Novel subtypes and unexpected heterogeneity of hepatitis E viral strains in wild boar captured in a small area in Central Italy
Abstract
Wild boar is the main sylvatic reservoir of the genotype 3 of hepatitis E virus (HEV). The occurrence of HEV-3 human cases has been linked to the consumption of raw or undercooked pig and wild boar meat and liver. The zoonotic transmission of HEV-3 has been confirmed by sequencing identical or strictly related viral strains in humans, wild boar and derived food. The HEV sequences classified within the HEV-3 genotype are highly variable, and although only one serotype has been identified so far, the observed differences allow for the further classification of the HEV-3 genotype into subtypes, named in alphabetical order. Compared to human and pig strains, an even higher heterogeneity is observed among strains infecting wild boar.
In the present study, the genetic variability of eight HEV-3 strains detected in wild boars sampled in a small geographical area in Central Italy (Lazio and Umbria regions) was investigated by full genome sequencing and phylogenetic analysis.
The strains were classified within the HEV-3a, HEV-3c, HEV-3f subtypes and within two new recently proposed subtypes. Results demonstrate – despite the relatively small geographic area of origin – an unexpected divergence within HEV-3 strains hosted by the investigated wild boar population and highlights the need for extensive sequencing of HEV in reservoirs to fully understand diversity, geographical distribution and evolution of this group of viruses.
1 INTRODUCTION
Hepatitis E infection is recognized worldwide as an emerging public health issue (Kamar et al., 2017). The virus (HEV) is a quasi-enveloped RNA virus with a single-stranded positive-sense genome with an extensive genomic diversity, which determines the classification into two genera: Orthohepevirus and Piscihepevirus. The Orthohepevirus is divided into four species (A, B, C and D) of which only two infect humans (A, C). Orthohepevirus A includes genetically distinct genotypes (HEV 1−8): HEV-1 and HEV-2 only infect humans; HEV-3 and HEV-4 are zoonotic. The infections are globally distributed in low-income and developed countries, causing large outbreaks mostly waterborne in the former and foodborne sporadic cases and small outbreaks in the latter. The two different epidemiological settings are associated with different genotypes of Orthohepevirus A species: HEV-1 and HEV-2 circulate in low-income countries; HEV-3 and HEV-4 mainly circulate in developed countries (Kamar et al., 2017; Dalton et al., 2018). HEV-3 and HEV-4 can be transmitted from animal to animal via the faecal-oral route or from animal to human by consumption of contaminated food of animal origin. Pigs and wild boar are the main reservoirs of HEV-3 and HEV-4. In Europe, HEV-3 is the most frequently detected genotype (Adlhoch et al., 2016). Due to the nucleotide diversity, the genotypes have been further classified into subtypes, named using alphabetical letters, based on their shared nucleotide and amino acid identity (Lu et al., 2006; Smith et al., 2013; Smith & Simmonds, 2018).
HEV-3 genotype strains are classified into 11 subtypes (a-m) and novel subtypes are constantly proposed. At least three full genomes clustering together and showing a high p-distance with all known subtypes by phylogenetic analyses are required for the classification into novel subtype (Smith et al., 2016; Smith & Simmonds, 2018; Nicot et al., 2021). In Europe, the subtypes HEV-3c, HEV-3e and HEV-3f are those most commonly detected in both humans and animals (Adlhoch et al., 2016; Dorn-In et al., 2017; Lapa et al., 2015; Nicot et al., 2018; Oeser et al., 2019; Suin et al., 2019) while the others are rarely detected (Kozyra et al., 2021; Nicot et al., 2018; Reuter et al., 2009). To date, few studies aimed at understanding if any differences, such as host adaptation, infectious dose and symptoms caused, among subtypes exist. There is evidence to suggest that subtypes may have a clinical significance and enable adaptation to new hosts (Abravanel et al., 2020; Subissi et al., 2019). To some extent, the subtypes also show a geographical and temporal distribution; therefore, their identification helps in evolutionary studies of HEV-3 and could also be useful in tracing time-space movement of strains.
In Europe, HEV-3c has recently emerged as predominant in humans, pigs and wild boar (Izopet et al., 2019; Wang et al., 2019).
In Italy, HEV-3f has been reported as the most common subtype affecting humans and pigs, followed by HEV-3c and HEV-3e (De Sabato et al., 2020b). A few subtypes have also been reported less frequently, such as HEV-3a, and others that remain unclassified (De Sabato et al., 2020b).
Heterogeneity of HEV-3 viral strains is the greatest in wild boar. HEV-3c and HEV-3i are widespread in wild boar in Central Europe (Anheyer-Behmenburg et al., 2017; Oliveira-Filho et al., 2014) and several other subtypes have been detected over European countries (Dorn-In et al., 2017; Jemeršić et al., 2019; Kukielka et al., 2016; Mesquita et al., 2016; Okano et al., 2014; Vina-Rodriguez et al., 2015). In Italy, several studies have reported HEV-3 to circulate in wild boar throughout the country (Aprea et al., 2018; Caruso et al., 2015; De Sabato et al., 2020a; Di Pasquale et al., 2019; Forzan et al., 2021; Martinelli et al., 2015; Mazzei et al., 2015; Pierini et al., 2021). Just as in pigs and humans, the wild boar strains belong to subtypes HEV-3f and HEV-3e but include also subtypes HEV-3c, HEV-3a and several unclassified subtypes (Aprea et al., 2018; Aprea et al., 2020; Arnaboldi et al., 2021; Caruso et al., 2015; De Sabato et al., 2018a; Di Pasquale et al., 2019; Lo Presti et al., 2020; Pierini et al., 2021; Serracca et al., 2015; Zecchin et al., 2019). Four recent studies have revealed the high prevalence of HEV-3 in wild boar hunted in Central Italy (De Sabato et al., 2018a; De Sabato et al., 2020a; Di Pasquale et al., 2019; Pierini et al., 2021); in addition, short genome region sequencing has revealed the genetic diversity of these HEV-3 strains to be also higher than expected. To more fully explore this variability, and to better understand the evolution of HEV-3 in this subpopulation of wild boar, 8 of the HEV-3 strains, collected in Central Italy (Lazio and Umbria, two neighbouring regions), were selected and subjected to full genome sequencing and detailed phylogenetic analysis.
2 METHODS
2.1 Sample collection
Eight wild boar livers (50 mg each) previously found to be positive for HEV-3 by real-time RT-PCR in previous studies (De Sabato et al., 2018a; De Sabato et al., 2020a; Di Pasquale et al., 2019; Pierini et al., 2021) were used in the present study (Table 1). Originally, the livers were obtained from adult wild boars hunted in the region of Lazio (province of Viterbo; North Lazio) in 2016 and 2017 and in the neighbouring region of Umbria (Perugia province) in 2018. RNA was extracted from liver using the Qiamp-Viral mini kit (Qiagen) as previously described (De Sabato et al., 2018b).
| Sequence | Collection date | Region | Province | Subtype | Reference |
|---|---|---|---|---|---|
| WB119VT2017 | 2017 | Lazio | Viterbo | HEV-3c | (De Sabato et al., 2020a) |
| WB171VT2017 | 2017 | Lazio | Viterbo | HEV-3n | (De Sabato et al., 2020a) |
| 132 | 2012 | Lazio | Viterbo | HEV-3* | (Di Pasquale et al., 2019) |
| WB01VT2016 | 2016 | Lazio | Viterbo | HEV-3a | (De Sabato et al., 2018) |
| WB03VT2016 | 2016 | Lazio | Viterbo | HEV-3f | (De Sabato et al., 2018) |
| WB110VT2017 | 2017 | Lazio | Viterbo | HEV-3f | (De Sabato et al., 2020a) |
| 55863/2018 | 2018 | Umbria | Perugia | HEV-3f | (Pierini et al., 2021) |
| 47272/2017 | 2017 | Umbria | Perugia | HEV-3f | (Pierini et al., 2021) |
- HEV-3*: novel subtype not assigned yet.
2.2 Complete genome sequencing
The total RNA was used to prepare libraries for Next Generation Sequencing (NGS). Six genomes were obtained following the sequence independent single primer amplification (SISPA) method and sequenced by Ion Torrent S5 (Thermo Fisher Scientific, Rodano, Italy) as previously described (De Sabato et al., 2018b) at Istituto Superiore di Sanità (FAST service). Two complete genomes were obtained at Istituto Zooprofilattico Sperimentale dell'Umbria e delle Marche ‘Togo Rosati’ as previously described (Pierini et al., 2021) by Istituto Zooprofilattico Sperimentale dell'Abruzzo e del Molise ‘G. Caporale’ (Teramo, Italy). The reads obtained by the NGS run were quality checked and analysed by Galaxy Aries (https://aries.iss.it) (Knijn et al., 2020). The complete genomes were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov): 132 (Acc. N° OK340737), WB03VT2016 (Acc. N° OK340738), WB01VT2016 (Acc. N° OK340739), WB110VT2017 (Acc. N° OK340740), WB171VT2017 (Acc. N° OK340741), WB119VT2017 (Acc. N° OK340742), 55863/2018 (Acc. N° OK429318), 47272/2017 (Acc. N° OK429319).
2.3 Data sets of sequences
To clarify the evolutionary correlation amongst HEV strains, seven sequence data sets (numbered I to VII) were built with sequences from this study. The first data set (I) used to develop the phylogenetic tree, was built with the eight full genomes of this study and 59 complete HEV genomes downloaded from the NCBI database; these include 18 HEV-3 subtype reference genomes (Smith et al., 2020), 40 HEV-3 complete genomes downloaded from the NCBI database, which share > 89.0% BLASTn nucleotide identity (nt. id.) with the Italian sequences of this study. An HEV-4 sequence (HE-JA30, Acc. N° LC02274) serves as outgroup. For Bayesian analysis, six other data sets were built to clarify the evolutionary correlation amongst Italian strains, to determine the most recent common ancestor (TMRCA), and to calculate the HEV evolutionary rate. For each of these six data sets (II–VII), the selection of HEV sequences was based on four criteria: (i) sequences originated from Italy, (ii) year of collection, (iii) classified as genotype HEV-3, and (iv) sourced from swine, wild boar or human. The second data set (II) comprises the 20 Italian HEV-3 full genome sequences available on the NCBI database and the eight full genome sequences reported upon in this study. Owing to the scarcity of full genome sequences for Italian strains of HEV, data sets III to VII were built using a 290 bp short genomic region within ORF2, the most sequenced and represented region on NCBI database; in order to strengthen the analysis of the evolutionary correlation that exist amongst Italian strains. Data set III comprises 41 HEV-3e and 93 HEV-3f sequences; data set IV 23 HEV-3a subtype sequences; data set V 56 HEV-3c subtype sequences; data set VI 32 HEV-3n subtype sequences and data set VII 27 unclassified sequences. For each data set nucleotide sequences were aligned using Mafft v7.475 (Katoh et al., 2019) and manually edited using Aliview (Larsson, 2014).
2.4 Likelihood mapping
The software TREE-PUZZLE was used to investigate the phylogenetic signal each data set contained through the analysis of four arbitrarily selected sequences referred to as quartets (Schmidt et al., 2002). The Likelihood mapping analysis involved 10,000 quartets.
2.5 Phylogenetic analysis
The first data set was used to build the Maximum Likelihood (ML) phylogenetic tree using IQ-TREE2 (v.1.6.10) with the best fit model indicated by the Model Finder and involving 1000 bootstrap replicates (Minh et al., 2020). The p-distances between strains were calculated using MEGAX software (Kumar et al., 2018).
2.6 Bayesian evolutionary rate estimate and dated trees
A root-to-tip regression analysis was undertaken using TempEst to establish the temporal signal of data sets II-VII. The GTR+G+I model was selected as the evolutionary model using JmodelTest, v. 2.1.7, while the mean evolutionary rate and the dated trees were made using a Bayesian Markov Chain Monte Carlo approach implemented in BEAST, v.1.10.4 (Suchard et al., 2018). Two coalescent priors (constant population size and exponential growth) and strict versus relaxed molecular clock models were tested by means of path sampling (PS) and stepping stone (SS) sampling. Convergence was assessed by estimating the effective sampling size (ESS) after a 10% burn-in, using Tracer software (http://tree.bio.ed.ac.uk/software/tracer/), and accepting ESS values of 200 or more. Uncertainty in the estimates was indicated by 95% highest posterior density (95% HPD) intervals. Posterior probability was used as statistical support for specific clades and clusters. The trees were summarized in Tree Annotator, and the tree with the maximum product of posterior probabilities (maximum clade credibility or MCC), after a 10% burn-in, chosen.
3 RESULTS
3.1 Phylogenetic analysis of eight complete HEV-3 genomes from wild boar
The tree-puzzle analysis showed that the percentage of unresolved quartets was lower than 33% for all data sets providing a phylogenetic signal sufficient for analysis. The Maximum Likelihood tree (data set I; Figure 1) showed the 8 complete genomes to cluster amongst five different HEV-3 subtypes: HEV-3c (n = 1), HEV-3f (n = 4), HEV-3a (n = 1), the recently proposed subtype HEV-3n (n = 1), and the novel unclassified subtype (named in this study HEV-3*; n = 1) (Table 1). The wild boar strain (WB119VT2017) that clustered within subtype HEV-3c, showed a 90.0% nt. id. with HEV-3c reference (wbGER27, FJ705359) and 93.5% nt. id. with the HEV isolate HEPAC-96 (MF444114) obtained from a human patient in France in 2008. Several strains of HEV-3c have been reported and sequenced from Italian wild boars. Despite this, only a few full genomes are available for comparison (n = 6) (last sequence download from NCBI database February 2022); all these sequences were detected in wild boar hunted in Abruzzo (e.g. 2018.AZ.6050.8.2, MT840359), a region in Central Italy, and show ≈90.0% nt. id. with the aforementioned WB119VT2017 (Figure 1).
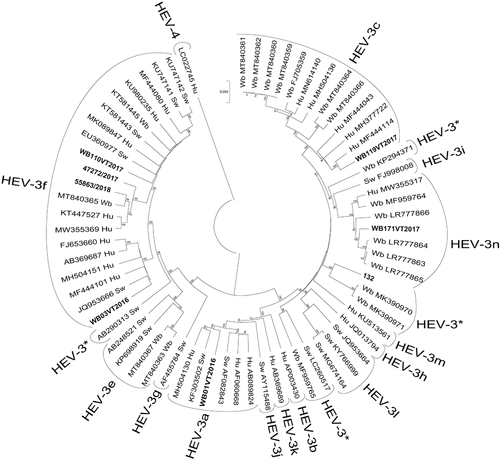
In the absence of complete genome sequences for Italian swine strains of HEV-3c, a BLASTn analysis was conducted based on the short 290 bp diagnostic fragment contained within ORF2 and involving sequences available in the NCBI database. This comparison highlighted a nucleotide identity of 95.2% with the HEV-3c swine strain SWHEV82BO2012 (KF888276) detected in the north of Italy in 2012.
As noted above, four wild boar strains (WB03VT2016, WB110VT2017, 55863/2018 and 47272/2017) clustered within the HEV-3f subtype (Figure 1); however, their sequences were extremely diverse, separating into two sub-clusters (> 87.0% nt. id.).
Among the Italian HEV-3f strains, 91% nt. id and 88% nt. id. were displayed by the WB03VT2016 and the groups of WB110VT2017, 55863/2018, 47272/2017, respectively with the HEV-3f reference (E116-YKH98C, AB369687). The first sub-cluster includes the strain (WB03VT2016) that shared the highest nt. id. of 91.2% with a swine strain obtained in France in 2008, namely HEV strain FR-SHEV3f (JQ953666); this strain showed a lower nucleotide identity (≈87.0%) with strains of the second sub-cluster.
The second sub-cluster of HEV-3f includes strains WB110VT2017, 55863/2018 and 47272/2017 and the HEV complete genome of another previously reported strain (2019.AZ.5758.1.5, MT840365) obtained from a wild boar in the region of Abruzzo in 2019. Amongst these, strain WB110VT2017 was distant from other strains and its nucleotide identity was the lowest (<90.0%) in comparison to all other HEV-3f strains, including Italian strains (Figure 1). Two wild boar strains (WB03VT2016 and WB110VT2017) showed high nucleotide identities (> 97% nt. id.) to HEV-3f strains isolated from patients hospitalized in the regions of Lazio and Abruzzo: isolate ISS_ID_201/2017 (MZ274237), isolate ISS_ID_340/2019 (MZ274263), and ISS_ID_258/2018 (MZ274246), INMI_1902_2019 (MN537876), respectively. Two strains (55863/2018 and 47272/2017) showed the highest nucleotide identity (96%) with a swine strain HEV/13RS985-10 (KF939868) and a human strain ISS_ID_305/2019 (MZ274253). Lower identities (< 93.5%) have been reported with other human strains isolated in Central Italy from patients hospitalized in Rome (INMI_1825_2018 MN444850; INMI_1823_2018 MN444849; INMI_1809_2018 MN444853; INMI_1203_2012 MN50947) and with several additional Italian swine strains detected in other regions (13RS1494-12, MK532924; SwHEVE12ITNA2012, KP965762; 13RS1494-11, MK532923; 13RS1494-9, MK532927; SwHEVPIR71ITA18, MN546867; HEV/13RS985-10, KF939868.1; 14RS333-8_50549-4, MK532935; ITswHEV1331, KF891380). Interestingly, all human strains correlated with wild boar strains, came from patients hospitalized in the regions of Abruzzo and Lazio, not far from the area where wild boar were hunted.
The complete genome of WB01VT2016 strain belonged to subtype HEV-3a showing 90% nt. id. with the HEV-3a reference (Meng, AF082843). The WB01VT2016 displayed 91.1% nt. id. with the HEV isolate_GiSw (KF303502) and 90.9% nt. id. with the HEV isolate HEV_34_Newcastle_UK_221214 (MH504130) reported in 2006 from a pig in Germany and from a human case in 2014 from the United Kingdom. In Italy, HEV-3a is not frequently encountered. The single wild boar HEV-3a strain reported in this study showed by BLASTn, comparing short ORF2 fragment, a 96.5% nt. id. with the HEV-3a human strain INMI_1736_2017 (MN444837) and 92.0% nt. id. with several strains reported from wild boar in Northern Italy (e.g. IZSLER_B6_32, MW263036).
The strain 132, isolated from a wild boar in 2012 (Lazio), clustered within a novel subtype of HEV-3* (reference strain MK390970) (Nicot et al., 2021; Smith et al., 2020); this novel subtype remains unassigned for the lack of the three full genomes needed for definitive classification. The cluster included three full genomes, all from wild boar, and detected in Italy: 132 and strains 17RS1920 (MK390971) proposed as references for the classification (Smith et al., 2020) and 17RS2551-4 (MK390970), showing a low 88.5% nt. id. (Figure 1).
Based on nucleotide sequences of short genome ORF2 fragment, strain 132 was similar (>93.0% nt. id.) to 5 other Italian strains from wild boar hunted in the same area (Di Pasquale et al., 2019) and with the human strain 122.16_F reported in 2012 from north-eastern Italy (KC782933). No higher identity was observed even by comparing the short ORF2 diagnostic fragments with the whole database of NCBI.
The strain WB171VT2017 formed a cluster (96.0 % nt.id.) with strains detected in 2016–2017 in wild boar hunted in the neighbouring Umbria region (e.g. 54563/UM/2016 LR777864) and includes a human strain reported in 2018 from France (92.3% nt. id., HESQL059; MW355317) and the reference strain of the recently proposed novel subtype HEV-3n (93% nt. id., WB/HEV/NA17ITA15, MF959764) first detected in southern Italy (Figure 1). Furthermore, using ORF2, the wild boar strain 47270.213/UM/2017 (LR777866) from Umbria and the human strain HESQL053 (MW355383) from France, together, supported the existence of tentatively named HEV-3n.
3.2 Bayesian evolutionary rate estimate and dated trees
Root-to-tip regression analysis of the temporal signal from data sets revealed a moderate association between genetic distances and sampling years.
The best fitting models, correlation coefficient, the estimated mean value of the evolutionary rate and the most common recent ancestor (TMRCA) for each data set are provided in Table 2. Due to the low correlation coefficient, the phylodinamic analysis was not performed on data set VII.
| Data set | Sequences | Correlation coefficient | Clock model | Tree prior | Mean evolutionary rate (substitutions/site/year) | TMRCAa (year) | ||
|---|---|---|---|---|---|---|---|---|
| Mean | 95% HPDb | Mean | 95% HPDb | |||||
| II | Full Genome | 0.41 | Strict clock | Exponential growth | 2.72 × 10−3 | 1.5 × 10−3 to 4.0 × 10−3 |
1894 | 1829–1948 |
| III | HEV-3e/3f | 0.51 | Strict clock | Exponential growth | 2.87 × 10−3 |
1.9 × 10−3 to 3.9 × 10−3 |
1957 | 1937–1975 |
| IV | HEV-3a | 0.46 | Uncorrelated relaxed clock | Costant size | 4.8 × 10−3 | 1.9 × 10−3 to 8.0 × 10−3 |
1999 | 1987–2009 |
| V | HEV-3c | 0.77 | Strict clock | Costant size | 9.9 × 10−3 | 3.3 × 10−3 to 1.4 × 10−2 |
2000 | 1991–2005 |
| VI | HEV-3n | 0.66 | Uncorrelated relaxed clock | Exponential growth | 3.3 × 10−3 | 9.0 × 10−3 to 6.6 × 10−2 |
2014 | 2011–2016 |
| VII | HEV-3* | 0.0001 | – | – | – | – | – | |
- a TMRCA: the most common recent ancestor.
- b HPD: highest posterior density.
- HEV-3*: novel subtype not assigned yet.
In the Bayesian tree, built using complete genome (data set II; Figure S1), two main clades were found: one included HEV-3e and HEV-3f sequences separated back to 1952 (95% HPD: 1918–1988) and the other HEV-3a, HEV-3c, HEV-3l, HEV-3n and unclassified strains originated back to 1936 (95% HPD: 1890–1971). The classification of Italian strains based on Maximum likelihood tree correlated with the Bayesian tree.
In data set III (Figure S2), built with HEV-3e and HEV-3f ORF2 short genomic regions, the WB110VT2017 clustered with wild boar strains previously reported (WB161VT2017, MK889004; WB125VT2017, MK889005, WB02VT2016, MG582608) in the same time period and with two human strains reported in 2018 and 2019 (INMI_1902_2019, MN537876; ISS_ID_258/2018, MZ274246). The WB03VT2016 clustered with a correlated wild boar strain WB02VT2016 (MG582608). Both wild boar and human strains were detected in the same region (Lazio).
Similarly, the 55863/2018 strain clustered with other wild boar strains reported from the same region (Umbria region, e.g., 59068.5/UM/2018, LR699563). However, 55863/2018 clustered also with a human strain from Lazio (ISS_ID_305/2019, MZ274253) and the cluster also included a swine HEV-3 strain (14RS333-8_50549-4, MK532935) detected in Northern Italy in 2014.
In data set IV (Figure S3), built with the few sequences of HEV-3a ORF2 available for Italian strains, two clusters were apparent. The WB01VT2016 strain clustered with two strains sequenced from wild boar hunted in the same area in 2012 (Hepatitis_E_virus_isolate_5, MH836549; Hepatitis_E_virus_isolate_116, MH836544) and with the strain INMI_1736_2017 (MN444837) isolated from a patient in 2017; all strains found in the same region (Lazio). The second HEV-3a cluster comprised all of the sequences isolated from wild boar in northern Italy (Figure S3).
In data set V (Figure S4), built with HEV-3c ORF2, two main clusters were apparent: the first grouping Italian swine sequences reported in northern Italy and the second including Italian human and wild boar sequences collected in central and northern Italy. The strain WB119VT2017 clustered with several wild boar and human strains reported in Central Italy (Lazio, Abruzzo and Umbria).
In the Bayesian tree built with HEV-3n ORF2, data set VI (Figure S5), two main clades were observed: the first one only formed by wild boar strains reported in northern Lazio (Viterbo province) between 2016/2017 and including the wild boar strain WB171VT2017 from this study; the second clade included strains isolated from wild boar in Umbria (land border with northern Lazio) in 2016/2017 and wild boar strains from southern Italy collected in 2017.
4 DISCUSSION
Four studies were conducted over 5 years with the aim of detecting HEV in wild boar in bordering areas located in Central Italy (Lazio and Umbria regions) (De Sabato et al., 2018a; De Sabato et al., 2020a; Di Pasquale et al., 2019; Pierini et al., 2021). The retrieved strains were widely heterogeneous and based on ORF2, belong to multiple HEV-3 subtypes. Following these preliminary results, complete genome sequencing and characterization of the strains was undertaken to further explore viral diversity, the evolutionary relationships amongst subtypes, and geographical distribution within the country.
The complete genomes for 8 HEV-3 strains, which displayed high p-distance values between each other and other HEV reference sequences, were obtained and subjected to phylogenetic analysis. The 13 subtype reference strains, along with 6 reference strains representing unassigned novel HEV subtypes, were used for this analysis, following subtype assignment criteria recommended by Smith et al. (2020).
Based on this, the maximum likelihood tree assigned the 8 complete HEV genomes to 5 subtypes, namely HEV-3f, HEV-3c, HEV-3a, and two novel recently recognized subtypes (Nicot et al., 2021; Pierini et al., 2021; Smith et al., 2020). These novel subtypes remain unnumbered in the continued absence of three full genomes needed to achieve definitive classification (Smith & Simmonds, 2018). Only recently as it emerged that these strains group with two full genome strains obtained from wild boar (Pierini et al., 2021) and for which subtype HEV-3n was proposed.
In Italy, HEV-3f is the subtype most frequently encountered in wild boar, in order followed by HEV-3c, HEV-3e, and other rarer subtypes (De Sabato et al., 2020b). The most common subtype in humans is HEV-3f, followed by HEV-3e (De Sabato et al., 2020b; Garbuglia et al., 2021) and last HEV3c; this last-mentioned subtype apparently is more prevalent elsewhere in Europe (Lhomme et al., 2015; Nicot et al., 2018).
A different subtype distribution is observed in pigs, where the main subtypes are HEV-3f, followed by HEV-3e and, less frequently, HEV-3c and HEV-3l (De Sabato et al., 2020b).
In this study, the estimated mean value of the evolutionary rate is 2.72 × 10−3 substitutions/site/year for the HEV complete genome (95% HPD: 1.51 × 10−3 to 4.06 × 10−3) and 2.87 × 10−3 to 9.9 × 10−3 for the HEV ORF2 capsid gene, which is similar in both analyses and to results reported upon previously (Montesano et al., 2016; Nakano et al., 2012; Zehender et al., 2014).
Bayesian trees built with the diagnostic fragment within the ORF2 capsid gene, the region most sequenced and represented in the NCBI database, revealed strains within subtypes HEV-3e and HEV-3f to form geographical clusters. Some clusters involved strains sequenced in a single study, and in terms of space and time, may introduce bias into Bayesian geographical analyses. Nevertheless, within HEV-3f some clusters included strains from pigs, wild boar and humans suggesting that correlated strains circulate in different regions and hosts in parts of Italy, and that transmission or co-circulation amongst swine and wild boar may occur. This possibility underlines the importance of biosecurity in terms of minimizing contact between wild boar and domestic pig populations. Furthermore, in the HEV-3f cluster, some groups that comprised both human (i.e. ISS_ID_201/2017, INMI_1902_2019; Garbuglia et al., 2021) and wild boar strains (i.e. WB03VT2016), showed geographical correlation. This result supports the hypothesis that human cases are definitively linked to the consumption of local produced wild boar foods, which is a typical local eating habits in Central Italy particularly in Abruzzo. Besides this, the four HEV-3f sequences included in this study, though assigned to one and the same subtype, clearly fall within different clades, revealing HEV-3f subtype sequences to be genetically heterogeneous (Smith et al., 2020).
Within subtype HEV-3a, based on ORF2, the wild boar strain WB01VT2016 showed a high nucleotide identity (96.0%) with the only human HEV-3a strain (MN444837) and obtained from a patient hospitalized in the same region (Lazio) (De Sabato et al., 2020b) and which a year later (in 2017) was detected also in wild boar. Without corroborative epidemiological data, this percentage of nucleotide identity is insufficient for definitively correlating the two strains; however, the result merits further investigation by sequencing and comparing complete genomes, since HEV-3a in Italy is rare in both wild boar (Di Pasquale et al., 2019) and humans (De Sabato et al., 2020b).
In this study, two of the full genomes obtained were assigned to novel subtypes HEV-3n and HEV-3*. Only a limited number of sequences are available for these recently recognized subtypes (Nicot et al., 2021; Pierini et al., 2021; Smith et al., 2020).
By comparing full genomes, the HEV-3n sequences, from this and a previous study (Pierini et al., 2021) formed a group including strains only detected in wild boar in Italy. By comparing ORF2 fragments, an additional human strain, detected in France in 2018 (Kozyra et al., 2021), was also included in the HEV-3n subtype cluster. Since, according to the Bayesian tree, the HEV-3n may have originated only recently (2014), the results suggest that the subtype may have spread to other countries, where it could be present, however, still with a low prevalence. The HEV-3n strains of this study were detected in Italian wild boar hunted in two neighbouring areas, the Lazio (Viterbo province) and Umbria regions (Perugia) in 2016 and 2017, indicating animal movement to occur between the two areas. However, the first identification of this novel subtype, subsequently established as reference strain (Smith & Simmonds, 2018), occurred in a wild boar hunted in southern Italy (Aprea et al., 2018), suggesting that this strain is widespread in Italy, circulating in more distant and non-adjoining regions.
The other novel subtype detected in this study is still unclassified (HEV-3*) and included the strain 132 and two wild boar strains from northern Italy, collected in 2017 (17RS1920, 17RS2551-4) (Zecchin et al., 2019). In the previous study (Di Pasquale et al., 2019), the strain 132 clustered with eleven other wild boar strains from the same geographical area. It is noteworthy that this unclassified strain shares a low percentage (<89.0%) of nucleotide identity with all other HEV sequences in the NCBI database, representing either complete genomes or the short ORF2 fragment. Based on phylogenetic and sequence analysis, strains correlated to strain 132 have only been described from Italian wild boar. This evidence suggests that this subtype could have originated recently in Italy and may not have spread yet to other countries. However, due to the absence of a supportive temporal signal, a phylogenetic analysis was not conducted.
The strain 132 is to some extent related (91%) to an Italian human strain reported in 2012 (KC782933) (Giordani et al., 2013) but whose source is uncertain, possibly linked to wild boar. The patient had butchered a wild boar hunted in Tuscany region, not far from the area where strain 132 was obtained in wild boar. Although these two strains are not identical and related epidemiological data are missing, the result may suggest the zoonotic transmission, due to the consumption of raw or undercooked wild boar products (Montone et al., 2019; Rivero-Juarez et al., 2017). The strict correlation of human and wild boar HEV-3 suggesting homemade local food products as source of infection, have also been observed in other EU country, confirming the role of wild boar as important reservoir of the zoonotic HEV-3 (Jemeršić et al., 2019).
Within the same geographical area, and over a number of years, the local wild boar population acts as host to a highly heterogeneous group of HEV-3 strains, some of which belong to rare subtypes or have been reported from Italy only. This picture around strain diversity possibly is incomplete due to the limited number of full genome sequences available. Nevertheless, the same result is obtained when analysing ORF2, the fragment that is most abundantly represented in the NCBI database. Since no intensive pig farms are present in the investigated areas, contact and transmission between pigs and wild boar is greatly limited; therefore, the wide heterogeneity observed confirms wild boar to be a significant reservoir of HEV-3.
Within the subtypes for which whole genomes are available, such as HEV-3c or HEV-3f (Aprea et al., 2020; Zecchin et al., 2019), no sequences that could be considered identical were obtained by us in this study. The absence of identical sequences may be explained by limited intermixing of wild boar populations in recent time, and which deals with an animal host that does not move far from its breeding grounds of choice, thereby ensuring the maintenance of intra-subtype variability. The occurrence of common subtypes such as HEV-3c and HEV-3f but also less common strains such as HEV-3a and HEV-3n (detected only in wild boar in Italy), supports the boar's autochthonous reservoir role in the maintenance of local strain heterogeneity. Among the subtypes analysed in this study some are rare or not detected in humans (HEV-3a, HEV-3n and HEV-3*) but are common in wild boar. This could be due to specific host adaptation for some subtypes or could be associated to a lower probability of transmission by wild boar derived food, which is not consumed frequently.
In wild reservoir confined in an area with no or limited contacts with farmed animals the variability of HEV-3 strains within the confined population is kept. This wild boar scenario is in sharp contrast to the situation on pig farms where the same HEV strain is identified repeatedly over the years, even after stamping out (Ianiro et al., 2021; Widen et al., 2011).
Our results show that whole genome sequencing and sharing of sequences in public databases is essential to accurate analysis to track the evolutionary history and geographic spread of HEV-3 subtypes and their role in human and animal infection. Comparing only short gene fragments may introduce bias into a classification. To date, only few Italian HEV whole genomes are available in the NCBI database, 17 from wild boar, 3 from swine and none from humans, making it difficult to correlate strains. The implementation of systematic whole genome sequencing for human and swine samples is therefore of paramount importance for a better understanding of virus circulation and evolution.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required for this specific study.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest associated with this study.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




