | 10 | 2 | 9 | 1 | 9 | 0.04 | 0.01 |
| LBR_MOUSE | Lamin-B receptor | 56 | 71395 | 9.43 | 4 | 1 | 4 | 1 | 4 | 0.09 | 0.06 |
| MARC2_MOUSE | Mitochondrial amidoxime reducing component 2 | 56 | 38170 | 8.95 | 4 | 2 | 3 | 2 | 3 | 0.18 | 0.24 |
| CMC1_MOUSE | Calcium-binding mitochondrial carrier protein Aralar1 | 56 | 74523 | 8.43 | 4 | 3 | 2 | 2 | 2 | 0.05 | 0.12 |
| BAX_MOUSE | Apoptosis regulator BAX | 56 | 21381 | 4.86 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.21 |
| BASP1_MOUSE | Brain acid soluble protein 1 | 56 | 22074 | 4.5 | 14 | 2 | 1 | 1 | 1 | 0.14 | 0.21 |
| HINT1_MOUSE | Histidine triad nucleotide-binding protein 1 | 56 | 13768 | 6.36 | 4 | 1 | 4 | 1 | 4 | 0.6 | 0.35 |
| SCYL1_MOUSE | N-terminal kinase-like protein | 56 | 89104 | 6.03 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.05 |
| QCR2_MOUSE | Cytochrome b-c1 complex subunit 2, mitochondrial | 55 | 48205 | 9.26 | 2 | 1 | 2 | 1 | 2 | 0.08 | 0.09 |
| TMF1_MOUSE | TATA element modulatory factor | 55 | 121729 | 4.83 | 4 | 1 | 4 | 1 | 4 | 0.07 | 0.03 |
| RENT1_MOUSE | Regulator of nonsense transcripts 1 | 55 | 123889 | 6.18 | 7 | 2 | 6 | 2 | 6 | 0.13 | 0.07 |
| IKIP_MOUSE | Inhibitor of nuclear factor kappa-B kinase-interacting protein | 55 | 42505 | 5.03 | 6 | 4 | 4 | 3 | 4 | 0.19 | 0.34 |
| HPRT_MOUSE | Hypoxanthine-guanine phosphoribosyltransferase | 53 | 24555 | 6.21 | 6 | 4 | 3 | 3 | 2 | 0.32 | 0.66 |
| ISC2A_MOUSE | Isochorismatase domain-containing protein 2A | 55 | 22403 | 8.25 | 1 | 1 | 1 | 1 | 1 | 0.13 | 0.2 |
| NU4M_MOUSE | NADH-ubiquinone oxidoreductase chain 4 | 55 | 51847 | 9.42 | 3 | 1 | 2 | 1 | 2 | 0.04 | 0.08 |
| CTL2_MOUSE | Choline transporter-like protein 2 | 55 | 80057 | 9.06 | 5 | 4 | 1 | 1 | 1 | 0.02 | 0.05 |
| JIP4_MOUSE | C-Jun-amino-terminal kinase-interacting protein 4 | 54 | 146129 | 5.05 | 8 | 2 | 6 | 2 | 6 | 0.09 | 0.06 |
| 5NT3B_MOUSE | 7-Methylguanosine phosphate-specific 5 ′-nucleotidase | 54 | 34403 | 5.82 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.13 |
| LAP2A_MOUSE | Lamina-associated polypeptide 2, isoforms alpha/zeta | 54 | 75122 | 8.29 | 4 | 1 | 2 | 1 | 2 | 0.05 | 0.06 |
| EM55_MOUSE | 55 kDa erythrocyte membrane protein | 54 | 52194 | 6.72 | 6 | 3 | 4 | 3 | 4 | 0.16 | 0.27 |
| KAP0_MOUSE | cAMP-dependent protein kinase type I-alpha regulatory subunit | 54 | 43158 | 5.27 | 5 | 2 | 3 | 1 | 3 | 0.15 | 0.1 |
| GDIR2_MOUSE | Rho GDP-dissociation inhibitor 2 | 54 | 22836 | 4.97 | 7 | 4 | 4 | 3 | 4 | 0.26 | 0.72 |
| NDUS2_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial | 54 | 52592 | 6.52 | 4 | 1 | 3 | 1 | 3 | 0.14 | 0.08 |
| SYHC_MOUSE | Histidine–tRNA ligase, cytoplasmic | 53 | 57396 | 5.79 | 4 | 3 | 4 | 3 | 4 | 0.17 | 0.24 |
| IVD_MOUSE | Isovaleryl-CoA dehydrogenase, mitochondrial | 54 | 46296 | 8.53 | 10 | 2 | 4 | 2 | 4 | 0.26 | 0.2 |
| DHRS1_MOUSE | Dehydrogenase/reductase SDR family member 1 | 54 | 33983 | 8.66 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.13 |
| SRP68_MOUSE | Signal recognition particle subunit SRP68 | 53 | 70530 | 8.77 | 5 | 1 | 5 | 1 | 5 | 0.13 | 0.06 |
| PREP_MOUSE | Presequence protease, mitochondrial | 53 | 117297 | 6.76 | 6 | 4 | 6 | 4 | 6 | 0.13 | 0.15 |
| GUAA_MOUSE | GMP synthase [glutamine-hydrolyzing] | 53 | 76675 | 6.29 | 3 | 2 | 2 | 1 | 2 | 0.09 | 0.06 |
| NT5C_MOUSE | 5 ′(3 ′)-Deoxyribonucleotidase, cytosolic type | 53 | 23062 | 5.31 | 2 | 1 | 2 | 1 | 2 | 0.2 | 0.2 |
| DNMT1_MOUSE | DNA (cytosine-5)-methyltransferase 1 | 53 | 183074 | 7.92 | 6 | 1 | 6 | 1 | 6 | 0.08 | 0.02 |
| GALK1_MOUSE | Galactokinase | 53 | 42268 | 5.17 | 2 | 1 | 2 | 1 | 2 | 0.07 | 0.1 |
| PSD12_MOUSE | 26S proteasome non-ATPase regulatory subunit 12 | 53 | 52861 | 6.66 | 11 | 3 | 8 | 3 | 8 | 0.25 | 0.27 |
| GLMP_MOUSE | Glycosylated lysosomal membrane protein | 52 | 43776 | 5.73 | 4 | 1 | 3 | 1 | 3 | 0.18 | 0.1 |
| UD17C_MOUSE | UDP-glucuronosyltransferase 1-7C | 52 | 59719 | 8.64 | 6 | 3 | 5 | 3 | 5 | 0.18 | 0.23 |
| DHPR_MOUSE | Dihydropteridine reductase | 52 | 25554 | 7.67 | 3 | 1 | 2 | 1 | 2 | 0.19 | 0.18 |
| NAA25_MOUSE | N-Alpha-acetyltransferase 25, NatB auxiliary subunit | 52 | 111637 | 6.1 | 2 | 1 | 2 | 1 | 2 | 0.04 | 0.04 |
| LDAH_MOUSE | Lipid droplet-associated hydrolase | 52 | 37349 | 8.5 | 4 | 2 | 3 | 1 | 3 | 0.22 | 0.12 |
| TR10B_MOUSE | Tumor necrosis factor receptor superfamily member 10B | 52 | 42138 | 6.75 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.1 |
| BAG2_MOUSE | BAG family molecular chaperone regulator 2 | 52 | 23459 | 6.01 | 7 | 1 | 5 | 1 | 5 | 0.38 | 0.19 |
| DNPEP_MOUSE | Aspartyl aminopeptidase | 52 | 52174 | 6.82 | 10 | 4 | 3 | 1 | 3 | 0.19 | 0.17 |
| ODO2_MOUSE | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | 52 | 48963 | 9.11 | 4 | 2 | 2 | 1 | 2 | 0.07 | 0.09 |
| TBCB_MOUSE | Tubulin-folding cofactor B | 52 | 27368 | 5.14 | 2 | 2 | 1 | 1 | 1 | 0.08 | 0.16 |
| PNKP_MOUSE | Bifunctional polynucleotide phosphatase/kinase | 51 | 57188 | 8.03 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.08 |
| ASAH1_MOUSE | Acid ceramidase | 51 | 44641 | 8.68 | 10 | 4 | 6 | 3 | 6 | 0.24 | 0.32 |
| VATE1_MOUSE | V-type proton ATPase subunit E 1 | 51 | 26141 | 8.44 | 4 | 2 | 3 | 1 | 3 | 0.3 | 0.17 |
| ITPA_MOUSE | Inosine triphosphate pyrophosphatase | 51 | 21883 | 5.6 | 4 | 1 | 2 | 1 | 2 | 0.15 | 0.21 |
| MCM2_MOUSE | DNA replication licensing factor MCM2 | 51 | 102013 | 5.49 | 6 | 2 | 4 | 1 | 4 | 0.09 | 0.04 |
| B2MG_MOUSE | Beta-2-microglobulin | 50 | 13770 | 8.55 | 2 | 1 | 2 | 1 | 2 | 0.31 | 0.35 |
| MRCKB_MOUSE | Serine/threonine-protein kinase MRCK beta | 50 | 194630 | 6.05 | 8 | 5 | 5 | 2 | 5 | 0.06 | 0.04 |
| TCOF_MOUSE | Treacle protein | 50 | 134921 | 9.35 | 3 | 2 | 2 | 1 | 2 | 0.04 | 0.03 |
| ACOT9_MOUSE | Acyl-coenzyme A thioesterase 9, mitochondrial | 50 | 50528 | 8.74 | 8 | 3 | 5 | 2 | 5 | 0.23 | 0.18 |
| RRP1_MOUSE | Ribosomal RNA processing protein 1 homolog A | 50 | 54743 | 5.04 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.08 |
| CHRD1_MOUSE | Cysteine and histidine-rich domain-containing protein 1 | 50 | 37327 | 8.13 | 4 | 1 | 3 | 1 | 3 | 0.18 | 0.12 |
| HMGCL_MOUSE | Hydroxymethylglutaryl-CoA lyase, mitochondrial | 50 | 34217 | 8.7 | 5 | 2 | 4 | 2 | 4 | 0.18 | 0.27 |
| ANFY1_MOUSE | Rabankyrin-5 | 49 | 128571 | 5.58 | 6 | 3 | 6 | 3 | 6 | 0.11 | 0.1 |
| MOGS_MOUSE | Mannosyl-oligosaccharide glucosidase | 49 | 91774 | 9.06 | 7 | 4 | 4 | 3 | 4 | 0.1 | 0.15 |
| SAMH1_MOUSE | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | 49 | 72604 | 8.17 | 7 | 3 | 6 | 3 | 6 | 0.18 | 0.19 |
| MPRIP_MOUSE | Myosin phosphatase Rho-interacting protein | 49 | 116337 | 5.9 | 6 | 1 | 4 | 1 | 4 | 0.07 | 0.04 |
| G6PD1_MOUSE | Glucose-6-phosphate 1-dehydrogenase X | 49 | 59225 | 6.06 | 17 | 4 | 9 | 2 | 9 | 0.32 | 0.15 |
| ATP5I_MOUSE | ATP synthase subunit e, mitochondrial | 49 | 8230 | 9.34 | 2 | 1 | 1 | 1 | 1 | 0.18 | 0.63 |
| RL14_MOUSE | 60S ribosomal protein L14 | 49 | 23549 | 11.03 | 4 | 1 | 4 | 1 | 4 | 0.23 | 0.19 |
| FABP4_MOUSE | Fatty acid-binding protein, adipocyte | 49 | 14641 | 8.53 | 2 | 1 | 2 | 1 | 2 | 0.27 | 0.32 |
| MCM7_MOUSE | DNA replication licensing factor MCM7 | 49 | 81160 | 5.98 | 6 | 2 | 6 | 2 | 6 | 0.16 | 0.11 |
| S2611_MOUSE | Sodium-independent sulfate anion transporter | 49 | 64068 | 7.56 | 4 | 3 | 2 | 1 | 2 | 0.08 | 0.07 |
| UBQL1_MOUSE | Ubiquilin-1 | 49 | 61937 | 4.86 | 10 | 4 | 4 | 2 | 4 | 0.13 | 0.14 |
| GSH0_MOUSE | Glutamate–cysteine ligase regulatory subunit | 49 | 30516 | 5.35 | 2 | 1 | 2 | 1 | 2 | 0.17 | 0.15 |
| PDK3_MOUSE | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 3, mitochondrial | 49 | 47893 | 8.89 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.09 |
| P3H3_MOUSE | Prolyl 3-hydroxylase 3 | 48 | 81650 | 6.21 | 7 | 2 | 5 | 1 | 5 | 0.14 | 0.05 |
| ARP10_MOUSE | Actin-related protein 10 | 48 | 46178 | 7.54 | 4 | 2 | 4 | 2 | 4 | 0.2 | 0.2 |
| ARL2_MOUSE | ADP-ribosylation factor-like protein 2 | 48 | 20851 | 5.67 | 4 | 2 | 1 | 1 | 1 | 0.14 | 0.22 |
| LAMA5_MOUSE | Laminin subunit alpha-5 | 48 | 403792 | 6.28 | 6 | 1 | 3 | 1 | 3 | 0.02 | 0.01 |
| IFI5B_MOUSE | Interferon-activable protein 205-B | 48 | 46948 | 8.2 | 11 | 2 | 8 | 2 | 8 | 0.27 | 0.19 |
| NXN_MOUSE | Nucleoredoxin | 48 | 48314 | 4.84 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.09 |
| TTC27_MOUSE | Tetratricopeptide repeat protein 27 | 47 | 96369 | 5.56 | 9 | 3 | 4 | 2 | 4 | 0.11 | 0.09 |
| GPAA1_MOUSE | Glycosylphosphatidylinositol anchor attachment 1 protein | 47 | 67905 | 8.61 | 2 | 1 | 2 | 1 | 2 | 0.08 | 0.06 |
| CP062_MOUSE | UPF0505 protein C16orf62 homolog | 47 | 109007 | 6.97 | 7 | 1 | 5 | 1 | 5 | 0.08 | 0.04 |
| CNOT1_MOUSE | CCR4-NOT transcription complex subunit 1 | 47 | 266637 | 6.65 | 8 | 2 | 8 | 2 | 8 | 0.07 | 0.03 |
| NDUS3_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | 47 | 30131 | 6.67 | 4 | 1 | 3 | 1 | 3 | 0.18 | 0.15 |
| CGNL1_MOUSE | Cingulin-like protein 1 | 47 | 148140 | 5.6 | 5 | 1 | 5 | 1 | 5 | 0.06 | 0.03 |
| MCES_MOUSE | mRNA cap guanine-N7 methyltransferase | 47 | 53258 | 6.12 | 4 | 2 | 3 | 1 | 3 | 0.09 | 0.08 |
| EXOC7_MOUSE | Exocyst complex component 7 | 47 | 79910 | 6.51 | 6 | 2 | 4 | 1 | 4 | 0.14 | 0.05 |
| GOGA3_MOUSE | Golgin subfamily A member 3 | 47 | 167118 | 5.29 | 7 | 1 | 7 | 1 | 7 | 0.1 | 0.03 |
| IR3IP_MOUSE | Immediate early response 3-interacting protein 1 | 47 | 9011 | 7.96 | 2 | 1 | 2 | 1 | 2 | 0.32 | 0.56 |
| CIP4_MOUSE | Cdc42-interacting protein 4 | 47 | 68447 | 5.66 | 1 | 1 | 1 | 1 | 1 | 0.02 | 0.06 |
| GALT2_MOUSE | Polypeptide N-acetylgalactosaminyltransferase 2 | 46 | 64473 | 8.8 | 9 | 1 | 6 | 1 | 6 | 0.17 | 0.07 |
| CD9_MOUSE | CD9 antigen | 46 | 25241 | 6.88 | 5 | 4 | 2 | 1 | 2 | 0.26 | 0.18 |
| NOP58_MOUSE | Nucleolar protein 58 | 46 | 60305 | 8.53 | 4 | 1 | 3 | 1 | 3 | 0.11 | 0.07 |
| NIPS2_MOUSE | Protein NipSnap homolog 2 | 46 | 32912 | 9.31 | 4 | 2 | 2 | 2 | 2 | 0.15 | 0.29 |
| AMPB_MOUSE | Aminopeptidase B | 45 | 72370 | 5.22 | 7 | 3 | 6 | 3 | 6 | 0.22 | 0.19 |
| NB5R1_MOUSE | NADH-cytochrome b5 reductase 1 | 45 | 34113 | 8.97 | 4 | 1 | 3 | 1 | 3 | 0.15 | 0.13 |
| VPP2_MOUSE | V-type proton ATPase 116 kDa subunit a isoform 2 | 45 | 98081 | 6.2 | 3 | 1 | 2 | 1 | 2 | 0.05 | 0.04 |
| IMPCT_MOUSE | Protein IMPACT | 45 | 36253 | 4.97 | 9 | 2 | 4 | 2 | 4 | 0.24 | 0.26 |
| MARE3_MOUSE | Microtubule-associated protein RP/EB family member 3 | 45 | 31946 | 5.33 | 2 | 1 | 2 | 1 | 2 | 0.17 | 0.14 |
| TMM43_MOUSE | Transmembrane protein 43 | 45 | 44755 | 6.85 | 3 | 2 | 3 | 2 | 3 | 0.14 | 0.2 |
| FA98B_MOUSE | Protein FAM98B | 45 | 45321 | 8.77 | 4 | 1 | 3 | 1 | 3 | 0.19 | 0.1 |
| ERP44_MOUSE | Endoplasmic reticulum resident protein 44 | 44 | 46823 | 5.09 | 2 | 1 | 2 | 1 | 2 | 0.1 | 0.09 |
| TSP2_MOUSE | Thrombospondin-2 | 44 | 129798 | 4.61 | 9 | 2 | 6 | 2 | 5 | 0.1 | 0.07 |
| HEM2_MOUSE | Delta-aminolevulinic acid dehydratase | 44 | 36000 | 6.32 | 11 | 7 | 3 | 1 | 3 | 0.2 | 0.12 |
| UROK_MOUSE | Urokinase-type plasminogen activator | 44 | 48236 | 8.53 | 5 | 1 | 3 | 1 | 3 | 0.17 | 0.09 |
| IF4G2_MOUSE | Eukaryotic translation initiation factor 4 gamma 2 | 44 | 102041 | 6.7 | 10 | 5 | 7 | 3 | 7 | 0.14 | 0.13 |
| RSU1_MOUSE | Ras suppressor protein 1 | 44 | 31531 | 8.86 | 10 | 2 | 5 | 1 | 5 | 0.39 | 0.14 |
| ITB5_MOUSE | Integrin beta-5 | 44 | 87851 | 5.81 | 8 | 3 | 8 | 3 | 8 | 0.22 | 0.15 |
| SEP10_MOUSE | Septin-10 | 44 | 52388 | 6.17 | 8 | 2 | 5 | 1 | 5 | 0.15 | 0.08 |
| EMC1_MOUSE | ER membrane protein complex subunit 1 | 44 | 111535 | 7 | 11 | 3 | 7 | 3 | 7 | 0.15 | 0.12 |
| A16L1_MOUSE | Autophagy-related protein 16-1 | 43 | 68130 | 5.96 | 6 | 1 | 2 | 1 | 2 | 0.06 | 0.06 |
| MINP1_MOUSE | Multiple inositol polyphosphate phosphatase 1 | 43 | 54503 | 7.21 | 3 | 2 | 2 | 1 | 2 | 0.09 | 0.08 |
| MTAP_MOUSE | S-Methyl-5 ′-thioadenosine phosphorylase | 43 | 31042 | 6.71 | 6 | 3 | 3 | 2 | 3 | 0.22 | 0.31 |
| EMAL2_MOUSE | Echinoderm microtubule-associated protein-like 2 | 43 | 70689 | 5.83 | 5 | 2 | 2 | 1 | 2 | 0.07 | 0.06 |
| ACADV_MOUSE | Very-long-chain-specific acyl-CoA dehydrogenase, mitochondrial | 43 | 70831 | 8.91 | 9 | 1 | 6 | 1 | 6 | 0.19 | 0.06 |
| AOFB_MOUSE | Amine oxidase [flavin-containing] B | 43 | 58520 | 8.52 | 7 | 2 | 5 | 2 | 5 | 0.16 | 0.15 |
| NRADD_MOUSE | Death domain-containing membrane protein NRADD | 43 | 24711 | 4.89 | 3 | 3 | 1 | 1 | 1 | 0.14 | 0.4 |
| DHSD_MOUSE | Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial | 43 | 17003 | 9.3 | 1 | 1 | 1 | 1 | 1 | 0.16 | 0.27 |
| CSN9_MOUSE | COP9 signalosome complex subunit 9 | 43 | 6193 | 3.6 | 1 | 1 | 1 | 1 | 1 | 0.35 | 0.89 |
| VP13C_MOUSE | Vacuolar protein sorting-associated protein 13C | 43 | 419824 | 6.37 | 8 | 1 | 8 | 1 | 8 | 0.05 | 0.01 |
| EMAL1_MOUSE | Echinoderm microtubule-associated protein-like 1 | 43 | 89624 | 6.56 | 10 | 2 | 8 | 2 | 8 | 0.21 | 0.1 |
| TIP_MOUSE | T cell immunomodulatory protein | 42 | 67422 | 5.4 | 6 | 1 | 2 | 1 | 2 | 0.09 | 0.06 |
| CLU_MOUSE | Clustered mitochondria protein homolog | 42 | 147975 | 5.69 | 2 | 2 | 2 | 2 | 2 | 0.03 | 0.06 |
| PSME1_MOUSE | Proteasome activator complex subunit 1 | 40 | 28655 | 5.73 | 5 | 3 | 4 | 3 | 4 | 0.26 | 0.54 |
| MFGM_MOUSE | Lactadherin | 42 | 51208 | 6.1 | 8 | 3 | 6 | 3 | 6 | 0.19 | 0.28 |
| E41L3_MOUSE | Band 4.1-like protein 3 | 42 | 103274 | 5.2 | 1 | 1 | 1 | 1 | 1 | 0.01 | 0.04 |
| LMAN2_MOUSE | Vesicular integral-membrane protein VIP36 | 42 | 40404 | 6.46 | 6 | 2 | 5 | 2 | 5 | 0.23 | 0.23 |
| PPM1F_MOUSE | Protein phosphatase 1F | 42 | 49580 | 5.16 | 6 | 3 | 5 | 3 | 5 | 0.28 | 0.29 |
| SERC3_MOUSE | Serine incorporator 3 | 42 | 52588 | 6.95 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.08 |
| ARHG1_MOUSE | Rho guanine nucleotide exchange factor 1 | 42 | 102741 | 5.43 | 6 | 2 | 6 | 2 | 6 | 0.11 | 0.08 |
| RS27A_MOUSE | Ubiquitin-40S ribosomal protein S27a | 34 | 17939 | 9.68 | 7 | 2 | 2 | 2 | 2 | 0.22 | 0.58 |
| MANEA_MOUSE | Glycoprotein endo-alpha-1,2-mannosidase | 42 | 53149 | 8.66 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.08 |
| MYO1B_MOUSE | Unconventional myosin-Ib | 41 | 128483 | 9.32 | 3 | 1 | 2 | 1 | 2 | 0.04 | 0.03 |
| PTCD3_MOUSE | Pentatricopeptide repeat domain-containing protein 3, mitochondrial | 41 | 77747 | 5.61 | 4 | 2 | 3 | 1 | 3 | 0.08 | 0.06 |
| COG2_MOUSE | Conserved oligomeric Golgi complex subunit 2 | 41 | 81988 | 5.84 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.05 |
| KTN1_MOUSE | Kinectin | 41 | 152498 | 5.67 | 7 | 2 | 7 | 2 | 7 | 0.1 | 0.06 |
| ARXS1_MOUSE | Adipocyte-related X-chromosome expressed sequence 1 | 41 | 20134 | 9.6 | 2 | 1 | 2 | 1 | 2 | 0.14 | 0.23 |
| RAB23_MOUSE | Ras-related protein Rab-23 | 41 | 26662 | 6.36 | 4 | 1 | 4 | 1 | 4 | 0.41 | 0.17 |
| SMRC1_MOUSE | SWI/SNF complex subunit SMARCC1 | 41 | 122813 | 5.5 | 5 | 1 | 5 | 1 | 4 | 0.08 | 0.03 |
| HGNAT_MOUSE | Heparan-alpha-glucosaminide N-acetyltransferase | 41 | 72458 | 8.58 | 2 | 2 | 1 | 1 | 1 | 0.05 | 0.06 |
| GGCT_MOUSE | Gamma-glutamylcyclotransferase | 41 | 21152 | 5.52 | 3 | 1 | 3 | 1 | 3 | 0.31 | 0.21 |
| PLPP1_MOUSE | Phospholipid phosphatase 1 | 41 | 31871 | 6.54 | 6 | 3 | 3 | 2 | 3 | 0.16 | 0.3 |
| RFOX2_MOUSE | RNA binding protein fox-1 homolog 2 | 40 | 47301 | 6.1 | 3 | 1 | 3 | 1 | 3 | 0.1 | 0.09 |
| DPP9_MOUSE | Dipeptidyl peptidase 9 | 40 | 97939 | 6.18 | 3 | 1 | 3 | 1 | 3 | 0.03 | 0.04 |
| CD63_MOUSE | CD63 antigen | 40 | 25749 | 6.69 | 3 | 2 | 1 | 1 | 1 | 0.05 | 0.17 |
| ARFG1_MOUSE | ADP-ribosylation factor GTPase-activating protein 1 | 40 | 45260 | 5.39 | 2 | 1 | 2 | 1 | 2 | 0.11 | 0.1 |
| NOP16_MOUSE | Nucleolar protein 16 | 40 | 21126 | 9.91 | 1 | 1 | 1 | 1 | 1 | 0.1 | 0.22 |
| SNRPA_MOUSE | U1 small nuclear ribonucleoprotein A | 40 | 31814 | 9.81 | 3 | 1 | 3 | 1 | 3 | 0.18 | 0.14 |
| KIF1C_MOUSE | Kinesin-like protein KIF1C | 40 | 122358 | 6.62 | 6 | 1 | 5 | 1 | 5 | 0.1 | 0.03 |
| LRC40_MOUSE | Leucine-rich repeat-containing protein 40 | 40 | 68033 | 6.43 | 3 | 1 | 3 | 1 | 3 | 0.1 | 0.06 |
| MAOM_MOUSE | NAD-dependent malic enzyme, mitochondrial | 40 | 65757 | 7.53 | 2 | 2 | 2 | 2 | 2 | 0.08 | 0.14 |
| LAMB1_MOUSE | Laminin subunit beta-1 | 40 | 196961 | 4.82 | 13 | 2 | 6 | 1 | 6 | 0.08 | 0.02 |
| GTR1_MOUSE | Solute carrier family 2, facilitated glucose transporter member 1 | 40 | 53949 | 9.05 | 5 | 3 | 4 | 2 | 4 | 0.13 | 0.17 |
| CKAP5_MOUSE | Cytoskeleton-associated protein 5 | 40 | 225492 | 8.17 | 11 | 2 | 9 | 2 | 9 | 0.08 | 0.04 |
| LYAR_MOUSE | Cell growth-regulating nucleolar protein | 40 | 43708 | 9.52 | 4 | 1 | 3 | 1 | 3 | 0.1 | 0.1 |
| SAAL1_MOUSE | Protein SAAL1 | 40 | 52735 | 4.4 | 4 | 1 | 4 | 1 | 4 | 0.2 | 0.08 |
| WDR12_MOUSE | Ribosome biogenesis protein WDR12 | 40 | 47317 | 5.36 | 2 | 1 | 1 | 1 | 1 | 0.08 | 0.09 |
| TBC15_MOUSE | TBC1 domain family member 15 | 39 | 76478 | 5.15 | 2 | 2 | 1 | 1 | 1 | 0.04 | 0.06 |
| PGLT1_MOUSE | Protein O-glucosyltransferase 1 | 39 | 46350 | 9.04 | 2 | 2 | 2 | 2 | 2 | 0.11 | 0.2 |
| ACOD2_MOUSE | Acyl-CoA desaturase 2 | 39 | 40890 | 9.14 | 2 | 1 | 2 | 1 | 2 | 0.12 | 0.11 |
| ARHG2_MOUSE | Rho guanine nucleotide exchange factor 2 | 39 | 111905 | 6.89 | 3 | 2 | 2 | 1 | 2 | 0.06 | 0.04 |
| NFKB2_MOUSE | Nuclear factor NF-kappa-B p100 subunit | 39 | 96772 | 5.93 | 4 | 3 | 2 | 2 | 2 | 0.06 | 0.09 |
| NU188_MOUSE | Nucleoporin NUP188 homolog | 39 | 196570 | 6.56 | 5 | 1 | 4 | 1 | 4 | 0.05 | 0.02 |
| MCA3_MOUSE | Eukaryotic translation elongation factor 1 epsilon-1 | 39 | 19846 | 8.6 | 4 | 2 | 3 | 1 | 3 | 0.33 | 0.23 |
| STING_MOUSE | Stimulator of interferon genes protein | 39 | 42802 | 7.12 | 6 | 3 | 4 | 2 | 4 | 0.23 | 0.21 |
| 5NT3A_MOUSE | Cytosolic 5 ′-nucleotidase 3A | 39 | 37228 | 6.21 | 2 | 1 | 2 | 1 | 2 | 0.15 | 0.12 |
| NDUV3_MOUSE | NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial | 39 | 11806 | 9.34 | 3 | 2 | 2 | 1 | 2 | 0.32 | 0.41 |
| MTX1_MOUSE | Metaxin-1 | 39 | 35601 | 5.82 | 2 | 2 | 1 | 1 | 1 | 0.04 | 0.12 |
| EMC3_MOUSE | ER membrane protein complex subunit 3 | 39 | 29960 | 6.33 | 4 | 3 | 2 | 1 | 2 | 0.16 | 0.15 |
| TULP3_MOUSE | Tubby-related protein 3 | 38 | 51199 | 5.88 | 3 | 1 | 2 | 1 | 2 | 0.12 | 0.08 |
| SEC63_MOUSE | Translocation protein SEC63 homolog | 38 | 87815 | 5.28 | 8 | 3 | 5 | 3 | 5 | 0.09 | 0.15 |
| TFAM_MOUSE | Transcription factor A, mitochondrial | 38 | 27970 | 9.71 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.16 |
| TMX4_MOUSE | Thioredoxin-related transmembrane protein 4 | 38 | 37108 | 4.3 | 2 | 1 | 1 | 1 | 1 | 0.05 | 0.12 |
| SP16H_MOUSE | FACT complex subunit SPT16 | 38 | 119749 | 5.5 | 7 | 3 | 4 | 2 | 4 | 0.08 | 0.07 |
| ACADS_MOUSE | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | 38 | 44861 | 8.68 | 2 | 2 | 1 | 1 | 1 | 0.08 | 0.1 |
| HEXB_MOUSE | Beta-hexosaminidase subunit beta | 38 | 61077 | 8.29 | 4 | 2 | 2 | 1 | 2 | 0.11 | 0.07 |
| GBP2_MOUSE | Guanylate-binding protein 2 | 38 | 66697 | 5.56 | 2 | 2 | 1 | 1 | 1 | 0.04 | 0.06 |
| SK2L2_MOUSE | Superkiller viralicidic activity 2-like 2 | 37 | 117561 | 6.01 | 10 | 2 | 8 | 1 | 8 | 0.16 | 0.04 |
| PFKAP_MOUSE | ATP-dependent 6-phosphofructokinase, platelet type | 37 | 85400 | 6.73 | 9 | 1 | 8 | 1 | 8 | 0.18 | 0.05 |
| T2FB_MOUSE | General transcription factor IIF subunit 2 | 37 | 28364 | 9.23 | 3 | 1 | 2 | 1 | 2 | 0.11 | 0.16 |
| SAM50_MOUSE | Sorting and assembly machinery component 50 homolog | 37 | 51831 | 6.34 | 3 | 1 | 3 | 1 | 3 | 0.12 | 0.08 |
| XRCC6_MOUSE | X-ray repair cross-complementing protein 6 | 37 | 69441 | 6.35 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.06 |
| KBL_MOUSE | 2-Amino-3-ketobutyrate coenzyme A ligase, mitochondrial | 37 | 44902 | 6.92 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.1 |
| PRI2_MOUSE | DNA primase large subunit | 37 | 58372 | 8.47 | 3 | 1 | 3 | 1 | 3 | 0.13 | 0.07 |
| TGM2_MOUSE | Protein-glutamine gamma-glutamyltransferase 2 | 37 | 77012 | 4.98 | 2 | 2 | 2 | 2 | 2 | 0.07 | 0.11 |
| SPART_MOUSE | Spartin | 37 | 72610 | 5.64 | 4 | 1 | 3 | 1 | 3 | 0.12 | 0.06 |
| SRRM1_MOUSE | Serine/arginine repetitive matrix protein 1 | 37 | 106798 | 11.87 | 4 | 1 | 2 | 1 | 2 | 0.04 | 0.04 |
| CSPG4_MOUSE | Chondroitin sulfate proteoglycan 4 | 37 | 252153 | 5.23 | 8 | 2 | 7 | 2 | 7 | 0.07 | 0.03 |
| DHX29_MOUSE | ATP-dependent RNA helicase DHX29 | 36 | 153879 | 8.13 | 3 | 1 | 3 | 1 | 3 | 0.03 | 0.03 |
| DECR_MOUSE | 2,4-Dienoyl-CoA reductase, mitochondrial | 36 | 36191 | 9.1 | 6 | 2 | 5 | 1 | 5 | 0.22 | 0.12 |
| CHKB_MOUSE | Choline/ethanolamine kinase | 36 | 45097 | 5.3 | 3 | 1 | 2 | 1 | 2 | 0.12 | 0.1 |
| RLA1_MOUSE | 60S acidic ribosomal protein P1 | 36 | 11468 | 4.28 | 2 | 2 | 1 | 1 | 1 | 0.14 | 0.43 |
| MBD3_MOUSE | Methyl-CpG-binding domain protein 3 | 36 | 32148 | 5.64 | 3 | 1 | 2 | 1 | 2 | 0.11 | 0.14 |
| IF6_MOUSE | Eukaryotic translation initiation factor 6 | 36 | 26494 | 4.63 | 2 | 1 | 2 | 1 | 2 | 0.26 | 0.17 |
| NISCH_MOUSE | Nischarin | 36 | 174903 | 5.05 | 3 | 2 | 3 | 2 | 3 | 0.04 | 0.05 |
| CD44_MOUSE | CD44 antigen | 36 | 85565 | 4.82 | 4 | 3 | 2 | 2 | 2 | 0.05 | 0.1 |
| NPM3_MOUSE | Nucleoplasmin-3 | 36 | 19011 | 4.71 | 1 | 1 | 1 | 1 | 1 | 0.19 | 0.24 |
| SNX2_MOUSE | Sorting nexin-2 | 35 | 58435 | 5.04 | 2 | 1 | 2 | 1 | 2 | 0.1 | 0.07 |
| MRP1_MOUSE | Multidrug resistance-associated protein 1 | 35 | 171075 | 7.03 | 5 | 1 | 5 | 1 | 5 | 0.07 | 0.02 |
| SERB_MOUSE | Phosphoserine phosphatase | 35 | 25080 | 5.81 | 4 | 2 | 3 | 2 | 3 | 0.24 | 0.39 |
| SMYD5_MOUSE | SET and MYND domain-containing protein 5 | 35 | 47065 | 5.09 | 2 | 1 | 1 | 1 | 1 | 0.06 | 0.09 |
| PGFRB_MOUSE | Platelet-derived growth factor receptor beta | 35 | 122728 | 4.99 | 8 | 2 | 6 | 2 | 6 | 0.09 | 0.07 |
| APAF_MOUSE | Apoptotic protease-activating factor 1 | 35 | 140913 | 5.98 | 6 | 1 | 3 | 1 | 3 | 0.04 | 0.03 |
| PLS3_MOUSE | Phospholipid scramblase 3 | 35 | 31782 | 5.97 | 6 | 2 | 3 | 2 | 3 | 0.11 | 0.3 |
| NMRL1_MOUSE | NmrA-like family domain-containing protein 1 | 35 | 34355 | 6.37 | 3 | 1 | 2 | 1 | 2 | 0.15 | 0.13 |
| EHD2_MOUSE | EH domain-containing protein 2 | 35 | 61136 | 6.08 | 10 | 3 | 6 | 3 | 6 | 0.21 | 0.23 |
| CRKL_MOUSE | Crk-like protein | 34 | 33809 | 6.26 | 9 | 2 | 4 | 1 | 4 | 0.23 | 0.13 |
| EI2BB_MOUSE | Translation initiation factor eIF-2B subunit beta | 34 | 38873 | 5.82 | 4 | 2 | 3 | 1 | 3 | 0.22 | 0.11 |
| BLVRB_MOUSE | Flavin reductase (NADPH) | 34 | 22183 | 6.49 | 7 | 1 | 4 | 1 | 4 | 0.38 | 0.2 |
| SRSF6_MOUSE | Serine/arginine-rich splicing factor 6 | 34 | 39002 | 11.46 | 2 | 1 | 1 | 1 | 1 | 0.07 | 0.11 |
| TRI47_MOUSE | Tripartite motif-containing protein 47 | 34 | 69868 | 6 | 2 | 2 | 1 | 1 | 1 | 0.05 | 0.06 |
| ICLN_MOUSE | Methylosome subunit pICln | 34 | 26005 | 3.98 | 2 | 1 | 2 | 1 | 2 | 0.15 | 0.17 |
| GNS_MOUSE | N-Acetylglucosamine-6-sulfatase | 34 | 61136 | 8.52 | 16 | 2 | 9 | 2 | 9 | 0.35 | 0.15 |
| PLBL2_MOUSE | Putative phospholipase B-like 2 | 34 | 66247 | 5.77 | 5 | 1 | 4 | 1 | 4 | 0.12 | 0.07 |
| TBCA_MOUSE | Tubulin-specific chaperone A | 34 | 12750 | 5.24 | 3 | 1 | 2 | 1 | 2 | 0.37 | 0.38 |
| SGPL1_MOUSE | Sphingosine-1-phosphate lyase 1 | 34 | 63636 | 9.2 | 6 | 1 | 5 | 1 | 5 | 0.11 | 0.07 |
| T126A_MOUSE | Transmembrane protein 126A | 34 | 21526 | 9.45 | 2 | 1 | 2 | 1 | 2 | 0.17 | 0.21 |
| DHR11_MOUSE | Dehydrogenase/reductase SDR family member 11 | 34 | 28256 | 5.91 | 3 | 2 | 2 | 1 | 2 | 0.17 | 0.16 |
| KTAP2_MOUSE | Keratinocyte-associated protein 2 | 34 | 14665 | 9.67 | 3 | 1 | 1 | 1 | 1 | 0.2 | 0.32 |
| LAMC1_MOUSE | Laminin subunit gamma-1 | 33 | 177185 | 5.08 | 3 | 1 | 3 | 1 | 3 | 0.04 | 0.02 |
| 3HIDH_MOUSE | 3-Hydroxyisobutyrate dehydrogenase, mitochondrial | 33 | 35417 | 8.37 | 3 | 1 | 2 | 1 | 2 | 0.19 | 0.12 |
| BGAL_MOUSE | Beta-galactosidase | 33 | 73074 | 7.15 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.06 |
| FKBP8_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP8 | 33 | 43501 | 5.08 | 2 | 1 | 2 | 1 | 2 | 0.11 | 0.1 |
| CASP3_MOUSE | Caspase-3 | 33 | 31454 | 6.45 | 4 | 1 | 3 | 1 | 3 | 0.24 | 0.14 |
| RT33_MOUSE | 28S ribosomal protein S33, mitochondrial | 33 | 12452 | 10.25 | 1 | 1 | 1 | 1 | 1 | 0.2 | 0.39 |
| ATX2L_MOUSE | Ataxin-2-like protein | 33 | 110580 | 8.94 | 3 | 1 | 3 | 1 | 3 | 0.07 | 0.04 |
| SELB_MOUSE | Selenocysteine-specific elongation factor | 33 | 63498 | 8.58 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.07 |
| TRI16_MOUSE | Tripartite motif-containing protein 16 | 33 | 62903 | 5.57 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.07 |
| ARSA_MOUSE | Arylsulfatase A | 33 | 53714 | 5.5 | 3 | 1 | 2 | 1 | 2 | 0.1 | 0.08 |
| BL1S6_MOUSE | Biogenesis of lysosome-related organelles complex 1 subunit 6 | 33 | 19670 | 5.91 | 2 | 1 | 1 | 1 | 1 | 0.11 | 0.23 |
| ECH1_MOUSE | Delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, mitochondrial | 33 | 36095 | 7.6 | 4 | 1 | 4 | 1 | 4 | 0.31 | 0.12 |
| DIAP3_MOUSE | Protein diaphanous homolog 3 | 32 | 133601 | 7.53 | 3 | 1 | 3 | 1 | 3 | 0.05 | 0.03 |
| ERG1_MOUSE | Squalene monooxygenase | 32 | 63730 | 8.67 | 5 | 1 | 2 | 1 | 2 | 0.1 | 0.07 |
| DDX46_MOUSE | Probable ATP-dependent RNA helicase DDX46 | 32 | 117376 | 9.3 | 4 | 1 | 3 | 1 | 3 | 0.05 | 0.04 |
| M4K4_MOUSE | Mitogen-activated protein kinase 4 | 32 | 140515 | 7.1 | 7 | 1 | 7 | 1 | 7 | 0.1 | 0.03 |
| ABCD3_MOUSE | ATP-binding cassette sub-family D member 3 | 32 | 75426 | 9.32 | 9 | 1 | 7 | 1 | 7 | 0.13 | 0.06 |
| PSMG1_MOUSE | Proteasome assembly chaperone 1 | 32 | 33083 | 6.05 | 4 | 2 | 2 | 1 | 2 | 0.12 | 0.29 |
| FRDA_MOUSE | Frataxin, mitochondrial | 32 | 22910 | 7.81 | 1 | 1 | 1 | 1 | 1 | 0.08 | 0.2 |
| THOP1_MOUSE | Thimet oligopeptidase | 32 | 77976 | 5.72 | 3 | 2 | 2 | 1 | 2 | 0.06 | 0.06 |
| CREL2_MOUSE | Cysteine-rich with EGF-like domain protein 2 | 32 | 38194 | 4.49 | 3 | 1 | 2 | 1 | 2 | 0.05 | 0.11 |
| SSRD_MOUSE | Translocon-associated protein subunit delta | 32 | 18924 | 5.5 | 4 | 1 | 4 | 1 | 4 | 0.31 | 0.24 |
| I2BP2_MOUSE | Interferon regulatory factor 2-binding protein 2 | 32 | 59255 | 9 | 6 | 1 | 5 | 1 | 5 | 0.18 | 0.07 |
| RL29_MOUSE | 60S ribosomal protein L29 | 31 | 17576 | 11.84 | 2 | 1 | 2 | 1 | 2 | 0.17 | 0.26 |
| B4GT1_MOUSE | Beta-1,4-galactosyltransferase 1 | 31 | 44383 | 9.45 | 3 | 1 | 2 | 1 | 2 | 0.07 | 0.1 |
| HTR5A_MOUSE | HEAT repeat-containing protein 5A | 31 | 219754 | 6.31 | 6 | 1 | 5 | 1 | 5 | 0.06 | 0.02 |
| SYFB_MOUSE | Phenylalanine–tRNA ligase beta subunit | 31 | 65655 | 6.69 | 4 | 2 | 3 | 1 | 3 | 0.09 | 0.07 |
| BAG5_MOUSE | BAG family molecular chaperone regulator 5 | 31 | 50911 | 5.75 | 3 | 1 | 3 | 1 | 3 | 0.14 | 0.09 |
| HSPB8_MOUSE | Heat shock protein beta-8 | 31 | 21520 | 4.92 | 1 | 1 | 1 | 1 | 1 | 0.13 | 0.21 |
| SNX7_MOUSE | Sorting nexin-7 | 31 | 44971 | 4.99 | 4 | 2 | 2 | 2 | 2 | 0.12 | 0.2 |
| TMTC3_MOUSE | Transmembrane and TPR repeat-containing protein 3 | 31 | 104131 | 8.75 | 5 | 2 | 4 | 1 | 4 | 0.06 | 0.04 |
| NNMT_MOUSE | Nicotinamide N-methyltransferase | 31 | 29579 | 5.3 | 4 | 3 | 2 | 2 | 2 | 0.16 | 0.32 |
| WIPI1_MOUSE | WD repeat domain phosphoinositide-interacting protein 1 | 30 | 48727 | 6.03 | 3 | 2 | 2 | 1 | 2 | 0.13 | 0.09 |
| K1468_MOUSE | LisH domain and HEAT repeat-containing protein KIAA1468 | 30 | 134502 | 5.2 | 7 | 1 | 5 | 1 | 5 | 0.09 | 0.03 |
| FKBP3_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP3 | 30 | 25132 | 9.29 | 6 | 2 | 6 | 2 | 6 | 0.42 | 0.39 |
| FHL2_MOUSE | Four and a half LIM domains protein 2 | 30 | 32051 | 7.31 | 2 | 1 | 2 | 1 | 2 | 0.15 | 0.14 |
| ARI4B_MOUSE | AT-rich interactive domain-containing protein 4B | 30 | 147553 | 4.98 | 4 | 1 | 3 | 1 | 3 | 0.03 | 0.03 |
| ANX11_MOUSE | Annexin A11 | 29 | 54045 | 7.53 | 2 | 1 | 1 | 1 | 1 | 0.05 | 0.08 |
| RM48_MOUSE | 39S ribosomal protein L48, mitochondrial | 29 | 23938 | 9.45 | 3 | 1 | 1 | 1 | 1 | 0.13 | 0.19 |
| SEN34_MOUSE | tRNA-splicing endonuclease subunit Sen34 | 29 | 34176 | 6.56 | 2 | 1 | 2 | 1 | 2 | 0.13 | 0.13 |
| HP1B3_MOUSE | Heterochromatin protein 1-binding protein 3 | 29 | 60829 | 9.71 | 5 | 2 | 4 | 2 | 4 | 0.16 | 0.15 |
| COX8A_MOUSE | Cytochrome c oxidase subunit 8A, mitochondrial | 29 | 7643 | 9.74 | 3 | 1 | 1 | 1 | 1 | 0.49 | 0.69 |
| LSM7_MOUSE | U6 snRNA-associated Sm-like protein LSm7 | 29 | 11629 | 5.1 | 4 | 2 | 1 | 1 | 1 | 0.24 | 0.42 |
| CND1_MOUSE | Condensin complex subunit 1 | 29 | 155567 | 5.98 | 2 | 1 | 2 | 1 | 2 | 0.03 | 0.03 |
| ARSB_MOUSE | Arylsulfatase B | 29 | 59609 | 6.78 | 2 | 1 | 1 | 1 | 1 | 0.05 | 0.07 |
| RBP2_MOUSE | E3 SUMO-protein ligase RanBP2 | 29 | 340907 | 5.82 | 7 | 1 | 7 | 1 | 7 | 0.04 | 0.01 |
| SRPK1_MOUSE | SRSF protein kinase 1 | 29 | 73043 | 5.82 | 2 | 1 | 1 | 1 | 1 | 0.02 | 0.06 |
| VAC14_MOUSE | Protein VAC14 homolog | 29 | 87992 | 5.75 | 3 | 2 | 2 | 1 | 2 | 0.05 | 0.05 |
| UBR2_MOUSE | E3 ubiquitin-protein ligase UBR2 | 29 | 199026 | 5.9 | 8 | 1 | 5 | 1 | 5 | 0.06 | 0.02 |
| NCEH1_MOUSE | Neutral cholesterol ester hydrolase 1 | 29 | 45711 | 6.56 | 3 | 1 | 1 | 1 | 1 | 0.07 | 0.1 |
| LMAN1_MOUSE | Protein ERGIC-53 | 29 | 57753 | 5.92 | 7 | 2 | 5 | 2 | 5 | 0.21 | 0.16 |
| DREB_MOUSE | Drebrin | 29 | 77239 | 4.45 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.06 |
| AURKB_MOUSE | Aurora kinase B | 29 | 39360 | 9.47 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.11 |
| STOM_MOUSE | Erythrocyte band 7 integral membrane protein | 29 | 31355 | 6.45 | 2 | 1 | 2 | 1 | 2 | 0.17 | 0.14 |
| SETD3_MOUSE | Histone-lysine N-methyltransferase setd3 | 29 | 67134 | 5.47 | 4 | 2 | 2 | 2 | 2 | 0.08 | 0.13 |
| DAAM1_MOUSE | Disheveled-associated activator of morphogenesis 1 | 29 | 123293 | 7.12 | 6 | 1 | 5 | 1 | 5 | 0.08 | 0.03 |
| IF2B2_MOUSE | Insulin-like growth factor 2 mRNA-binding protein 2 | 28 | 65543 | 7.81 | 5 | 1 | 4 | 1 | 4 | 0.1 | 0.07 |
| GSDMD_MOUSE | Gasdermin-D | 28 | 53204 | 5.03 | 4 | 2 | 3 | 2 | 3 | 0.17 | 0.17 |
| ACON_MOUSE | Aconitate hydratase, mitochondrial | 28 | 85410 | 8.08 | 8 | 2 | 5 | 2 | 5 | 0.12 | 0.1 |
| IF4B_MOUSE | Eukaryotic translation initiation factor 4B | 28 | 68799 | 5.47 | 3 | 1 | 2 | 1 | 2 | 0.09 | 0.06 |
| XRCC5_MOUSE | X-ray repair cross-complementing protein 5 | 28 | 83004 | 5.04 | 5 | 1 | 5 | 1 | 5 | 0.16 | 0.05 |
| GALE_MOUSE | UDP-glucose 4-epimerase | 28 | 38200 | 6.27 | 5 | 2 | 2 | 1 | 2 | 0.14 | 0.11 |
| SYMPK_MOUSE | Symplekin | 28 | 142194 | 5.76 | 11 | 1 | 6 | 1 | 6 | 0.07 | 0.03 |
| NAA10_MOUSE | N-Alpha-acetyltransferase 10 | 28 | 26503 | 5.41 | 8 | 3 | 3 | 1 | 3 | 0.28 | 0.37 |
| GALD1_MOUSE | Glutamine amidotransferase-like class 1 domain-containing protein 1 | 28 | 23263 | 6.58 | 1 | 1 | 1 | 1 | 1 | 0.09 | 0.19 |
| ZO1_MOUSE | Tight junction protein ZO-1 | 28 | 194622 | 6.17 | 6 | 1 | 6 | 1 | 6 | 0.07 | 0.02 |
| PIGS_MOUSE | GPI transamidase component PIG-S | 27 | 61671 | 6.45 | 5 | 2 | 3 | 1 | 3 | 0.13 | 0.07 |
| NRP1_MOUSE | Neuropilin-1 | 27 | 102935 | 5.6 | 3 | 1 | 3 | 1 | 3 | 0.07 | 0.04 |
| LEMD2_MOUSE | LEM domain-containing protein 2 | 27 | 57471 | 9.18 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.08 |
| RTCA_MOUSE | RNA 3 ′-terminal phosphate cyclase | 27 | 39229 | 8.06 | 4 | 1 | 2 | 1 | 2 | 0.13 | 0.11 |
| SULF1_MOUSE | Extracellular sulfatase Sulf-1 | 27 | 100858 | 9.17 | 4 | 1 | 3 | 1 | 3 | 0.04 | 0.04 |
| UFC1_MOUSE | Ubiquitin-fold modifier-conjugating enzyme 1 | 27 | 19469 | 6.9 | 5 | 1 | 3 | 1 | 3 | 0.28 | 0.24 |
| IMPA3_MOUSE | Inositol monophosphatase 3 | 27 | 38592 | 6.03 | 2 | 1 | 2 | 1 | 2 | 0.14 | 0.11 |
| RINT1_MOUSE | RAD50-interacting protein 1 | 27 | 90036 | 5.02 | 5 | 1 | 4 | 1 | 4 | 0.06 | 0.05 |
| KCY_MOUSE | UMP-CMP kinase | 26 | 22151 | 5.68 | 4 | 1 | 4 | 1 | 4 | 0.28 | 0.2 |
| TRIP6_MOUSE | Thyroid receptor-interacting protein 6 | 26 | 50901 | 7.03 | 4 | 1 | 2 | 1 | 2 | 0.09 | 0.09 |
| PLXB2_MOUSE | Plexin-B2 | 26 | 206099 | 5.58 | 7 | 1 | 6 | 1 | 6 | 0.07 | 0.02 |
| UFL1_MOUSE | E3 UFM1-protein ligase 1 | 26 | 89464 | 6.24 | 4 | 1 | 3 | 1 | 3 | 0.1 | 0.05 |
| SMRC2_MOUSE | SWI/SNF complex subunit SMARCC2 | 26 | 132522 | 5.41 | 4 | 1 | 4 | 1 | 3 | 0.07 | 0.03 |
| PPOX_MOUSE | Protoporphyrinogen oxidase | 26 | 50839 | 9.07 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.09 |
| PDLI7_MOUSE | PDZ and LIM domain protein 7 | 26 | 50087 | 8.82 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.09 |
| CLN3_MOUSE | Battenin | 26 | 47627 | 5.25 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.09 |
| TRM61_MOUSE | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A | 26 | 31619 | 6.3 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.14 |
| TBRG4_MOUSE | Protein TBRG4 | 26 | 71468 | 8.55 | 2 | 1 | 2 | 1 | 2 | 0.04 | 0.06 |
| AN13A_MOUSE | Ankyrin repeat domain-containing protein 13A | 26 | 67136 | 4.99 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.06 |
| TMED3_MOUSE | Transmembrane emp24 domain-containing protein 3 | 26 | 25449 | 5.62 | 2 | 1 | 2 | 1 | 2 | 0.16 | 0.18 |
| NPL4_MOUSE | Nuclear protein localization protein 4 homolog | 26 | 67974 | 6.01 | 4 | 1 | 4 | 1 | 4 | 0.13 | 0.06 |
| SAP18_MOUSE | Histone deacetylase complex subunit SAP18 | 26 | 17584 | 9.38 | 2 | 1 | 2 | 1 | 2 | 0.24 | 0.26 |
| SUMF2_MOUSE | Sulfatase-modifying factor 2 | 26 | 34715 | 6.61 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.13 |
| SYMC_MOUSE | Methionine–tRNA ligase, cytoplasmic | 25 | 101366 | 6.78 | 10 | 1 | 8 | 1 | 8 | 0.15 | 0.04 |
| NOL6_MOUSE | Nucleolar protein 6 | 25 | 129146 | 6.33 | 3 | 1 | 3 | 1 | 3 | 0.07 | 0.03 |
| CRIP2_MOUSE | Cysteine-rich protein 2 | 25 | 22712 | 8.94 | 4 | 1 | 4 | 1 | 4 | 0.39 | 0.2 |
| GSKIP_MOUSE | GSK3B-interacting protein | 25 | 15632 | 4.3 | 3 | 1 | 1 | 1 | 1 | 0.17 | 0.3 |
| STAR5_MOUSE | StAR-related lipid transfer protein 5 | 25 | 23907 | 5.97 | 3 | 1 | 3 | 1 | 3 | 0.18 | 0.19 |
| LARP4_MOUSE | La-related protein 4 | 25 | 79713 | 6.02 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.05 |
| OFUT1_MOUSE | GDP-fucose protein O-fucosyltransferase 1 | 25 | 44660 | 8.67 | 1 | 1 | 1 | 1 | 1 | 0.05 | 0.1 |
| RHG10_MOUSE | Rho GTPase-activating protein 10 | 25 | 89309 | 6.75 | 6 | 1 | 4 | 1 | 4 | 0.1 | 0.05 |
| SRA1_MOUSE | Steroid receptor RNA activator 1 | 25 | 25541 | 6.01 | 2 | 1 | 2 | 1 | 2 | 0.17 | 0.18 |
| COR1B_MOUSE | Coronin-1B | 25 | 53878 | 5.54 | 9 | 1 | 6 | 1 | 6 | 0.18 | 0.08 |
| NIF3L_MOUSE | NIF3-like protein 1 | 25 | 41719 | 6.28 | 4 | 2 | 1 | 1 | 1 | 0.05 | 0.1 |
| LYOX_MOUSE | Protein-lysine 6-oxidase | 25 | 46671 | 8.73 | 3 | 1 | 1 | 1 | 1 | 0.08 | 0.09 |
| GSH1_MOUSE | Glutamate–cysteine ligase catalytic subunit | 25 | 72525 | 5.59 | 8 | 2 | 3 | 1 | 3 | 0.09 | 0.06 |
| GIT2_MOUSE | ARF GTPase-activating protein GIT2 | 25 | 78717 | 7.64 | 5 | 1 | 5 | 1 | 5 | 0.12 | 0.05 |
| BL1S2_MOUSE | Biogenesis of lysosome-related organelles complex 1 subunit 2 | 24 | 16289 | 4.82 | 3 | 2 | 1 | 1 | 1 | 0.19 | 0.29 |
| IBP7_MOUSE | Insulin-like growth factor-binding protein 7 | 24 | 28951 | 8.71 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.15 |
| PLIN3_MOUSE | Perilipin-3 | 24 | 47233 | 5.45 | 8 | 2 | 4 | 2 | 4 | 0.24 | 0.19 |
| TM9S2_MOUSE | Transmembrane 9 superfamily member 2 | 24 | 75280 | 7.21 | 5 | 2 | 2 | 1 | 2 | 0.09 | 0.06 |
| CAP2_MOUSE | Adenylyl cyclase-associated protein 2 | 24 | 52829 | 6 | 3 | 1 | 2 | 1 | 2 | 0.06 | 0.08 |
| LPP_MOUSE | Lipoma-preferred partner homolog | 24 | 65848 | 7.19 | 7 | 1 | 5 | 1 | 5 | 0.17 | 0.07 |
| STXB3_MOUSE | Syntaxin-binding protein 3 | 24 | 67899 | 8.28 | 5 | 1 | 3 | 1 | 3 | 0.12 | 0.06 |
| EDC4_MOUSE | Enhancer of mRNA-decapping protein 4 | 23 | 152389 | 5.51 | 5 | 1 | 4 | 1 | 4 | 0.06 | 0.03 |
| CO6A2_MOUSE | Collagen alpha-2(VI) chain | 23 | 110266 | 6.01 | 2 | 1 | 2 | 1 | 2 | 0.04 | 0.04 |
| ATG4B_MOUSE | Cysteine protease ATG4B | 23 | 44347 | 4.93 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.1 |
| GGPPS_MOUSE | Geranylgeranyl pyrophosphate synthase | 23 | 34685 | 6.03 | 2 | 1 | 1 | 1 | 1 | 0.09 | 0.13 |
| TT39B_MOUSE | Tetratricopeptide repeat protein 39B | 23 | 70248 | 6.22 | 3 | 1 | 2 | 1 | 2 | 0.06 | 0.06 |
| VPS11_MOUSE | Vacuolar protein sorting-associated protein 11 homolog | 22 | 107650 | 6.54 | 3 | 1 | 2 | 1 | 2 | 0.05 | 0.04 |
| KANK2_MOUSE | KN motif and ankyrin repeat domain-containing protein 2 | 22 | 90190 | 5.38 | 2 | 1 | 2 | 1 | 2 | 0.06 | 0.05 |
| ANO6_MOUSE | Anoctamin-6 | 22 | 106186 | 6.34 | 2 | 1 | 1 | 1 | 1 | 0.01 | 0.04 |
| ACAD8_MOUSE | Isobutyryl-CoA dehydrogenase, mitochondrial | 22 | 44990 | 8.46 | 1 | 1 | 1 | 1 | 1 | 0.06 | 0.1 |
| BASI_MOUSE | Basigin | 22 | 42418 | 5.56 | 3 | 1 | 1 | 1 | 1 | 0.04 | 0.1 |
| RO60_MOUSE | 60 kDa SS-A/Ro ribonucleoprotein | 22 | 60085 | 8.11 | 4 | 1 | 3 | 1 | 3 | 0.09 | 0.07 |
| AHSA2_MOUSE | Activator of 90 kDa heat shock protein ATPase homolog 2 | 22 | 37624 | 6.1 | 2 | 1 | 2 | 1 | 2 | 0.11 | 0.12 |
| CND2_MOUSE | Condensin complex subunit 2 | 21 | 82251 | 4.81 | 2 | 1 | 2 | 1 | 2 | 0.05 | 0.05 |
| TM214_MOUSE | Transmembrane protein 214 | 21 | 76381 | 9.41 | 3 | 1 | 2 | 1 | 2 | 0.06 | 0.06 |
| MPC1_MOUSE | Mitochondrial pyruvate carrier 1 | 21 | 12446 | 9.67 | 3 | 1 | 1 | 1 | 1 | 0.19 | 0.39 |
| PIGT_MOUSE | GPI transamidase component PIG-T | 21 | 65663 | 8.68 | 1 | 1 | 1 | 1 | 1 | 0.03 | 0.07 |
| UBP4_MOUSE | Ubiquitin carboxyl-terminal hydrolase 4 | 21 | 108274 | 5.42 | 6 | 2 | 3 | 1 | 3 | 0.09 | 0.04 |
| DNLI1_MOUSE | DNA ligase 1 | 21 | 102226 | 6.43 | 1 | 1 | 1 | 1 | 1 | 0.02 | 0.04 |
| FUCO_MOUSE | Tissue alpha-L-fucosidase | 20 | 52247 | 6.47 | 1 | 1 | 1 | 1 | 1 | 0.04 | 0.08 |
| CEBPZ_MOUSE | CCAAT/enhancer-binding protein zeta | 20 | 120187 | 5.53 | 5 | 1 | 3 | 1 | 3 | 0.08 | 0.04 |
| E2AK2_MOUSE | Interferon-induced, double-stranded RNA-activated protein kinase | 20 | 58243 | 8.76 | 4 | 1 | 3 | 1 | 3 | 0.12 | 0.07 |
| AFG31_MOUSE | AFG3-like protein 1 | 20 | 86992 | 9 | 1 | 1 | 1 | 1 | 1 | 0.02 | 0.05 |
| RICTR_MOUSE | Rapamycin-insensitive companion of mTOR | 20 | 191449 | 6.8 | 9 | 1 | 5 | 1 | 5 | 0.05 | 0.02 |
| NOC4L_MOUSE | Nucleolar complex protein 4 homolog | 19 | 58638 | 6.29 | 3 | 1 | 1 | 1 | 1 | 0.04 | 0.07 |
| SOAT1_MOUSE | Sterol O-acyltransferase 1 | 19 | 63757 | 9.15 | 6 | 1 | 4 | 1 | 4 | 0.15 | 0.07 |
| SH3G1_MOUSE | Endophilin-A2 | 19 | 41492 | 5.53 | 2 | 1 | 1 | 1 | 1 | 0.04 | 0.11 |
| ILVBL_MOUSE | Acetolactate synthase-like protein | 19 | 68113 | 8.96 | 2 | 1 | 2 | 1 | 2 | 0.09 | 0.06 |
| LASP1_MOUSE | LIM and SH3 domain protein 1 | 19 | 29975 | 6.61 | 6 | 1 | 5 | 1 | 5 | 0.19 | 0.15 |
| NUMA1_MOUSE | Nuclear mitotic apparatus protein 1 | 18 | 235487 | 5.68 | 10 | 1 | 10 | 1 | 10 | 0.08 | 0.02 |
| DPP8_MOUSE | Dipeptidyl peptidase 8 | 18 | 102121 | 5.51 | 3 | 1 | 2 | 1 | 2 | 0.04 | 0.04 |
| MTX2_MOUSE | Metaxin-2 | 18 | 29739 | 5.44 | 1 | 1 | 1 | 1 | 1 | 0.11 | 0.15 |
| RM15_MOUSE | 39S ribosomal protein L15, mitochondrial | 18 | 33521 | 10.08 | 2 | 1 | 2 | 1 | 2 | 0.1 | 0.13 |
| PM34_MOUSE | Peroxisomal membrane protein PMP34 | 17 | 34391 | 10.11 | 1 | 1 | 1 | 1 | 1 | 0.07 | 0.13 |
| PPP5_MOUSE | Serine/threonine-protein phosphatase 5 | 17 | 56840 | 5.83 | 4 | 1 | 4 | 1 | 4 | 0.17 | 0.08 |
| DOK1_MOUSE | Docking protein 1 | 16 | 52419 | 6.16 | 4 | 1 | 3 | 1 | 3 | 0.14 | 0.08 |
| LEG9_MOUSE | Galectin-9 | 16 | 40010 | 9.41 | 3 | 1 | 1 | 1 | 1 | 0.06 | 0.11 |
| PGM2_MOUSE | Phosphoglucomutase-2 | 16 | 68704 | 5.78 | 2 | 1 | 2 | 1 | 2 | 0.08 | 0.06 |
| HSP7E_MOUSE | Heat shock 70 kDa protein 14 | 16 | 54616 | 5.63 | 4 | 1 | 4 | 1 | 4 | 0.17 | 0.08 |
| PUM2_MOUSE | Pumilio homolog 2 | 15 | 114243 | 6.61 | 3 | 1 | 3 | 1 | 3 | 0.08 | 0.04 |
- aProtein score is calculated from the score of the peptide attributed to the protein. bpI is (predicted) isoelectric point. cNumber of matches is spectrum number matched to protein#1. dNumber of significant matches is spectrum number that matches protein and exceeds the identification criteria. eNumber of sequences is number of peptides matched to protein#2. fNumber of significant sequences is number of peptides exceeding the identification criteria matched to proteins. gNumber of unique sequences is a unique#3 number of peptides matched to proteins. hSequence coverage is the ratio of the total number of matched peptide residues to the total length of the protein. iExponentially modified protein abundance index (http://www.matrixscience.com/help/quant_empai_help.html). #1When multiple spectra are matched to the same peptide, the way of counting into one is called "peptide number". #2When multiple spectra are matched to the same peptide, the method of counting is called "spectrum number". #3"Unique" is a peptide that is not matched to other proteins and has been assigned only to the relevant protein.
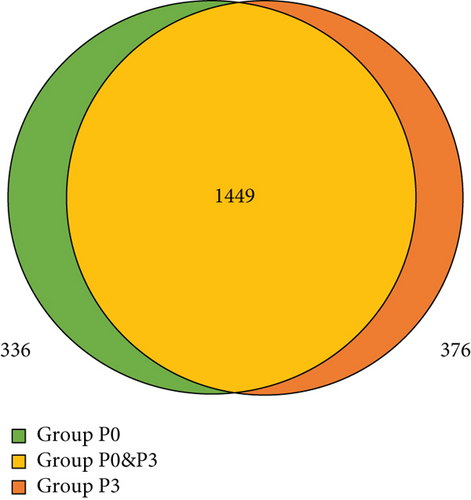
The amount of protein quantified in this paper is a theoretical value estimated based on the emPAI function of the Scaffold software program. The ratio of the number of measured peptides to the number of theoretical peptides is linearly related to the logarithm of the protein concentration, and the number obtained by subtracting 1 from the index of the peptide number ratio was defined as the emPAI. The larger the emPAI value, the greater the amount of protein. We recently published a paper on the correlation between different emPAI values (>0, >1, >2, >3, >5, and >10) and the results of protein expression analyses. The results showed that, for an emPAI value > 10, the presence of protein can be detected with a high probability, even if the number of samples is n = 1 [25]. Proteins quantified using emPAI were listed from the top in the tables showing the GO analysis results (Tables 1 and 2) in descending order of concentration. Since the data of this study were derived from a single protein expression analysis obtained from mMSC-ATs of three mice, the data reliability must be carefully considered.
3.4. The Biological Processes, Cellular Components, and Molecular Function of Proteins Identified from mMSC-ATs (P0)
The biological processes of proteins were analyzed using the Mascot software program with the SwissProt 2017 database.
3.4.1. Biological Processes
Antiviral protein was detected as a protein component of mMSC-ATs (P0) (rich in components of SVF). Isg15, Npc2, Ripk3, Fmr1, Dag1, Vps16, Gas6, Stmn1, Vapb, Tri25, Eea1, Asc, Ifm2, Bst2, Apoe, Ltor5, Oas1a, and Nect2 were detected as factors related to the viral process (Figure 3).
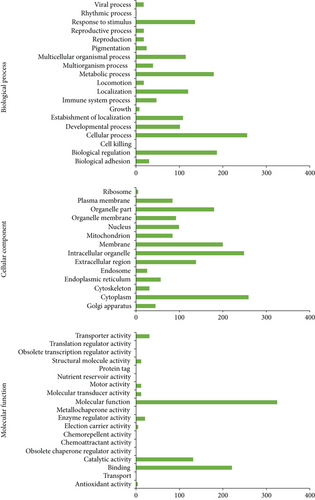
Stml2 (CD4-positive, alpha-beta T cell activation), CD36, CD180 (B cell proliferation), CD47 (opsonization), and CD166 (adaptive immune response) were detected as factors related to the CD cell surface markers related to the immune system. Cats, Ha1b, Erap1, Mdr1a, Psb8, and Ha11 were detected as factors related to the MHC class antigen and thereby related to the immune system. Csf1 (osteoclast differentiation), CD109 (osteoclast fusion), and Rab35 (antigen processing and presentation) were detected as factors related to the bone immune system. Isg15, Tri25, Ifm2, Bst2, Oas1a, and Asc were detected as factors related to the antiviral immune system. Tgbr3, Plek (hematopoietic progenitor cell differentiation), Ripk3 (T cell differentiation in thymus), Gas6 (macrophage cytokine production), Sfxn1 (erythrocyte differentiation), Ada (T cell activation, B cell differentiation), Itam (activated T cell proliferation), Pfd1 (B cell activation), Sbds (leukocyte chemotaxis), Aimp1 (leukocyte migration), Armc6 (hematopoietic progenitor cell differentiation), Glrx5 (hemopoiesis), Hdac7 (B cell activation, B cell differentiation), Psn1 (T cell activation), Ada10 (monocyte activation), and Tpd52 (B cell differentiation) were detected as factors related to hematopoiesis. A4, Ada15, Ptms (immune system process), Fcgrn (antigen processing and presentation), Il4ra, Ilf2, Ic1 (complement activation), Olr1, and Snp23 (histamine secretion by mast cell) were detected as factors related to the immune response. Stat1 was detected as an interferon-related immune response factor. Those proteins were detected as factors related to the immune system process (Figure 3).
Csf1 (mammary duct terminal end bud growth), Tgbr3 (cardiac muscle cell proliferation), A4 (synaptic growth at neuromuscular junction), Ada15 (tissue regeneration), CD166 (axon extension involved in axon guidance, neuron projection extension), Tbl1r (multicellular organism growth), Ninj1 (tissue regeneration), and Sbds (inner cell mass cell proliferation) were detected as factors related to growth in biological processes (Figure 3).
3.4.2. Cellular Components
Stml2 (T cell receptor complex), Stx7 (immunological synapse), Ha1b (MHC class I protein complex), Ha11 (MHC class I protein complex), CD166 (T cell receptor complex), At2b1 (apical plasma membrane), Gna11 (heterotrimeric G-protein complex), Home3 (postsynaptic membrane), Ampn, Rdh11, Nsf1c, Bag3, Csf1, Vatc1, Necp2, CD109, CD36, Ece1, Tgbr3, A4, Ripk3, Gdn, Erln2, Aaat, Suca, Snp23, Prio, Ly6a, Plp2, Plek, Tpbg, Fmr1, Plin4, Ldlr, Pdc10, Dag1, Rap2a, Vatg1, Pp2ba, Rab35, Il1ap, Eea1, Fcgrn, Ada, Cdv3, Erap1, Ifm2, Bst2, CD180, Il4ra, Itam, Osmr, P2rx4, Serc1, Tradd, CD97, Rab4b, Rab8a, Sucb2, Apoe, Bap31, Cdipt, Cn37, Crk, Dnjb4, Ggt5, Mdr1a, Piez1, Praf1, Psn1, S39ae, Stx2, Tnr12, Vmp1, Fen1, CD47, Olr1, Pygl, Wwox, Ctl1, Ly6c1, Nect2, and Ttyh2 (plasma membrane) were detected as factors related to growth in the plasma membrane of the cellular component (Figure 3).
3.4.3. Molecular Functions
SRXN1 (antioxidant activity), TXD17 (peroxidase activity), PGFS (thioredoxin peroxidase activity), and APOE (antioxidant activity) are proteins that are expressed on the plasma membrane. These proteins have been reported to be factors related to both growth and the antioxidant activity according to a classification of the molecular functions (Figure 3). The proteins of mMSC-ATs (P0) are listed in Table 3 with their molecular functions described in detail.
| Exclusive P0 group | |||
|---|---|---|---|
| UniProt/Swiss-Prot ID | Description | Molecular function | |
| FABP5_MOUSE | Fatty acid-binding protein, epidermal | Fatty acid binding, transporter activity | |
| AMPN_MOUSE | Aminopeptidase N | Metal ion binding, metalloaminopeptidase activity, metallopeptidase activity, peptide binding, zinc ion binding | |
| AOFA_MOUSE | Amine oxidase [flavin-containing] A | Oxidoreductase activity, primary amine oxidase activity, protein binding | |
| HXK3_MOUSE | Hexokinase-3 | ATP binding, fructokinase activity, glucokinase activity, glucose binding, kinase activity, mannokinase activity, nucleotide binding | |
| CTGF_MOUSE | Connective tissue growth factor | Fibronectin binding, growth factor activity, heparin binding, insulin-like growth factor binding, integrin binding, protein C-terminus binding | |
| THIL_MOUSE | Acetyl-CoA acetyltransferase, mitochondrial | Acetyl-CoA C-acetyltransferase activity, carbon-carbon lyase activity, coenzyme binding, enzyme binding, ligase activity, forming carbon-carbon bonds, metal ion binding, protein homodimerization activity, transferase activity, transferring acyl groups | |
| ATPD_MOUSE | ATP synthase subunit delta, mitochondrial | ATPase activity, hydrogen ion transmembrane transporter activity, proton-transporting ATP synthase activity, rotational mechanism | |
| CATS_MOUSE | Cathepsin S | Collagen binding, cysteine-type endopeptidase activity, cysteine-type peptidase activity, fibronectin binding, hydrolase activity, laminin binding, proteoglycan binding | |
| CYR61_MOUSE | Protein CYR61 | Extracellular matrix binding, growth factor binding, heparin binding, insulin-like growth factor binding, integrin binding | |
| ETFB_MOUSE | Electron transfer flavoprotein subunit beta | Electron carrier activity | |
| SFXN3_MOUSE | Sideroflexin-3 | Ion transmembrane transporter activity, molecular function | |
| STML2_MOUSE | Stomatin-like protein 2, mitochondrial | GTPase binding, cardiolipin binding, lipid binding | |
| RDH11_MOUSE | Retinol dehydrogenase 11 | NADP-retinol dehydrogenase activity, retinol dehydrogenase activity | |
| NSF1C_MOUSE | NSFL1 cofactor p47 | ATPase binding, lipid binding, phospholipid binding, ubiquitin binding | |
| BAG3_MOUSE | BAG family molecular chaperone regulator 3 | Adenyl-nucleotide exchange factor activity, cadherin binding involved in cell-cell adhesion, chaperone binding, protein complex binding | |
| CSF1_MOUSE | Macrophage colony-stimulating factor 1 | Cytokine activity, growth factor activity, macrophage colony-stimulating factor receptor binding, protein homodimerization activity | |
| LYZ2_MOUSE | Lysozyme C-2 | Hydrolase activity, identical protein binding, lysozyme activity | |
| VATC1_MOUSE | V-type proton ATPase subunit C 1 | Hydrogen-exporting ATPase activity, phosphorylative mechanism | |
| UBP2L_MOUSE | Ubiquitin-associated protein 2-like | RNA binding | |
| RL28_MOUSE | 60S ribosomal protein L28 | RNA binding, structural constituent of ribosome | |
| ACSL4_MOUSE | Long-chain-fatty-acid–CoA ligase 4 | ATP binding, arachidonate-CoA ligase activity, decanoate–CoA ligase activity, ligase activity, long-chain fatty acid-CoA ligase activity, nucleotide binding, very-long-chain fatty acid-CoA ligase activity | |
| THIKA_MOUSE | 3-Ketoacyl-CoA thiolase A, peroxisomal | Acetate CoA-transferase activity, acetyl-CoA C-acetyltransferase activity, acetyl-CoA C-acyltransferase activity, palmitoyl-CoA oxidase activity | |
| FA49B_MOUSE | Protein FAM49B | Protein binding | |
| CO5A2_MOUSE | Collagen alpha-2(V) chain | SMAD binding, extracellular matrix structural constituent, metal ion binding | |
| DCTN2_MOUSE | Dynactin subunit 2 | Motor activity, spectrin binding | |
| SUCB1_MOUSE | Succinate–CoA ligase [ADP-forming] subunit beta, mitochondrial | ATP binding, ligase activity, metal ion binding, nucleotide binding, succinate-CoA ligase (ADP-forming) activity | |
| SYUG_MOUSE | Gamma-synuclein | Protein binding | |
| GUAD_MOUSE | Guanine deaminase | Guanine deaminase activity, hydrolase activity, metal ion binding, zinc ion binding | |
| ISG15_MOUSE | Ubiquitin-like protein ISG15 | Protein binding, protein tag | |
| STK24_MOUSE | Serine/threonine-protein kinase 24 | ATP binding, cadherin binding involved in cell-cell adhesion, metal ion binding, nucleotide binding, signal transducer, downstream of receptor, with serine/threonine kinase activity, transferase activity | |
| STX7_MOUSE | Syntaxin-7 | SNAP receptor activity, chloride channel inhibitor activity, syntaxin binding | |
| EFHD2_MOUSE | EF-hand domain-containing protein D2 | Cadherin binding involved in cell-cell adhesion, calcium ion binding, metal ion binding | |
| NECP2_MOUSE | Adaptin ear-binding coat-associated protein 2 | Molecular function | |
| ATP5J_MOUSE | ATP synthase-coupling factor 6, mitochondrial | ATPase activity, hydrogen ion transmembrane transporter activity | |
| ERH_MOUSE | Enhancer of rudimentary homolog | RNA binding, methyl-CpG binding | |
| HAP28_MOUSE | 28 kDa heat- and acid-stable phosphoprotein | RNA binding, platelet-derived growth factor binding | |
| TOM20_MOUSE | Mitochondrial import receptor subunit TOM20 homolog | P-P-bond-hydrolysis-driven protein transmembrane transporter activity, mitochondrion targeting sequence binding, protein channel activity, unfolded protein binding | |
| CD109_MOUSE | CD109 antigen | Serine-type endopeptidase inhibitor activity, transforming growth factor beta binding | |
| CD36_MOUSE | Platelet glycoprotein 4 | High-density lipoprotein particle binding, lipid binding, lipoteichoic acid receptor activity, low-density lipoprotein particle binding, low-density lipoprotein receptor activity, protein binding | |
| ECE1_MOUSE | Endothelin-converting enzyme 1 | Metal ion binding, metalloendopeptidase activity, metallopeptidase activity, protein homodimerization activity | |
| STX12_MOUSE | Syntaxin-12 | SNAP receptor activity, SNARE binding, protein binding | |
| TOM1_MOUSE | Target of Myb protein 1 | Clathrin binding | |
| CO3A1_MOUSE | Collagen alpha-1(III) chain | SMAD binding, extracellular matrix structural constituent, integrin binding, metal ion binding, platelet-derived growth factor binding | |
| NPC2_MOUSE | Epididymal secretory protein E1 | Cholesterol binding, enzyme binding | |
| HA1B_MOUSE | H-2 class I histocompatibility antigen, K-B alpha chain | RNA binding, beta-2-microglobulin binding, peptide antigen binding, receptor binding | |
| VKOR1_MOUSE | Vitamin K epoxide reductase complex subunit 1 | Oxidoreductase activity, quinone binding, vitamin-K-epoxide reductase (warfarin-sensitive) activity | |
| CUL3_MOUSE | Cullin-3 | POZ domain binding, cyclin binding, protein binding, protein heterodimerization activity, protein homodimerization activity, ubiquitin protein ligase activity, ubiquitin protein ligase binding | |
| HA11_MOUSE | H-2 class I histocompatibility antigen, D-B alpha chain | RNA binding, beta-2-microglobulin binding, peptide antigen binding, protein binding | |
| ACAD9_MOUSE | Acyl-CoA dehydrogenase family member 9, mitochondrial | Acyl-CoA dehydrogenase activity, electron carrier activity, fatty-acyl-CoA binding, flavin adenine dinucleotide binding, oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor | |
| TGBR3_MOUSE | Transforming growth factor beta receptor type 3 | PDZ domain binding, SMAD binding, coreceptor activity, glycosaminoglycan binding, protein binding, transforming growth factor beta binding, transforming growth factor beta receptor activity, type III, transforming growth factor beta-activated receptor activity, type II transforming growth factor beta receptor binding | |
| A4_MOUSE | Amyloid-beta A4 protein | DNA binding, PTB domain binding, enzyme binding, growth factor receptor binding, heparin binding, identical protein binding, peptidase activator activity, peptidase inhibitor activity, serine-type endopeptidase inhibitor activity, transition metal ion binding | |
| RBMS2_MOUSE | RNA-binding motif, single-stranded-interacting protein 2 | RNA binding, nucleotide binding | |
| PAPS2_MOUSE | Bifunctional 3 ′-phosphoadenosine 5 ′-phosphosulfate synthase 2 | ATP binding, adenylylsulfate kinase activity, catalytic activity, nucleotide binding, nucleotidyltransferase activity, sulfate adenylyltransferase (ATP) activity | |
| SPA3N_MOUSE | Serine protease inhibitor A3N | Peptidase inhibitor activity, serine-type endopeptidase inhibitor activity | |
| DDX17_MOUSE | Probable ATP-dependent RNA helicase DDX17 | ATP binding, ATP-dependent RNA helicase activity, RNA binding, estrogen receptor binding, nucleotide binding, transcription coactivator activity | |
| RIPK3_MOUSE | Receptor-interacting serine/threonine-protein kinase 3 | ATP binding, NF-kappaB-inducing kinase activity, identical protein binding, nucleotide binding, protein complex binding | |
| MCAT_MOUSE | Mitochondrial carnitine/acylcarnitine carrier protein | Acyl carnitine transmembrane transporter activity | |
| GDN_MOUSE | Glia-derived nexin | Glycosaminoglycan binding, heparin binding, receptor binding, serine-type endopeptidase inhibitor activity | |
| ERLN2_MOUSE | Erlin-2 | Cholesterol binding, protein binding, ubiquitin protein ligase binding | |
| ARPC5_MOUSE | Actin-related protein 2/3 complex subunit 5 | Actin filament binding, structural constituent of cytoskeleton | |
| AAAT_MOUSE | Neutral amino acid transporter B(0) | Neutral amino acid transmembrane transporter activity, symporter activity | |
| NAGK_MOUSE | N-Acetyl-D-glucosamine kinase | ATP binding, N-acetylglucosamine kinase activity, N-acylmannosamine kinase activity, kinase activity, nucleotide binding | |
| STAT1_MOUSE | Signal transducer and activator of transcription 1 | DNA binding, RNA polymerase II core promoter proximal region sequence-specific DNA binding, RNA polymerase II core promoter sequence-specific DNA binding, cadherin binding involved in cell-cell adhesion, double-stranded DNA binding, enzyme binding, nuclear hormone receptor binding, protein homodimerization activity, signal transducer activity, transcription factor activity, RNA polymerase II core promoter sequence-specific, transcription factor activity, sequence-specific DNA binding, tumor necrosis factor receptor binding | |
| CCD47_MOUSE | Coiled-coil domain-containing protein 47 | RNA binding, calcium ion binding | |
| NDUB7_MOUSE | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 | NADH dehydrogenase (ubiquinone) activity | |
| F13A_MOUSE | Coagulation factor XIII A chain | Metal ion binding, protein-glutamine gamma-glutamyltransferase activity | |
| SUCA_MOUSE | Succinate–CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | GTP binding, RNA binding, cofactor binding, ligase activity, nucleotide binding, succinate-CoA ligase (ADP-forming) activity, succinate-CoA ligase (GDP-forming) activity | |
| FACR1_MOUSE | Fatty acyl-CoA reductase 1 | Fatty-acyl-CoA reductase (alcohol-forming) activity, long-chain-fatty-acyl-CoA reductase activity, oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | |
| EFTS_MOUSE | Elongation factor Ts, mitochondrial | RNA binding, translation elongation factor activity | |
| TM165_MOUSE | Transmembrane protein 165 | Molecular function | |
| ADA15_MOUSE | Disintegrin and metalloproteinase domain-containing protein 15 | SH3 domain binding, integrin binding, metal ion binding, metalloendopeptidase activity, metallopeptidase activity, peptidase activity, protein binding | |
| SNP23_MOUSE | Synaptosomal-associated protein 23 | SNAP receptor activity, syntaxin binding | |
| PRIO_MOUSE | Major prion protein | ATP-dependent protein binding, chaperone binding, copper ion binding, identical protein binding, ion channel binding, lamin binding, microtubule binding | |
| LY6A_MOUSE | Lymphocyte antigen 6A-2/6E-1 | ||
| PLP2_MOUSE | Proteolipid protein 2 | Chemokine binding | |
| TSNAX_MOUSE | Translin-associated protein X | A2A adenosine receptor binding, RNA binding, metal ion binding, protein complex binding, sequence-specific DNA binding, single-stranded DNA binding | |
| T106A_MOUSE | Transmembrane protein 106A | Molecular function | |
| UBFD1_MOUSE | Ubiquitin domain-containing protein UBFD1 | RNA binding, cadherin binding involved in cell-cell adhesion | |
| PLEK_MOUSE | Pleckstrin | Phosphatidylinositol-3,4-bisphosphate binding, protein homodimerization activity, protein kinase C binding | |
| TPBG_MOUSE | Trophoblast glycoprotein | ||
| ILEUA_MOUSE | Leukocyte elastase inhibitor A | Serine-type endopeptidase inhibitor activity | |
| TIMP2_MOUSE | Metalloproteinase inhibitor 2 | Enzyme activator activity, integrin binding, metal ion binding, metalloendopeptidase inhibitor activity, peptidase inhibitor activity, protease binding | |
| FMR1_MOUSE | Synaptic functional regulator FMR1 | G-quadruplex RNA binding, RNA stem-loop binding, RNA strand annealing activity, chromatin binding, dynein complex binding, ion channel binding, mRNA 3 ′-UTR binding, mRNA 5 ′-UTR binding, methylated histone binding, miRNA binding, microtubule binding, poly(G) binding, poly(U) RNA binding, protein heterodimerization activity, protein homodimerization activity, ribosome binding, sequence-specific mRNA binding, siRNA binding, translation initiation factor binding, translation repressor activity | |
| DCUP_MOUSE | Uroporphyrinogen decarboxylase | Carboxy-lyase activity, uroporphyrinogen decarboxylase activity | |
| PLIN4_MOUSE | Perilipin-4 | ||
| RHEB_MOUSE | GTP-binding protein Rheb | GTP binding, metal ion binding, nucleotide binding, protein kinase binding | |
| TPD52_MOUSE | Tumor protein D52 | Calcium ion binding, protein binding, protein heterodimerization activity, protein homodimerization activity | |
| LDLR_MOUSE | Low-density lipoprotein receptor | Calcium ion binding, glycoprotein binding, identical protein binding, low-density lipoprotein particle binding, low-density lipoprotein receptor activity, protease binding, very-low-density lipoprotein particle receptor activity | |
| PDC10_MOUSE | Programmed cell death protein 10 | Protein N-terminus binding, protein homodimerization activity, protein kinase binding | |
| UFM1_MOUSE | Ubiquitin-fold modifier 1 | Molecular function | |
| SRXN1_MOUSE | Sulfiredoxin-1 | ATP binding, antioxidant activity, nucleotide binding, oxidoreductase activity, oxidoreductase activity, acting on a sulfur group of donors, sulfiredoxin activity | |
| GAPR1_MOUSE | Golgi-associated plant pathogenesis-related protein 1 | Protein homodimerization activity | |
| CSN5_MOUSE | COP9 signalosome complex subunit 5 | Hydrolase activity, metal ion binding, metallopeptidase activity, protein binding, thiol-dependent ubiquitin-specific protease activity, transcription coactivator activity | |
| OCAD1_MOUSE | OCIA domain-containing protein 1 | Molecular function | |
| DCAKD_MOUSE | Dephospho-CoA kinase domain-containing protein | ATP binding, dephospho-CoA kinase activity, nucleotide binding | |
| CD166_MOUSE | CD166 antigen | Protein binding | |
| ATG3_MOUSE | Ubiquitin-like-conjugating enzyme ATG3 | Atg12 transferase activity, Atg8 ligase activity, enzyme binding, ligase activity, ubiquitin-like protein transferase activity | |
| DAG1_MOUSE | Dystroglycan | SH2 domain binding, actin binding, alpha-actinin binding, calcium ion binding, dystroglycan binding, protein binding, protein complex binding, structural constituent of muscle, tubulin binding, vinculin binding | |
| RAP2A_MOUSE | Ras-related protein Rap-2a | GTP binding, GTPase activity, nucleotide binding, protein binding | |
| MPEG1_MOUSE | Macrophage-expressed gene 1 protein | ||
| RBX1_MOUSE | E3 ubiquitin-protein ligase RBX1 | NEDD8 transferase activity, cullin family protein binding, eukaryotic initiation factor 4E binding, ligase activity, protein complex binding, ubiquitin protein ligase activity, ubiquitin protein ligase binding, ubiquitin-ubiquitin ligase activity, zinc ion binding | |
| VPS16_MOUSE | Vacuolar protein sorting-associated protein 16 homolog | Actin binding | |
| PKHO2_MOUSE | Pleckstrin homology domain-containing family O member 2 | Molecular function | |
| GAS6_MOUSE | Growth arrest-specific protein 6 | Binding, bridging, calcium ion binding, cysteine-type endopeptidase inhibitor activity involved in apoptotic process, phosphatidylserine binding, protein tyrosine kinase activator activity, receptor agonist activity, receptor binding, receptor tyrosine kinase binding, voltage-gated calcium channel activity | |
| GMPPA_MOUSE | Mannose-1-phosphate guanyltransferase alpha | Nucleotidyltransferase activity, transferase activity | |
| COMD4_MOUSE | COMM domain-containing protein 4 | Molecular function | |
| MECR_MOUSE | Enoyl-[acyl-carrier-protein] reductase, mitochondrial | Ligand-dependent nuclear receptor binding, oxidoreductase activity, trans-2-enoyl-CoA reductase (NADPH) activity, zinc ion binding | |
| PTMS_MOUSE | Parathymosin | Zinc ion binding | |
| RMD3_MOUSE | Regulator of microtubule dynamics protein 3 | Molecular function | |
| SRSF1_MOUSE | Serine/arginine-rich splicing factor 1 | RNA binding, RS domain binding, mRNA binding, nucleotide binding, protein kinase B binding | |
| TMCO1_MOUSE | Calcium load-activated calcium channel | Calcium channel activity | |
| TMX1_MOUSE | Thioredoxin-related transmembrane protein 1 | Disulfide oxidoreductase activity, protein disulfide isomerase activity | |
| VATG1_MOUSE | V-type proton ATPase subunit G 1 | ATPase activity, ATPase binding, hydrogen-exporting ATPase activity, phosphorylative mechanism, hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | |
| STMN1_MOUSE | Stathmin | Protein binding, tubulin binding | |
| H14_MOUSE | Histone H1.4 | DNA binding, RNA binding, chromatin DNA binding, protein binding | |
| PROS_MOUSE | Vitamin K-dependent protein S | Calcium ion binding | |
| PP2BA_MOUSE | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Calcium-dependent protein serine/threonine phosphatase activity, calmodulin binding, calmodulin-dependent protein phosphatase activity, drug binding, enzyme binding, hydrolase activity, metal ion binding, protein dimerization activity, protein heterodimerization activity | |
| VAPB_MOUSE | Vesicle-associated membrane protein-associated protein B | FFAT motif binding, beta-tubulin binding, cadherin binding involved in cell-cell adhesion, enzyme binding, microtubule binding, protein heterodimerization activity, protein homodimerization activity | |
| TBL1R_MOUSE | F-box-like/WD repeat-containing protein TBL1XR1 | DNA binding, beta-catenin binding, histone binding, protein N-terminus binding, protein binding, transcription corepressor activity, transcription regulatory region DNA binding | |
| RAB35_MOUSE | Ras-related protein Rab-35 | GDP binding, GTP binding, GTPase activity, nucleotide binding, phosphatidylinositol-4,5-bisphosphate binding, protein binding | |
| BID_MOUSE | BH3-interacting domain death agonist | Protein heterodimerization activity, ubiquitin protein ligase binding | |
| GLTP_MOUSE | Glycolipid transfer protein | Glycolipid binding, glycolipid transporter activity | |
| IL1RA_MOUSE | Interleukin-1 receptor antagonist protein | Cytokine activity, interleukin-1 Type I receptor antagonist activity, interleukin-1 Type II receptor antagonist activity, interleukin-1, Type I receptor binding, interleukin-1, Type II receptor binding | |
| PDXK_MOUSE | Pyridoxal kinase | ATP binding, kinase activity, lithium ion binding, magnesium ion binding, nucleotide binding, potassium ion binding, protein homodimerization activity, pyridoxal kinase activity, pyridoxal phosphate binding, sodium ion binding, zinc ion binding | |
| TRI25_MOUSE | E3 ubiquitin/ISG15 ligase TRIM25 | RNA binding, acid-amino acid ligase activity, cadherin binding involved in cell-cell adhesion, ligase activity, metal ion binding, ubiquitin protein ligase activity involved in ERAD pathway, ubiquitin-protein transferase activity, zinc ion binding | |
| VATD_MOUSE | V-type proton ATPase subunit D | ATPase activity, coupled to transmembrane movement of substances | |
| IL1AP_MOUSE | Interleukin-1 receptor accessory protein | Interleukin-1 receptor activity, interleukin-33 receptor activity, protein tyrosine kinase binding, signal transducer activity | |
| SFXN1_MOUSE | Sideroflexin-1 | Ion transmembrane transporter activity | |
| EEA1_MOUSE | Early endosome antigen 1 | 1-Phosphatidylinositol binding, GTP-dependent protein binding, metal ion binding, protein homodimerization activity | |
| CBR2_MOUSE | Carbonyl reductase [NADPH] 2 | Carbonyl reductase (NADPH) activity, oxidoreductase activity, protein binding, protein self-association | |
| FKB14_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP14 | FK506 binding, calcium ion binding, isomerase activity, peptidyl-prolyl cis-trans isomerase activity | |
| DEGS1_MOUSE | Sphingolipid delta(4)-desaturase DES1 | Sphingolipid delta-4 desaturase activity | |
| FCGRN_MOUSE | IgG receptor FcRn large subunit p51 | IgG binding, IgG receptor activity, antigen binding, beta-2-microglobulin binding, peptide antigen binding | |
| ACSF2_MOUSE | Acyl-CoA synthetase family member 2, mitochondrial | ATP binding, ligase activity, molecular function, nucleotide binding | |
| ADA_MOUSE | Adenosine deaminase | Adenosine deaminase activity, deaminase activity, hydrolase activity, purine nucleoside binding, zinc ion binding | |
| ASC_MOUSE | Apoptosis-associated speck-like protein containing a CARD | BMP receptor binding, Pyrin domain binding, cysteine-type endopeptidase activity involved in apoptotic process, enzyme binding, interleukin-6 receptor binding, ion channel binding, myosin I binding, peptidase activator activity involved in apoptotic process, protease binding, protein dimerization activity, protein homodimerization activity, tropomyosin binding | |
| TXD17_MOUSE | Thioredoxin domain-containing protein 17 | Peroxidase activity, protein-disulfide reductase activity | |
| MXRA8_MOUSE | Matrix remodeling-associated protein 8 | Molecular function | |
| COX6C_MOUSE | Cytochrome c oxidase subunit 6C | Cytochrome c oxidase activity, molecular function | |
| COASY_MOUSE | Bifunctional coenzyme A synthase | ATP binding, dephospho-CoA kinase activity, nucleotide binding, pantetheine-phosphate adenylyltransferase activity, transferase activity | |
| CHCH2_MOUSE | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2 | Sequence-specific DNA binding, transcription factor binding | |
| SHOC2_MOUSE | Leucine-rich repeat protein SHOC-2 | Protein phosphatase 1 binding, protein phosphatase binding | |
| CDV3_MOUSE | Protein CDV3 | Molecular function | |
| ERAP1_MOUSE | Endoplasmic reticulum aminopeptidase 1 | Endopeptidase activity, interleukin-6 receptor binding, metalloaminopeptidase activity, metalloexopeptidase activity, peptide binding, zinc ion binding | |
| AL3A2_MOUSE | Fatty aldehyde dehydrogenase | 3-Chloroallyl aldehyde dehydrogenase activity, aldehyde dehydrogenase (NAD) activity, aldehyde dehydrogenase [NAD(P)+] activity, long-chain-alcohol oxidase activity | |
| ABHEB_MOUSE | Protein ABHD14B | Hydrolase activity | |
| GLYG_MOUSE | Glycogenin-1 | Glycogenin glucosyltransferase activity, metal ion binding, transferase activity, transferase activity, transferring glycosyl groups | |
| BPNT1_MOUSE | 3 ′(2 ′),5 ′-Bisphosphate nucleotidase 1 | 3 ′(2 ′),5 ′-Bisphosphate nucleotidase activity, hydrolase activity, magnesium ion binding, metal ion binding | |
| IFM2_MOUSE | Interferon-induced transmembrane protein 2 | Molecular function | |
| PFD6_MOUSE | Prefoldin subunit 6 | Chaperone binding, unfolded protein binding | |
| COMD8_MOUSE | COMM domain-containing protein 8 | Molecular function | |
| S10AD_MOUSE | Protein S100-A13 | RAGE receptor binding, calcium ion binding, copper ion binding, fibroblast growth factor binding, lipid binding, metal ion binding, protein homodimerization activity, zinc ion binding | |
| T176A_MOUSE | Transmembrane protein 176A | Protein binding | |
| SPD2B_MOUSE | SH3 and PX domain-containing protein 2B | SH2 domain binding, phosphatidylinositol-3,5-bisphosphate binding, phosphatidylinositol-3-phosphate binding, phosphatidylinositol-4-phosphate binding, phosphatidylinositol-5-phosphate binding, superoxide-generating NADPH oxidase activator activity | |
| RM46_MOUSE | 39S ribosomal protein L46, mitochondrial | Hydrolase activity, structural constituent of ribosome | |
| ADAS_MOUSE | Alkyldihydroxyacetonephosphate synthase, peroxisomal | FAD binding, alkylglycerone-phosphate synthase activity, flavin adenine dinucleotide binding, oxidoreductase activity, acting on CH-OH group of donors | |
| APMAP_MOUSE | Adipocyte plasma membrane-associated protein | Arylesterase activity, hydrolase activity, acting on ester bonds, strictosidine synthase activity | |
| ARK72_MOUSE | Aflatoxin B1 aldehyde reductase member 2 | Oxidoreductase activity, oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor, phenanthrene-9,10-epoxide hydrolase activity | |
| BST2_MOUSE | Bone marrow stromal antigen 2 | RNA binding, metalloendopeptidase inhibitor activity, protein homodimerization activity, signal transducer activity | |
| CASP1_MOUSE | Caspase-1 | Cysteine-type endopeptidase activity, cysteine-type endopeptidase activity involved in apoptotic process, cysteine-type peptidase activity, protein binding | |
| CD180_MOUSE | CD180 antigen | Receptor activity | |
| CK054_MOUSE | Ester hydrolase C11orf54 homolog | Hydrolase activity, acting on ester bonds, metal ion binding, zinc ion binding | |
| COX7C_MOUSE | Cytochrome c oxidase subunit 7C, mitochondrial | Cytochrome c oxidase activity | |
| CPSF7_MOUSE | Cleavage and polyadenylation specificity factor subunit 7 | RNA binding, nucleotide binding | |
| DCAF8_MOUSE | DDB1- and CUL4-associated factor 8 | Molecular function | |
| DHB11_MOUSE | Estradiol 17-beta-dehydrogenase 11 | Estradiol 17-beta-dehydrogenase activity, steroid dehydrogenase activity | |
| ERGI1_MOUSE | Endoplasmic reticulum-Golgi intermediate compartment protein 1 | Molecular function | |
| ERGI3_MOUSE | Endoplasmic reticulum-Golgi intermediate compartment protein 3 | Molecular function | |
| FKB15_MOUSE | FK506-binding protein 15 | Actin binding, peptidyl-prolyl cis-trans isomerase activity | |
| GSTA1_MOUSE | Glutathione S-transferase A1 | Glutathione transferase activity, transferase activity | |
| IL4RA_MOUSE | Interleukin-4 receptor subunit alpha | Cytokine receptor activity, protein binding | |
| ILF2_MOUSE | Interleukin enhancer-binding factor 2 | ATP binding, DNA binding, RNA binding, double-stranded RNA binding, transferase activity | |
| ITAM_MOUSE | Integrin alpha-M | Glycoprotein binding, heparan sulfate proteoglycan binding, heparin binding, metal ion binding, opsonin binding | |
| LSG1_MOUSE | Large subunit GTPase 1 homolog | GTP binding, GTPase activity, nucleotide binding | |
| NDUA2_MOUSE | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 | NADH dehydrogenase (ubiquinone) activity | |
| NINJ1_MOUSE | Ninjurin-1 | ||
| OSMR_MOUSE | Oncostatin-M-specific receptor subunit beta | Cytokine binding, cytokine receptor activity, growth factor binding, oncostatin-M receptor activity | |
| P2RX4_MOUSE | P2X purinoceptor 4 | ATP binding, cadherin binding, extracellular ATP-gated cation channel activity, ion channel activity, purinergic nucleotide receptor activity | |
| PFD1_MOUSE | Prefoldin subunit 1 | Protein binding involved in protein folding, unfolded protein binding | |
| PGFS_MOUSE | Prostamide/prostaglandin F synthase | Prostaglandin-F synthase activity, thioredoxin peroxidase activity | |
| PR2C2_MOUSE | Prolactin-2C2 | Growth factor activity, hormone activity | |
| PRRX1_MOUSE | Paired mesoderm homeobox protein 1 | DNA binding, HMG box domain binding, RNA polymerase II transcription coactivator activity, sequence-specific DNA binding | |
| RL35_MOUSE | 60S ribosomal protein L35 | RNA binding, mRNA binding, structural constituent of ribosome | |
| S10A1_MOUSE | Protein S100-A1 | ATPase binding, S100 protein binding, calcium ion binding, protein homodimerization activity | |
| SBDS_MOUSE | Ribosome maturation protein SBDS | Microtubule binding, rRNA binding, ribosome binding | |
| SDF2_MOUSE | Stromal cell-derived factor 2 | Dolichyl-phosphate-mannose-protein mannosyltransferase activity | |
| SERC1_MOUSE | Serine incorporator 1 | L-Serine transmembrane transporter activity | |
| TIAR_MOUSE | Nucleolysin TIAR | AU-rich element binding, RNA binding, nucleotide binding | |
| TRADD_MOUSE | Tumor necrosis factor receptor type 1-associated DEATH domain protein | Binding, bridging, death domain binding, identical protein binding, kinase binding, protein complex binding, signal transducer activity, tumor necrosis factor receptor binding | |
| TRNT1_MOUSE | CCA tRNA nucleotidyltransferase 1, mitochondrial | ATP binding, ATP:3 ′-cytidine-cytidine-tRNA adenylyltransferase activity, CTP:3 ′-cytidine-tRNA cytidylyltransferase activity, CTP:tRNA cytidylyltransferase activity, nucleotide binding, tRNA binding, tRNA nucleotidyltransferase activity | |
| VPS52_MOUSE | Vacuolar protein sorting-associated protein 52 homolog | Syntaxin binding | |
| ZFYV1_MOUSE | Zinc finger FYVE domain-containing protein 1 | 1-Phosphatidylinositol binding, metal ion binding, phosphatidylinositol-3,4,5-trisphosphate binding, phosphatidylinositol-3,4-bisphosphate binding | |
| ZWINT_MOUSE | ZW10 interactor | Protein N-terminus binding, protein binding | |
| FBLN3_MOUSE | EGF-containing fibulin-like extracellular matrix protein 1 | Calcium ion binding, epidermal growth factor receptor binding, epidermal growth factor-activated receptor activity, growth factor activity | |
| NDUB9_MOUSE | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 | NADH dehydrogenase (ubiquinone) activity | |
| MIC10_MOUSE | MICOS complex subunit Mic10 | Molecular function | |
| NPL_MOUSE | N-Acetylneuraminate lyase | N-Acetylneuraminate lyase activity, lyase activity | |
| UCRI_MOUSE | Cytochrome b-c1 complex subunit Rieske, mitochondrial | 2 iron, 2 sulfur cluster binding, metal ion binding, oxidoreductase activity, acting on diphenols and related substances as donors, protein complex binding, ubiquinol-cytochrome c reductase activity | |
| CD97_MOUSE | CD97 antigen | G-protein coupled receptor activity, calcium ion binding, signal transducer activity | |
| WDR26_MOUSE | WD repeat-containing protein 26 | Molecular function | |
| XIRP2_MOUSE | Xin actin-binding repeat-containing protein 2 | Actin binding, alpha-actinin binding, protein binding, zinc ion binding | |
| DYR_MOUSE | Dihydrofolate reductase | NADP binding, NADPH binding, dihydrofolate reductase activity, dihydrofolic acid binding, folic acid binding, mRNA binding, methotrexate binding, oxidoreductase activity | |
| AIMP1_MOUSE | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1 | GTPase binding, cytokine activity, protein homodimerization activity, tRNA binding | |
| SYTC2_MOUSE | Probable threonine–tRNA ligase 2, cytoplasmic | ATP binding, RNA binding, nucleotide binding, threonine-tRNA ligase activity | |
| RAB4B_MOUSE | Ras-related protein Rab-4B | GTP binding, nucleotide binding | |
| RAB8A_MOUSE | Ras-related protein Rab-8A | GDP binding, GTP binding, GTPase activity, Rab GTPase binding, myosin V binding, nucleotide binding, protein binding, protein kinase binding | |
| AT2B1_MOUSE | Plasma membrane calcium-transporting ATPase 1 | ATP binding, PDZ domain binding, calcium-transporting ATPase activity, calmodulin binding, hydrolase activity, metal ion binding, nucleotide binding | |
| GNA11_MOUSE | Guanine nucleotide-binding protein subunit alpha-11 | G-protein beta/gamma-subunit complex binding, GTP binding, GTPase activity, alkylglycerophosphoethanolamine phosphodiesterase activity, guanyl nucleotide binding, metal ion binding, signal transducer activity, type 2A serotonin receptor binding | |
| SUCB2_MOUSE | Succinate–CoA ligase [GDP-forming] subunit beta, mitochondrial | ATP binding, GDP binding, GTP binding, ligase activity, metal ion binding, nucleotide binding, protein heterodimerization activity, succinate-CoA ligase (GDP-forming) activity, succinate-semialdehyde dehydrogenase (NAD+) activity | |
| GIPC1_MOUSE | PDZ domain-containing protein GIPC1 | GTPase activator activity, PDZ domain binding, actin binding, cadherin binding involved in cell-cell adhesion, myosin binding, protein homodimerization activity, receptor binding | |
| ACPM_MOUSE | Acyl carrier protein, mitochondrial | Molecular function | |
| ALG11_MOUSE | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity, transferase activity, transferase activity, transferring glycosyl groups | |
| AN32E_MOUSE | Acidic leucine-rich nuclear phosphoprotein 32 family member E | Histone binding, phosphatase inhibitor activity | |
| APOE_MOUSE | Apolipoprotein E | Antioxidant activity, beta-amyloid binding, cholesterol binding, cholesterol transporter activity, heparin binding, lipid transporter activity, lipoprotein particle binding, low-density lipoprotein particle receptor binding, metal chelating activity, phosphatidylcholine-sterol O-acyltransferase activator activity, phospholipid binding, protein binding, protein homodimerization activity, tau protein binding, very-low-density lipoprotein particle receptor binding | |
| ARMC6_MOUSE | Armadillo repeat-containing protein 6 | Molecular function | |
| BAP31_MOUSE | B cell receptor-associated protein 31 | MHC class I protein binding, protein complex binding | |
| CC90B_MOUSE | Coiled-coil domain-containing protein 90B, mitochondrial | Molecular function | |
| CCD22_MOUSE | Coiled-coil domain-containing protein 22 | Cullin family protein binding, protein binding | |
| CDIPT_MOUSE | CDP-diacylglycerol–inositol 3-phosphatidyltransferase | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity, alcohol binding, carbohydrate binding, diacylglycerol binding, manganese ion binding | |
| CGAT1_MOUSE | Chondroitin sulfate N-acetylgalactosaminyltransferase 1 | Glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity, glucuronosyltransferase activity, glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity, metal ion binding, peptidoglycan glycosyltransferase activity | |
| CN37_MOUSE | 2 ′,3 ′-Cyclic-nucleotide 3 ′-phosphodiesterase | 2 ′,3 ′-Cyclic-nucleotide 3 ′-phosphodiesterase activity, RNA binding, cyclic nucleotide binding, hydrolase activity | |
| CO4A2_MOUSE | Collagen alpha-2(IV) chain | Extracellular matrix structural constituent | |
| CPSF6_MOUSE | Cleavage and polyadenylation specificity factor subunit 6 | RNA binding, mRNA binding, nucleotide binding | |
| CRK_MOUSE | Adapter molecule crk | SH2 domain binding, SH3/SH2 adaptor activity, enzyme binding, ephrin receptor binding, protein binding, bridging, protein phosphorylated amino acid binding | |
| CTBP2_MOUSE | C-terminal-binding protein 2 | NAD binding, chromatin binding, oxidoreductase activity, oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor, protein homodimerization activity, retinoic acid receptor binding, transcription coactivator activity, transcription corepressor activity | |
| DAD1_MOUSE | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit DAD1 | Dolichyl-diphosphooligosaccharide-protein glycotransferase activity, transferase activity, transferring glycosyl groups | |
| DERL2_MOUSE | Derlin-2 | Molecular function | |
| DHSO_MOUSE | Sorbitol dehydrogenase | D-Xylulose reductase activity, L-iditol 2-dehydrogenase activity, NAD binding, identical protein binding, zinc ion binding | |
| DNJB4_MOUSE | DnaJ homolog subfamily B member 4 | Chaperone binding, unfolded protein binding | |
| DYLT1_MOUSE | Dynein light chain Tctex-type 1 | G-protein beta-subunit binding, GTP-dependent protein binding, identical protein binding, motor activity | |
| DYLT3_MOUSE | Dynein light chain Tctex-type 3 | Identical protein binding, motor activity | |
| EFL1_MOUSE | Elongation factor-like GTPase 1 | GTP binding, GTPase activity, nucleotide binding, ribosome binding | |
| EH1L1_MOUSE | EH domain-binding protein 1-like protein 1 | Molecular function | |
| EMC8_MOUSE | ER membrane protein complex subunit 8 | Molecular function | |
| ETHE1_MOUSE | Persulfide dioxygenase ETHE1, mitochondrial | Dioxygenase activity, iron ion binding, metal ion binding, sulfur dioxygenase activity | |
| FKB11_MOUSE | Peptidyl-prolyl cis-trans isomerase FKBP11 | FK506 binding, isomerase activity, peptidyl-prolyl cis-trans isomerase activity | |
| FNTB_MOUSE | Protein farnesyltransferase subunit beta | Drug binding, farnesyltranstransferase activity, isoprenoid binding, peptide binding, protein farnesyltransferase activity, zinc ion binding | |
| FUND2_MOUSE | FUN14 domain-containing protein 2 | Molecular function | |
| GGA1_MOUSE | ADP-ribosylation factor-binding protein GGA1 | ||
| GGT5_MOUSE | Glutathione hydrolase 5 proenzyme | Gamma-glutamyltransferase activity, glutathione hydrolase activity | |
| GINM1_MOUSE | Glycoprotein integral membrane protein 1 | Molecular function | |
| GLRX5_MOUSE | Glutaredoxin-related protein 5, mitochondrial | 2 iron, 2 sulfur cluster binding, electron carrier activity, metal ion binding, protein disulfide oxidoreductase activity | |
| HDAC7_MOUSE | Histone deacetylase 7 | 14-3-3 protein binding, NAD-dependent histone deacetylase activity (H3-K14 specific), activating transcription factor binding, chromatin binding, metal ion binding, protein kinase C binding, protein kinase binding, repressing transcription factor binding, transcription corepressor activity | |
| HEBP1_MOUSE | Heme-binding protein 1 | Heme binding | |
| HIG1A_MOUSE | HIG1 domain family member 1A, mitochondrial | Molecular function | |
| HOME3_MOUSE | Homer protein homolog 3 | G-protein coupled glutamate receptor binding, protein C-terminus binding, protein domain specific binding | |
| HRG1_MOUSE | Heme transporter HRG1 | Heme transporter activity, molecular function | |
| HS2ST_MOUSE | Heparan sulfate 2-O-sulfotransferase 1 | Heparan sulfate 2-O-sulfotransferase activity, transferase activity | |
| IC1_MOUSE | Plasma protease C1 inhibitor | Peptidase inhibitor activity, serine-type endopeptidase inhibitor activity | |
| IDHG1_MOUSE | Isocitrate dehydrogenase [NAD] subunit gamma 1, mitochondrial | ATP binding, NAD binding, isocitrate dehydrogenase (NAD+) activity, magnesium ion binding, oxidoreductase activity | |
| LRN4L_MOUSE | LRRN4 C-terminal-like protein | Molecular function | |
| LTOR5_MOUSE | Ragulator complex protein LAMTOR5 | Guanyl-nucleotide exchange factor activity, protein complex scaffold | |
| LXN_MOUSE | Latexin | Enzyme inhibitor activity, heparin binding, metalloendopeptidase inhibitor activity | |
| MA2B2_MOUSE | Epididymis-specific alpha-mannosidase | Alpha-mannosidase activity, carbohydrate binding, hydrolase activity, mannosidase activity, zinc ion binding | |
| MDR1A_MOUSE | Multidrug resistance protein 1A | ATP binding, ATPase activity, coupled, ATPase activity, coupled to transmembrane movement of substances, ceramide-translocating ATPase activity, nucleotide binding, phosphatidylcholine-translocating ATPase activity, phosphatidylethanolamine-translocating ATPase activity, xenobiotic-transporting ATPase activity | |
| MIC13_MOUSE | MICOS complex subunit MIC13 | Molecular function | |
| MMAB_MOUSE | Cob(I)yrinic acid a,c-diamide adenosyltransferase, mitochondrial | ATP binding, cob(I)yrinic acid a,c-diamide adenosyltransferase activity, nucleotide binding, transferase activity | |
| MMSA_MOUSE | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial | RNA binding, aldehyde dehydrogenase (NAD) activity, malonate-semialdehyde dehydrogenase (acetylating) activity, methylmalonate-semialdehyde dehydrogenase (acylating) activity, oxidoreductase activity | |
| MPI_MOUSE | Mannose-6-phosphate isomerase | Isomerase activity, mannose-6-phosphate isomerase activity, zinc ion binding | |
| NAGA_MOUSE | N-Acetylglucosamine-6-phosphate deacetylase | N-Acetylglucosamine-6-phosphate deacetylase activity, hydrolase activity, metal ion binding | |
| NDUS4_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | NADH dehydrogenase (ubiquinone) activity | |
| NDUS5_MOUSE | NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 | Molecular function | |
| NHLC3_MOUSE | NHL repeat-containing protein 3 | Molecular function | |
| NU160_MOUSE | Nuclear pore complex protein Nup160 | Nucleocytoplasmic transporter activity | |
| NUP50_MOUSE | Nuclear pore complex protein Nup50 | Ran GTPase binding | |
| OAS1A_MOUSE | 2 ′-5 ′-Oligoadenylate synthase 1A | 2 ′-5 ′-Oligoadenylate synthetase activity, ATP binding, double-stranded RNA binding, metal ion binding, nucleotide binding, protein binding, transferase activity | |
| PEDF_MOUSE | Pigment epithelium-derived factor | Serine-type endopeptidase inhibitor activity | |
| PEX14_MOUSE | Peroxisomal membrane protein PEX14 | Beta-tubulin binding, microtubule binding, protein N-terminus binding, receptor binding, transcription corepressor activity | |
| PEX19_MOUSE | Peroxisomal biogenesis factor 19 | ATPase binding, peroxisome membrane class-1 targeting sequence binding, protein N-terminus binding, protein binding | |
| PIEZ1_MOUSE | Piezo-type mechanosensitive ion channel component 1 | Cation channel activity, mechanically-gated ion channel activity | |
| PRAF1_MOUSE | Prenylated Rab acceptor protein 1 | Identical protein binding, proline-rich region binding, protein C-terminus binding | |
| PRAF2_MOUSE | PRA1 family protein 2 | Molecular function | |
| PREB_MOUSE | Prolactin regulatory element-binding protein | ARF guanyl-nucleotide exchange factor activity, DNA binding, GTPase binding, Rab guanyl-nucleotide exchange factor activity, Sar guanyl-nucleotide exchange factor activity, transcription factor activity, sequence-specific DNA binding | |
| PSN1_MOUSE | Presenilin-1 | PDZ domain binding, aspartic-type endopeptidase activity, beta-catenin binding, cadherin binding, calcium channel activity, endopeptidase activity | |
| RAB31_MOUSE | Ras-related protein Rab-31 | GDP binding, GTP binding, nucleotide binding | |
| RDH13_MOUSE | Retinol dehydrogenase 13 | Oxidoreductase activity | |
| RT11_MOUSE | 28S ribosomal protein S11, mitochondrial | mRNA 5 ′-UTR binding, small ribosomal subunit rRNA binding, structural constituent of ribosome | |
| S2546_MOUSE | Solute carrier family 25 member 46 | Molecular function | |
| S35F6_MOUSE | Solute carrier family 35 member F6 | Molecular function | |
| S39AE_MOUSE | Zinc transporter ZIP14 | Ferrous iron transmembrane transporter activity, metal ion transmembrane transporter activity, zinc ion transmembrane transporter activity | |
| SCFD2_MOUSE | Sec1 family domain-containing protein 2 | Molecular function | |
| SSRP1_MOUSE | FACT complex subunit SSRP1 | DNA binding, RNA binding, chromatin binding | |
| STX2_MOUSE | Syntaxin-2 | SNAP receptor activity, SNARE binding, calcium-dependent protein binding, protein dimerization activity | |
| SYVN1_MOUSE | E3 ubiquitin-protein ligase synoviolin | ATPase binding, chaperone binding, ligase activity, ubiquitin protein ligase activity, ubiquitin protein ligase activity involved in ERAD pathway, ubiquitin-specific protease binding, unfolded protein binding, zinc ion binding | |
| TENS3_MOUSE | Tensin-3 | Molecular function | |
| TI8AB_MOUSE | Putative mitochondrial import inner membrane translocase subunit Tim8 A-B | Metal ion binding | |
| TMX2_MOUSE | Thioredoxin-related transmembrane protein 2 | Molecular function | |
| TNR12_MOUSE | Tumor necrosis factor receptor superfamily member 12A | Protein binding | |
| TOIP2_MOUSE | Torsin-1A-interacting protein 2 | ATPase activator activity, ATPase binding | |
| TOM5_MOUSE | Mitochondrial import receptor subunit TOM5 homolog | Protein transporter activity | |
| TPC13_MOUSE | Trafficking protein particle complex subunit 13 | Molecular function | |
| UBP19_MOUSE | Ubiquitin carboxyl-terminal hydrolase 19 | Hsp90 protein binding, Lys48-specific deubiquitinase activity, metal ion binding, thiol-dependent ubiquitin-specific protease activity, ubiquitin protein ligase binding | |
| UFSP2_MOUSE | Ufm1-specific protease 2 | UFM1 hydrolase activity, cysteine-type peptidase activity, thiolester hydrolase activity | |
| VMP1_MOUSE | Vacuole membrane protein 1 | Molecular function | |
| TIMP1_MOUSE | Metalloproteinase inhibitor 1 | Cytokine activity, growth factor activity, metal ion binding, metalloendopeptidase inhibitor activity, protease binding | |
| TM9S4_MOUSE | Transmembrane 9 superfamily member 4 | Molecular function | |
| PSB8_MOUSE | Proteasome subunit beta type-8 | Endopeptidase activity, peptidase activity, threonine-type endopeptidase activity | |
| QCR7_MOUSE | Cytochrome b-c1 complex subunit 7 | Ubiquinol-cytochrome c reductase activity | |
| ADA10_MOUSE | Disintegrin and metalloproteinase domain-containing protein 10 | SH2 domain binding, SH3 domain binding, hydrolase activity, metal ion binding, metalloendopeptidase activity, metallopeptidase activity, protein binding, protein homodimerization activity, protein kinase binding | |
| KCT2_MOUSE | Keratinocyte-associated transmembrane protein 2 | Molecular function | |
| NDRG2_MOUSE | Protein NDRG2 | ||
| YIF1B_MOUSE | Protein YIF1B | Molecular function | |
| TIM8B_MOUSE | Mitochondrial import inner membrane translocase subunit Tim8 B | Metal ion binding | |
| DJC24_MOUSE | DnaJ homolog subfamily C member 24 | ATPase activator activity, ferrous iron binding, zinc ion binding | |
| VWA8_MOUSE | von Willebrand factor A domain-containing protein 8 | ATP binding, ATPase activity, nucleotide binding | |
| FEN1_MOUSE | Flap endonuclease 1 | 5 ′-3 ′ exonuclease activity, 5 ′-flap endonuclease activity, DNA binding, RNA-DNA hybrid ribonuclease activity, catalytic activity, exonuclease activity, flap endonuclease activity, magnesium ion binding, manganese ion binding | |
| TPD54_MOUSE | Tumor protein D54 | RNA binding | |
| MCL1_MOUSE | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | BH3 domain binding, protein heterodimerization activity, protein homodimerization activity | |
| PCBP4_MOUSE | Poly(rC)-binding protein 4 | DNA binding, RNA binding, mRNA 3 ′-UTR binding | |
| GCR_MOUSE | Glucocorticoid receptor | RNA polymerase II core promoter proximal region sequence-specific DNA binding, glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity, lipid binding, metal ion binding, protein complex binding, protein dimerization activity, sequence-specific DNA binding, steroid hormone binding, transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding, zinc ion binding | |
| RT16_MOUSE | 28S ribosomal protein S16, mitochondrial | Structural constituent of ribosome | |
| CD47_MOUSE | Leukocyte surface antigen CD47 | Protein binding, thrombospondin receptor activity | |
| UBP24_MOUSE | Ubiquitin carboxyl-terminal hydrolase 24 | Peptidase activity, thiol-dependent ubiquitinyl hydrolase activity | |
| ROMO1_MOUSE | Reactive oxygen species modulator 1 | Molecular function | |
| GOT1B_MOUSE | Vesicle transport protein GOT1B | Signal transducer activity | |
| OLR1_MOUSE | Oxidized low-density lipoprotein receptor 1 | Carbohydrate binding, low-density lipoprotein receptor activity | |
| SRSF2_MOUSE | Serine/arginine-rich splicing factor 2 | RNA binding, nucleotide binding, pre-mRNA binding, protein kinase C binding | |
| COMD2_MOUSE | COMM domain-containing protein 2 | Molecular function | |
| PYGL_MOUSE | Glycogen phosphorylase, liver form | AMP binding, ATP binding, bile acid binding, carbohydrate binding, catalytic activity, drug binding, glycogen phosphorylase activity, nucleotide binding, protein homodimerization activity, purine nucleobase binding, pyridoxal phosphate binding, vitamin binding | |
| NCF2_MOUSE | Neutrophil cytosol factor 2 | Rac GTPase binding, protein C-terminus binding, superoxide-generating NADPH oxidase activity | |
| YIF1A_MOUSE | Protein YIF1A | Molecular function | |
| PEPL1_MOUSE | Probable aminopeptidase NPEPL1 | Aminopeptidase activity, manganese ion binding, metalloexopeptidase activity | |
| WWOX_MOUSE | WW domain-containing oxidoreductase | RNA polymerase II transcription coactivator activity, enzyme binding, oxidoreductase activity | |
| PTH_MOUSE | Probable peptidyl-tRNA hydrolase | RNA binding, aminoacyl-tRNA hydrolase activity | |
| PYRD_MOUSE | Dihydroorotate dehydrogenase (quinone), mitochondrial | FMN binding, dihydroorotate dehydrogenase activity, drug binding, ubiquinone binding | |
| CTL1_MOUSE | Choline transporter-like protein 1 | Choline transmembrane transporter activity | |
| LY6C1_MOUSE | Lymphocyte antigen 6C1 | ||
| OTU6B_MOUSE | OTU domain-containing protein 6B | Thiol-dependent ubiquitinyl hydrolase activity | |
| NECT2_MOUSE | Nectin-2 | Cell adhesion molecule binding, identical protein binding, protein heterodimerization activity, protein homodimerization activity, receptor activity, receptor binding | |
| TTYH2_MOUSE | Protein tweety homolog 2 | Chloride channel activity, molecular function | |
| LMOD1_MOUSE | Leiomodin-1 | Actin binding, tropomyosin binding | |
| UD16_MOUSE | UDP-glucuronosyltransferase 1-6 | Glucuronosyltransferase activity, protein heterodimerization activity, protein homodimerization activity, transferase activity, transferring hexosyl groups | |
| MYL6B_MOUSE | Myosin light chain 6B | Calcium ion binding, motor activity, structural constituent of muscle | |
3.5. Relationship of the Quantitative Value (Normalized emPAI) per Housekeeping Gene of mMSC-ATs (P0 and P3)
The quantitative values of the proteins expressed in both mMSC-ATs (P0) and mMSC-ATs (P3) were represented with a scatter plot (y-axis = P3, x-axis = P0). The average quantitative value of P3-expressed proteins decreased to 69.4% compared to P0-expressed proteins (Atp5f1, B2m, Hprt1, Rplp1, Ppia, Rps18, Pgk1, Tfrc, Ywhaz, and Gapdh; Supplementary Figure 1). The quantitative values of Tubb5, Flnb, Tln1, Col1a1, Iqgap1, Hspa5, Flnc, Thbs1, Fn1, Prdx1, Rnh1, Col1a2, Vcl, Lcp1, and Fabp5 were higher at P0 than at P3. The quantitative value of Act2, Serponh1, Hsp90ab1, Hsp90aa1, Actbl2, Vdac1, S100a11, Anxa2, S100a6, Pgam1, argininosuccinate synthase (Ass1, which is regulated by hypoxia-inducible factor 1α (Hif1α)), Plec, Kpnb1, Gsn, Marcks, Eif5a, and Tpm4 was higher at P3 than at P0 (Figure 4).
We previously examined the protein components expressed by human MSC-ATs (hMSC-ATs) cultured in the clinical medium not containing FBS and hMSC-ATs cultured in DMEM containing FBS [25]. Based on the results of the protein expression analysis, the expression of TLN1, FLNC, and ASS1 was higher in hMSC-ATs cultured in DMEM containing FBS than in those cultured in the clinical culture medium. Therefore, the increased expression of Tln1, Flnc, and Ass1 protein at P3 compared with P0 is not caused by FBS. Regarding the change in the expression of Ass1, the activation level of Hif1α was considered to be higher at P0 than at P3, because the oxygen concentration is lower in vivo than in vitro [29, 30]. Therefore, the results of this study reflect not only the effect of cell division frequency but also the influence of oxygen concentration. When cultured cells are planted in a living body in a low-oxygen environment, the hypoxic response may be activated. Therefore, in order to interpret the results of this study more accurately, we must obtain data on the protein expression of MSCs cultured under hypoxic and high-oxygen conditions.
4. Conclusions
The functions of proteins classified by the GO analysis were quantified using the LC-MS/MS measurement system for the amount of proteins and components contained in primary cultured cells of mMSC-ATs and cells passaged three times. The ability of mMSC-ATs to differentiate into expression markers of cells, fat, and osteoblasts did not change, even after three passages. However, the protein expression decreased to 69.4%. The proteins whose expression levels decreased after three passages included Ass1 among the Hif-related proteins. Furthermore, it was revealed that 336 kinds of proteins are specifically expressed in primary cultured MSC-ATs. In conclusion, MSC-ATs used as therapeutic cells retained their cell properties after three passages but showed a decreased protein expression on LC-MS/MS.
Conflicts of Interest
The authors declare no conflicts of interest in association with the present study.
Authors’ Contributions
YN, SN, CS, and HN were assigned for the study design and study conducts. YN, SN, CS, TK, and HN were assigned for data collection, data analysis, and data interpretation. TK, NK, IS, MW, and JF were assigned for the provision of materials. YN, SN, and HN drafted the manuscript. YN, SN, and HN revised the content of the manuscript. All authors approved the final version of the manuscript. YN takes responsibility for the integrity of all of the data analyses.
Acknowledgments
We thank Naomi Kakazu (University of the Ryukyus) for clerical assistance and Saki Uema, Yuka Onishi, Maki Higa, Yuki Kawahira, and Saori Adaniya (University of the Ryukyus) for providing technical support. This work was supported by the Research Laboratory Center, Faculty of Medicine, and the Institute for Animal Experiments, Faculty of Medicine, University of the Ryukyus. This work was supported in part by the Japan Society for the Promotion of Science (JSPS; KAKENHI Grant numbers JP16H05404, JP16K10435, and JP18K08545), Japan Agency for Medical Research and Development, the Naito Foundation, and Okinawa Science and Technology Promotion Center (OSTC).
Open Research
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.




