Independent acquisition of short insertions at the RIR1 site in the spike N-terminal domain of the SARS-CoV-2 BA.2 lineage
Abstract
Although the major SARS-CoV-2 omicron lineages share over 30 non-synonymous substitutions in the spike glycoprotein, they show several unique mutations that were acquired after their ancestral split. One of the most intriguing mutations associated with BA.1 is the presence of the inserted tripeptide Glu-Pro-Glu within the N-terminal domain, at a site that had previously independently acquired short insertions in several other SARS-CoV-2 lineages. Although the functional implications of the small nucleotide sequences found at this insertion hotspot, named RIR1, are still unclear, we have previously hypothesized that they may play a compensatory role in counterbalancing minor fitness deficits associated with other co-occurring spike non-synonymous mutations. Here, we show that similar insertion events have independently occurred at RIR1 at least 20 times in early 2022 within the BA.2 lineage, being occasionally associated with significant community transmission. One of these omicron sublineages, characterized by a Ser-Gly-Arg insertion in position 212, has been responsible for over 4000 documented COVID-19 cases worldwide between January and July 2022, for the most part concentrated in Denmark, where it reached a national prevalence close to 4% (10% in the Nordjylland region) in mid-May. Although the concurrent spread of the BA.2.12.1, BA.4 and BA.5 lineages led to the rapid decline of this BA.2 sublineage, the independent acquisition of several other RIR1 insertions on a BA.2 genomic background suggests that these events may provide a slight fitness advantage. Therefore, they should be carefully monitored in the upcoming months in other emerging omicron-related lineages, including BA.5.
1 INTRODUCTION
Since its first emergence in humans during the Wuhan outbreak at the end of 2019, the genome of SARS-CoV-2 has been acquiring new mutations compared with the Wuhan-Hu-1 reference sequence at a rate close to 9 × 10−4 substitutions/site/year, based on GISAID data (Shu & McCauley, 2017). Nevertheless, in line with observations previously collected in other betacoronaviruses, such mutations do not display a uniform distribution, being disproportionately located within the spike glycoprotein, which has a key role in regulating viral entry through the interaction with receptors expressed on host cell membranes (Boni et al., 2020; Guo et al., 2020). Most specifically, the overwhelming majority of S gene mutations target the S1 subunit, which includes a N-terminal domain (NTD) and the C-terminal receptor-binding domain (RBD), responsible for ACE2 binding (Hoffmann et al., 2020; Li et al., 2003). As a major target of neutralizing antibodies (Ju et al., 2020; Walls et al., 2020), S1 has been characterized by strong positive selection and by a very high rate of non-synonymous substitutions during the first 2 years of the pandemics (i.e. nearly 2 × 10−2 amino acid/year, based on GISAID data (Shu & McCauley, 2017)). Starting from late 2020, several variants of interest (VOIs) and variants of concern (VOCs), characterized by the presence of non-synonymous spike mutations, emerged in different geographical locations, quickly outcompeting pre-existing viral lineages. Some of these mutations, such as N501Y, most likely increased the intrinsic transmissibility of the early VOCs alpha, beta and gamma, by increasing ACE2-binding affinity (Teruel et al., 2021; Zhu et al., 2021). However, other mutations associated with these VOCs targeted important antibody epitopes, highlighting the role played by growing population immunity in selecting variants characterized by immune escape properties (Harvey et al., 2021; Sabino et al., 2021). A few months later, the global spread of delta rapidly brought the other variants to the verge of extinction, revealing that the combination between mutations conferring antibody escape (e.g. L452R) (Planas et al., 2021) and enhanced intrinsic transmissibility (e.g. P681R) (Saito et al., 2021) could result in the generation of novel viral variants with highly enhanced fitness.
In late November 2021, the fifth VOC, named omicron, started to quickly spread in South Africa, showing a concerning association with reinfections (Pulliam et al., 2022). Omicron replaced delta with unprecedented speed, becoming the dominant SARS-CoV-2 lineage worldwide by early 2022. Unlike previous variants, which never showed the ability to fully escape the neutralizing activity of vaccine-elicited or convalescent sera, omicron displayed significant antigenic divergence compared with previous variants, suggesting its classification as a distinct SARS-CoV-2 serotype (Simon-Loriere & Schwartz, 2022). These immunological properties derive from the presence of a very high number of S1 mutations, several of which have been shown to mediate ACE2 recognition and/or immune escape (Cameroni et al., 2022; Mannar et al., 2022). Curiously, many of these mutations occur at sites subjected to strong purifying selection both in previous SARS-CoV-2 variants and in bat coronaviruses, suggesting that they may cooperatively interact to mitigate slight fitness deficits associated with individual substitution, with a significant structural and functional impact (Martin et al., 2022a). For example they may, to some extent, also explain the preferential use of the TMPRSS2-independent and cathepsin-mediated endosomal cell entry route (Pia & Rowland-Jones, 2022), as well as the lower fusogenicity and altered cell tropism displayed by this VOC (Meng et al., 2022).
Omicron includes different major lineages, which may derive from a chronic infection (Tegally et al., 2022) and rapidly outcompeted each other, gaining regional or global prevalence during early 2022. While BA.1 and BA.2 were the first ones to cause major outbreaks in humans, at the time of the writing of this manuscript, BA.5, endowed with multiple immune escape mutations, is quickly emerging as the dominant omicron lineage in multiple geographic locations (Cao et al., 2022; Hachmann et al., 2022; Wang et al., 2022). Although the major omicron lineages share several S1 mutations, they display a remarkable sequence divergence, which is roughly comparable with the divergence observed among earlier VOCs. One of the most puzzling mutations only found in BA.1 is a short insertion of three codons, which encodes the tripeptide Glu-Pro-Glu at position 215, within the spike NTD. Nearly all VOCs and VOIs carry small NTD deletions compared with Wuhan-Hu-1, which may either confer enhanced antibody escape or act as permissive mutations, counterbalancing the fitness cost of otherwise deleterious RBD mutations (Meng et al., 2021). These small deletions independently occurred on multiple occasions during SARS-CoV-2 evolution at Recurrent Deletion Regions (RDRs), marking one of the most significant signatures of convergent evolution documented in SARS-CoV-2. On the other hand, the occurrence of spike insertions has been much rarer and consequently subjected to far less study. We have previously shown that insertions of short sequence stretches, usually encoding three or four amino acids, had been independently acquired over 50 times at the very same NTD site, named Recurrent Insertion Region 1 (RIR1). Before the emergence of BA.1, RIR1 insertions have been documented in alpha, gamma and delta, where they did not lead to significant community spread, as well as in two other lineages (A.2.5 and B.1.214.2) that had a considerable diffusion in 2021 (Gerdol et al., 2022). Although the functional role of RIR1 insertions is presently unclear, their convergent acquisition by multiple viral lineages most certainly suggests the need for increased monitoring, due to the possibility of associated fitness advantages.
While BA.1 has been rapidly replaced by other omicron lineages characterized by higher intrinsic transmissibility or immune escape during early 2022, we here show that a few BA.2 sublineages have independently acquired RIR1 insertions. One of these, carrying a Ser-Gly-Arg insertion at position 212 and hereby named BA.2+ins(L), marks an interesting case due to the slight growth advantage it displayed over the parental BA.2 lineage in Denmark, before the rise of BA.5 led to its decline.
2 MATERIALS AND METHODS
All SARS-CoV-2 genome sequence data deposited in GISAID (Shu & McCauley, 2017) up to 16 July 2022 were screened, looking for sequences belonging to the BA.2 lineage and showing insertions between codons 210 and 220 in the S gene. Genomes displaying unidentified amino acids (i.e. X) in such insertions were flagged as the likely product of mis-assembly and discarded. The quality of all retrieved genomes was evaluated with NextClade CLI 2.3.0, removing all sequences marked as ‘bad’ or ‘mediocre’. Moreover, genomes assigned to the BA.2 lineage but carrying an ‘EPE’ insertion in position 215, previously described as one of the lineage-defining mutations of the sister lineage BA.1, were flagged as likely misclassified and were therefore discarded. All resulting genomes were grouped based on the inserted nucleotide sequence, whose exact position and phase of insertion were defined based on the alignment with the reference SARS-CoV-2 genome sequence (Wuhan-Hu-1, NCBI accession ID: NC_045512.2) and a reference BA.2 genome (EPI_ISL_8128502). To remove possible sequencing and bioinformatics artefacts, only the insertions independently obtained by at least two different laboratories (based on GISAID metadata) were considered for further analysis. Following the nomenclature scheme proposed in our previous work (Gerdol et al., 2022), each of these groups was labelled with progressive Roman numerals, following their chronological order of identification, starting from insertion L.
The molecular surveillance data from Denmark were subjected to further scrutiny, due to the local spread of one of the BA.2 sublineages carrying an insertion at RIR1, that is BA.2+ins(L). All genome data and associated metadata, deposited up to 16 July 2022, were downloaded from GISAID. This allowed to calculate the share of BA.2 genomes carrying insertion L, both at a national and at a regional scale (i.e. by separately taking into account Syddanmark, Sjælland, Nordjylland, Midtjylland and Hovestaden), up to 7 July 2022. The frequencies of observation of BA.2-ins(L) relative to all SARS-CoV-2 sequenced genomes reported the 7-day moving average, with 95% confidence intervals.
3 RESULTS AND DISCUSSION
In total, we identified 20 independent events of insertion at RIR1 associated with the BA.2 lineage. These were chronologically ordered, from the least to the most recent one, and assigned Roman numerals, from insertion L to insertion LXX, based on the naming scheme we have previously proposed (Gerdol et al., 2022) (Table 1).
| Designation | Insertion | Insertion type | GISAID entries | Other spike mutations not shared with BA.2a | Earliest detection | Latest detectionb |
|---|---|---|---|---|---|---|
| L | 212:SGR | in frame (codon 213 phase 0) | 4002 | / | 7 January 2022 | 5 July 2022 |
| LI | 212:NNR | out-of-frame (codon 212 phase 2) | 2 | G75D, L212F | 4 February 2022 | 7 February 2022 |
| LIII | 212:TVGG | in frame (codon 213 phase 0) | 914 | T1231S | 5 February 2022 | 4 July 2022 |
| LIV | 211:LTPT/212:TPTL | ambiguous; in frame (codon 212 phase 0/codon 213 phase 0) | 128 | / | 19 February 2022 | 16 June 2022 |
| LV | 212:MAEL | out-of-frame (codon 212 phase 2) | 68 | Y144del, L212F | 4 March 2022 | 5 June 2022 |
| LVI | 212:TGNTL | in frame (codon 213 phase 0) | 44 | / | 14 March 2022 | 20 June 2022 |
| LVII | 212:SEE | in frame (codon 213 phase 0) | 3 | / | 25 March 2022 | 23 May 2022 |
| LVIII | 212:QGK | in frame (codon 213 phase 0) | 31 | / | 28 March 2022 | 24 May 2022 |
| LIX | 212:QEG | in frame (codon 213 phase 0) | 2 | / | 5 April 2022 | 8 April 2022 |
| LX | 212:ETA | ambiguous (codon 213 phase 0/phase I) | 58 | / | 6 April 2022 | 20 June 2022 |
| LXI | 212:IRQ | in frame (codon 213 phase 0) | 50 | / | 11 April 2022 | 11 June 2022 |
| LXII | 212:EIVS | ambiguous (codon 213 phase 0/phase I) | 8 | / | 13 April 2022 | 28 April 2022 |
| LXIII | 212:AE | out-of-frame (codon 212 phase 2) | 4 | / | 14 April 2022 | 15 May 2022 |
| LXIV | 211:FNTY/212:NTYL | ambiguous (codon 211 phase 2/codon 212 phase 0/1/2) | 3 | / | 16 April 2022 | 19 May 2022 |
| LXV | 212:RDG | out-of-frame (codon 212 phase 2) | 2 | L212F | 17 April 2022 | 20 April 2022 |
| LXVI | 212:TME | in frame (codon 213 phase 0) | 2 | P1143S | 21 April 2022 | 25 April 2022 |
| LXVII | 211:REPD | out-of-frame (codon 211 phase 1) | 3 | N211T | 5 May 2022 | 5 July 2022 |
| LXVIII | 212:SEAG | out-of-frame (codon 212 phase 2) | 6 | L212F | 11 May 2022 | 11 June 2022 |
| LXIX | 212:SKI | out-of-frame (codon 212 phase 2) | 2 | L212F | 16 May 2022 | 16 May 2022 |
| LXX | 212:GVER/213:VERG | ambiguous; out-of-frame (codon 212 phase 2)/in-frame (codon 213 phase 0) | 2 | / | 14 May 2022 | 1 June 2022 |
- a Only mutations associated with at least 50% of the sequenced genomes are reported.
- b Latest detection, as of 16 July 2022.
The RIR1 insertions associated with BA.2 involved the acquisition of either two (a single case), three (ten cases), four (eight cases) or five (a single case) codons. However, their position was somewhat different compared with those that arose in other pre-omicron variants throughout 2020 and 2021 (Figure 1a). Indeed, one insertion (i.e. LXVII) occurred within codon 211 at phase I, six (i.e. LI, LV, LXIII, LXV, LXVIII and LXIX) within codon 212 at phase II and eight others (i.e. L, LIII, LVI, LVII, LVIII, LIX, LXI and LXVI) within codon 213 at phase 0. None of the 49 RIR1 insertions described in our previous work was observed in these locations (Gerdol et al., 2022). The exact placement of the four remaining BA.2-associated RIR1 insertions could not be unambiguously ascertained due to the possibility of multiple alternative sequence alignments with the BA.2 reference.
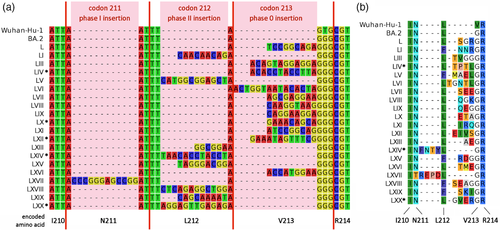
The inserted di-, tri-, tetra- or pentapeptides in BA.2 were mostly placed between Leu212 and Val213 (albeit this residue is replaced by Gly in BA.2), and never observed between Arg214 and Asp215, the most common placement for pre-omicron RIR1 insertions (Figure 1b). Because of the out-of-frame nature of several insertions, in five cases (insertions LI, LV, LXV, LXVIII and LXIX) Leu212 was replaced with Phe and in a single case (insertion LXVII) Asn211 was replaced with Thr (Figure 1b). As of note, due to of the presence of the Val213Gly substitution in BA.2, the placement of most of these insertions by GISAID was often offset by a single amino acid (e.g. insertion L is reported as ins213GRG instead of ins212SGR). Consequently, a non-synonymous substitution was often incorrectly called in position 213 (e.g. Val213Ser instead of Val213Gly for insertion L).
As previously mentioned, only three out of the 49 RIR1 insertions described in our previous work were associated with a significant community spread, either locally or globally: insertion III (lineage A.2.5), insertion IV (lineage B.1.214.2) and insertion XLI (lineage BA.1). The other 46 RIR1 insertions remained confined to small clusters counting no more than a few dozen sequenced cases, pointing out that not all RIR1 insertions were associated with the acquisition of increased fitness over previous variants. Due to the lack of structural overlap between RIR1 and important NTD antibody epitopes, we suggested that RIR1 insertions could mitigate slight negative fitness costs associated with the presence of non-synonymous RBD mutations affecting immune escape (Gerdol et al., 2022), as previously hypothesized for the H69/V70 deletion found in many VOCs and VOIs (Meng et al., 2021). The limited community spread of most SARS-CoV-2 variants bearing RIR1 insertions, together with their previously documented acquisition during in vitro experiments (Committee for Medicinal Products for Human Use, 2021; Shiliaev et al., 2021), may suggest a relevant role of intra-host selection in their emergence.
Although BA.2 is currently rapidly declining worldwide due to the recent rise of BA.5, a few sublineages carrying insertions at RIR1 have been responsible for several hundred documented cases in early 2022 (Table 1). With the single exception of BA.2.41 (i.e. the sublineage carrying insertion LIII), all these lack an official PANGO lineage designation. Five of these, whose continued detection for several months indicates sustained community transmission (Figure 2e), will be briefly discussed below.
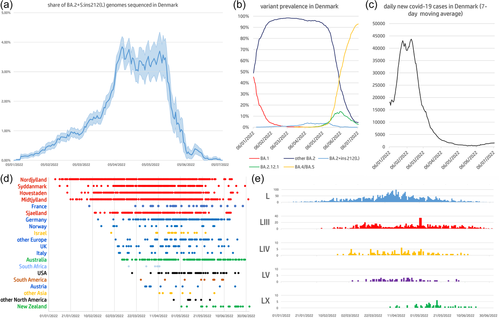
The most widespread BA.2 sublineage carrying an insertion at RIR1, with over 4000 sequenced genomes worldwide, is BA.2+ins(L), first detected on 7 January 2022 and characterized by the presence of an in-frame TCCGGCAGA insertion between codons 212 and 213, which determines the acquisition of the tripeptide Ser-Gly-Arg in position 212 (Figure 1). The overwhelming majority of the genomes belonging to this sublineage derive from Denmark (79.5%), the country where it apparently first originated, followed by Germany (5.4%), Australia (4.3%), France (2.2%), the United States (1.9%) and the United Kingdom (1.1%). As of 16 July 2022, cases have, however, been detected in all continents, with reports from Austria, Belgium, Brazil, Canada, Croatia, Czech Republic, Estonia, Hong Kong, Israel, Italy, Japan, Luxembourg, the Netherlands, New Zealand, Norway, Pakistan, Peru, Portugal, Singapore, Slovenia, South Africa, Spain, Sweden, Switzerland and Turkey (Figure 2d). The international spread of BA.2+ins(L) was likely the result of exportation from Denmark, where this sublineage displayed a moderate (10%–15%) weekly growth advantage over the parental BA.2 lineage from January to early April 2022. However, this trend was followed by a stationary phase (at a national prevalence comprised between 3% and 4%), which lasted approximately until mid-May (Figure 2a), coincident with the minimum in the daily number of nation-wide new cases, which fell below 1000 for the first time since October 2021 (Figure 2c). Later, the sudden rise of other competing omicron sublineages (BA.2.12.1 and BA.4/BA.5 in particular) led the rapid decline of all BA.2 sublineages, including BA.2+ins(L) (Figure 2b).
The national spread of BA.2+ins(L) was initially driven by the Nordjylland region, where this sublineage was first detected, undergoing a significant expansion compared with BA.2 from mid-January up to its peak of prevalence, on 11 May, when this BA.2 sublineage accounted for approximately 9.9% of all cases recorded in the region. Midtjylland, the only Denmark region with land borders with Nordjylland, followed a similar trend: after the detection of the first cases in late January, the share of BA.2+ins(L) genomes progressively increased over time, reaching the peak at approximately 6.3% of all cases on 13 May. The first few BA.2+ins(L) cases were recorded in Syddanmark, the southernmost continental region in Denmark, and in Hovestaden. This sublineage was first recorded in Sjælland, the largest insular region of Denmark, on 12 February.
In the second half of May and during June 2022, the observation of BA.2+ins(L) became more and more sporadic in all Danish regions, to the point that only one out of over 5000 genomes sequenced in early July belongs to this lineage, even though a number of cases continue to be detected abroad (Figure 2d). Even though the emergence of BA.2.12.1, BA.4 and BA.5 in an epidemiological context with high population immunity and low incidence of infections led to the rapid demise of BA.2+ins(L), the moderate growth advantage displayed by this sublineage over the parental BA.2 for nearly 3 months in multiple Danish regions warrants further investigation. Indeed, it would be interesting to clarify whether this trend was driven by biological factors (e.g. an alteration of the structure and function of the spike protein) or rather by other epidemiological factors, such as founder effects, as previously observed for other SARS-CoV-2 lineages in the past (Hodcroft et al., 2021).
The second most widespread BA.2 sublineage characterized by an insertion at RIR1 is BA.2+ins(LIII)/BA.2.41, with nearly 1000 sequenced genomes (Figure 2e; Table 1). First detected on 5 February 2022 in England, BA.2.41 displays a longer insertion compared with BA.2+ins(L), that is ACAGTAGGAGGA, which is also found in-frame between codons 212 and 213, and encodes the tetrapeptide Thr-Val-Gly-Gly (Figure 1). About 60% of the 914 sequenced genomes carrying this insertion sequenced so far (as of 16 July 2022) have been detected in England, where this sublineage is still occasionally sequenced, even though it never exceeded 1% prevalence over 5 months of community transmission. However, signs of international spread of BA.2.41 are marked by the identification of this sublineage in in Australia, Austria, Belgium, Canada, Czech Republic, Denmark, France, Georgia, Germany, Ireland, Israel, Italy, the Netherlands, Norway, Portugal, Spain, Trinidad & Tobago, Turkey and the United States.
Another noteworthy lineage is represented by BA.2+ins(LIV), characterized by an ACACCTACCTTA insertion, which occurs in an ambiguous site and could therefore be interpreted as either ins:211LTPT or ins:212:TPTL (Figure 1). This sublineage, first detected on 19 February 2022, mostly spread in Germany (where 63% of the genomes have been sequenced). However, its continued community transmission for at least 4 months (Figure 2e) led to its detection in seven other European countries, with two single cases detected outside the continent, in Israel and Canada.
Two additional relevant BA.2 sublineages carrying insertions at RIR1 mainly spread in the United States. BA.2+ins(LV), whose CATGGCGGAGCT out-of-frame insertion within codon 212 encodes the tetrapeptide Met-Ala-Glu-Leu (Figure 1), has been first identified on 4 March 2022, and has been detected other 67 times since, over a period of 3 months (Table 1; Figure 2e). Unlike the three aforementioned sublineages, insertion LV has been only spotted on four occasions abroad, pointing out limited exportation.
BA.2+ins(LX), carrying an in-frame GAAACAGCA insertion between codons 212 and 213, encoding the tripeptide Glu-Thr-Ala (Figure 1), was first identified on 6 April 2022 and has been so far responsible for 58 cases (Table 1; Figure 2e). Like insertion LV, insertion LX did not lead to significant clusters of infections abroad, with just three cases reported outside of the American continent.
Curiously, most BA.2-associated RIR1 insertions did not bear other relevant S1 non-synonymous substitutions targeting the RBD or known important epitopes recognized by neutralizing antibodies (Table 1), questioning their previously hypothesized ‘compensatory’ role in stabilizing otherwise slightly deleterious mutations with significant effect on immune escape (Gerdol et al., 2022). For example, the 212:SGR spike insertion was the only lineage-defining non-synonymous mutation found in BA.2+ins(L). A notable exception was the presence of Y144del in BA.2+ins(LV), a deletion which occurs in RDR2 and is associated with antibody escape (McCarthy et al., 2021). BA.2+ins(LIII) also displayed the presence of Thr1213Ser, a S2 non-synonymous conservative substitution rarely observed in SARS-CoV-2 prior to the emergence of BA.2, which helped its designation as BA.2.41.
The independent spread and continued community transmission of these sublineages suggest that the acquisition of RIR1 insertions on a BA.2 genomic background may occasionally provide a moderate fitness increase, even though the precise mechanism by which this would occur is still unclear. It is worth mentioning that several of the mutations shared by the different omicron lineages, if taken individually, would theoretically have a detrimental impact on viral fitness. Nevertheless, they are thought to cooperatively interact by positive epistasis, mitigating the negative fitness costs associated with their presence, being possibly responsible for the remarkable structural and functional alterations of the omicron spike protein (Martin et al., 2022b). Insertion XLI (i.e. ins215EPE), which is included among the mutations found in BA.1 but not shared with BA.2 and BA.5, undoubtedly emerged after the split between these lineages and its presence may therefore provide a fitness advantage in combination with any of the other spike mutations associated with BA.1, but absent in BA.2 or BA.5. Nevertheless, none of these mutations was observed in any of the BA.2 sublineages carrying RIR1 insertions (Table 1). Interestingly, the only study carried out so far which has explicitly tested the functional role of ins214EPE in BA.1 suggests that this insertion may improve spike expression (Javanmardi et al., 2022), which may be consistent with the significant structural alterations determined by RIR1 insertions on the spike protein trimer (Gerdol et al., 2022). Omicron displays enhanced inter-domain and inter-subunit packing, as well as a higher accessibility for ACE2 binding, driven by its higher predisposition towards an open configuration (Ye et al., 2022), and it would be of high interest to investigate whether the presence of RIR1 insertions has a significant impact on these properties. Undoubtedly, further structural studies would be required to clarify the effects of the acquisition of RIR1 insertions on a BA.2 genetic background in terms of the impact on the structural conformation and stability of spike trimers. Moreover, continued monitoring of similar insertions would be important for other emerging omicron lineages, such as BA.5, which as of 16 July 2022 only displays a single sequenced genome with an RIR1 insertion (i.e. EPI_ISL_13773295, carrying an in-frame AGAACAACCTAC insertion between codons 212 and 213, encoding the tetrapeptide Arg-Thr-Thr-Tyr).
ACKNOWLEDGEMENTS
The authors would like to acknowledge the tremendous efforts made by the clinicians, researchers and public health authorities that allowed the collection of SARS-CoV-2 genome data and made sequence data available in a timely manner though GISAID, as well as the great efforts made by the developers of Nextstrain to assist researchers in SARS-CoV-2 evolution studies.
Open Access Funding provided by Universita degli Studi di Trieste within the CRUI-CARE Agreement.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
FUNDING INFORMATION
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as this is a review article with no original research data.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.




