Genomic characteristics and recombination patterns of swine hepatitis E virus in China
Amina Nawal Bahoussi and Yan-Yan Guo equally contributed to the article.
Abstract
Zoonotic hepatitis E, mainly caused by swine hepatitis E virus (sHEV), is endemic in China, causing great economic disruption and public health threats. Although recombination is critical for the evolution of viruses, there is a limited assessment of its occurrence among sHEVs. Herein, we analysed all available sHEV full-length genomes isolated in China during the past two decades (40 isolates) compared to 72 other sHEV strains isolated in different countries and determined that sHEV genotype 4 (sHEV4) dominates China. Eight potential natural recombination events were identified, four of which occurred in China and were mainly between sHEV4 strains, indicating the distinct character of China sHEV. One intergenotype recombination event was found in China, alarming the emergence of a new sHEV lineage that could become a critical threat to human health.
1 INTRODUCTION
The human–animal coexistence and anthropogenic activities are among the main reasons for the worldwide spread and emergence of zoonotic diseases. Zoonoses are characterized by the ability of their pathogens to cross species barriers and jump from animals to infect humans and cover about 60% of the globally emerging infectious diseases, including Hepatitis E (Jones et al., 2008).
Hepatitis E, caused by Hepatitis E virus (HEV), is the leading cause of worldwide acute enterically-transmitted hepatitis and a substantial global health threat (Kamar et al., 2012). HEV is transmitted chiefly through the faecal–oral route, whereas blood transfusion (Hoad et al., 2017) and direct interaction with animals (Meng et al., 2002) have also been involved. HEV was first reported as an unknown source of a waterborne outbreak in India (Wong et al., 1980), then identified in 1983 as a non-A, non-B hepatitis virus (Balayan et al., 1983).
The first animal HEV was isolated from domestic pigs in 1997 in the United States and called swine HEV (sHEV) (Meng et al., 1997). sHEV was distinct but genetically closer to human HEV (hHEV), with the identity of over 80% at both protein and genomic sequence levels (Meng et al., 1997). Subsequently, the experimental study further demonstrated that a hHEV variant (US-2 strain) isolated from the United States could infect experimentally specific pathogen-free pigs, and in a reciprocal experiment, an sHEV strain isolated from pigs could infect nonhuman primate chimpanzees and rhesus monkeys (Meng et al., 1998). Therefore, sHEV was suggested as a zoonotic pathogen for which pigs could be the reservoir (Meng et al., 1998). Since then, many countries in America, Asia, Africa and Europe have started reporting the emergence of sHEV infection, including Canada (Ward et al., 2008), South Korea (Choi et al., 2003), Japan (Motoya et al., 2019), Nigeria (Owolodun et al., 2014) and Germany (Baechlein et al., 2010).
HEV is a small quasi-enveloped RNA virus containing a positive-sense, single-stranded RNA genome (Takahashi et al., 2010). HEV is a member of the species Orthohepevirus A, genus Orthohepevirus in the family Hepeviridae and currently comprises eight genetically distinct genotypes (HEV1-8) and 36 subtypes (Smith et al., 2020). HEV1 and HEV2 are typically restricted to humans and have emerged sporadically and endemically in developing countries in Africa and Asia (Hoofnagle et al., 2012). In contrast, HEV3 and HEV4 seemed to be a significant source of zoonotic transmission of HEV and were isolated in both humans and a variety of animals, including wild boars (Kozyra et al., 2021), rats (De Sabato et al., 2020) and rabbits (Bigoraj et al., 2020). The autochthonous human infection of HEV3 and HEV4 viruses has been reported in a wide range of countries including Ghana (Yeboah et al., 2021), South Korea (Jeong et al., 2017), Brazil (Lopes Dos Santos et al., 2010), New-Zealand (Kamar et al., 2012), and China (Y. Shu et al., 2019). HEV5 to HEV8 were isolated from a variety of animals. For example, HEV7 was isolated from dromedary camels in Dubai, United Arab Emirates (DcHEV) (Woo et al., 2014), and HEV8 from Bactrian Camels in Xinjiang, China (BcHEV) (Woo et al., 2016). However, viruses isolated from those animals showed limited global health relevance to human health compared with sHEV (Smith et al., 2014).
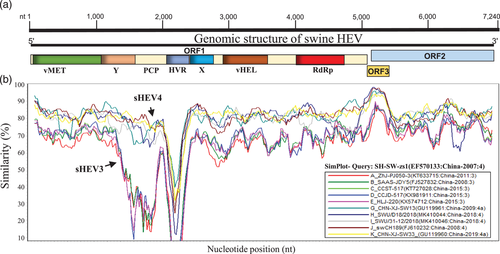
The RNA genome of HEV is ∼7.2 kb in length, flanked with an m7G cap at 5′ proximal terminus and a polyA tail at 3′ proximal terminus. It contains three open reading frames (ORFs; ORF1, 2 and 3) (Wißing et al., 2021). ORF1 encodes a large polyprotein of about 1693 amino acid residues (Purdy et al., 2012), which functions as the viral replicase during virus replication (Wißing et al., 2021). This polyprotein is composed of a series of functional domains, namely viral methyltransferase (vMET), Y-domain (Y), papain-like-protease (PCP), the hypervariable region (HVR), X-domain (X), viral helicase (vHEL) and the RNA-dependent RNA polymerase (RdRp), for most of which, the functions remain not well understood (Wißing et al., 2021). HVR is encoded by the most divergent genomic fragment in ORF1 region and is crucial for viral replication (Purdy et al., 2012). The sequence heterogenicity of HVR is significantly associated with the number of hosts (Purdy et al., 2012). ORF2 encodes the capsid protein that is crucial for the assembly of viral particles (Li et al., 1997) and mediates the HEV binding and infection of cells, making it an effective target for vaccine development (Gordeychuk et al., 2022; Jimenez de Oya et al., 2012; Mazalovska & Kouokam, 2020). ORF3 encodes a cytoskeleton-associated phosphoprotein (Zafrullah et al., 1997) and is responsible for efficiently releasing the virion from HEV-infected cells (Ding et al., 2017). The HEV ORF2 and ORF3 overlap in the viral genome and are translated from the same bicistronic subgenomic mRNA (Graff et al., 2006). ORF1 shares with ORF3 a junction region containing a cis-reactive element (CRE), which is crucial for synthesizing subgenomic RNA (Cao et al., 2018).
In China, HEV was first recognized during the large human HEV1 outbreak that occurred between 1986 and 1988 in Xinjiang Uighur autonomous region (Aye et al., 1992). Later, in 2000, China reported a full genomic sequence of a distinct novel strain isolated from a patient's stool and belonged to HEV4 (Wang et al., 2000). Since then, HEV4 was increasingly detected in humans and swine (Wang, 2003). Pigs have been reported as a principal reservoir of HEV4, which was circulating freely between 0.3% of healthy humans and 9.6% of asymptomatic pigs, in two swine farming districts in eastern China (Zheng et al., 2006). Moreover, a cross-sectional seroepidemiologic study conducted between 2003 and 2004 in eight rural communities in Guangxi Province in southern China exhibited that HEV infection is endemic (R.-C. Li et al., 2006). Besides, swine HEV3 was reported for the first time in a suburban pig farm in Shanghai in late 2006 (Ning et al., 2008). Swine HEV infection has been widely reported in multiple provinces in China and among health workers, pig farmers and pork products consumers (Liang et al., 2014; Zhu et al., 2014). The persistence of sHEV in pig farms and the high human–animal close contact increase the spillover of zoonotic HEV disease and potentially impact public health in China (Zhou et al., 2019). Therefore, we find it crucial to assess the distribution and genetic characteristics of the emerging sHEV to better understand its pathogenesis and transmission routes, thus addressing the knowledge gaps, minimizing the risk of future HEV emergence and promoting the containment of sHEV within farming industries and among human and pig populations. Herein, we focus on screening the molecular characteristics and genetic diversity of China's sHEV compared to that of other countries in different geographical regions, including Asia, America and Europe. We elucidated the sHEV evolutionary history using the phylogenetic and recombination analysis of sHEV genomic sequences isolated during the past two decades and provided valuable information for the public health management, prevention and control strategies of hepatitis E.
2 MATERIALS AND METHODS
2.1 Dataset
All available sHEV full-length genomic sequences isolated in China (40 isolates), in addition to 72 other strains isolated in 16 different countries involved as references, were retrieved from the NCBI GenBank database (in total 112 viruses between 2001 and 2019). Virus strains were identified in this report by their GenBank ID, name, country, year of collection and genotype, in a format as [virus name (GenBank ID: country-collection date: genotype in GenBank)].
2.2 Phylogenetic analysis
Phylogenetic trees were constructed using the neighbor-joining method (Saitou & Nei, 1987) and maximum-likelihood method in Molecular Evolutionary Genetics Analysis software version X (MEGA X) (Kumar et al., 2018). We computed the evolutionary distances using the Maximum Composite Likelihood method (Tamura, Nei, & Kumar, 2004). The internal node numbers indicate the bootstrap values as a percentage of trees obtained from 1000 replicates.
2.3 Similarity analysis
Genomic similarities between the sHEV strains collected in China were determined using SimPlot version 3.5.1 (Lole et al., 1999).
2.4 Recombination analysis
The occurrence of potential recombination events in the sHEV full-length genome was explored using the RDP4 software package (Martin et al., 2015). The recombination events were identified by each of the seven algorithms, including RDP, GENECONV, Bootscan, MaxChi, Chimaera, SiScan and 3seq embedded in the RDP4 package. Phylogenetic trees were generated using the neighbor-joining method based on the indicated nucleotide genomic regions of sHEV strains involved in the recombination.
3 RESULTS AND DISCUSSION
3.1 Swine HEV4 dominates China
To review the genetic characteristics and how China sHEV evolved during the past two decades, we conducted a phylogenetic analysis of the entire sHEV genomes by using neighbor-joining method (Figure 1) and maximum-likelihood method (Figure S1). Both methods produced consistent results. As shown in Figure 1 and Figure S1, the phylogenetic analysis identified two major clades of sHEV (sHEV3 and sHEV4). Among a total of 40 China sHEV isolates, 35 strains clustered into sHEV4, and only five isolates, including ZhJ-PJ050-3 (GenBank ID: KT633715), SAAS-JDY5 (GenBank ID: FJ527832), CCST-517 (GenBank ID: KT727028), CCJD-517(GenBank ID: KX981911) and HLJ-220 (GenBank ID: KX574712) clustered into sHEV3, which are also genetically closer to each other. In contrast, all strains isolated from other countries fell into sHEV3, except two isolates from Japan and one from India, which fell into sHEV4, indicating the distinct genetic characteristics of sHEV China strains from the worldwide emerging sHEV. Moreover, based on the genetic distance, each of the two sHEV genotypes was further divided into multiple subgroups, where five sub-genotypes from sHEV3 (3f, 3e, 3g, 3l and 3j) and three subgenotypes from sHEV4 (4i, 4h and 4e/b) have been identified (Figure 1).
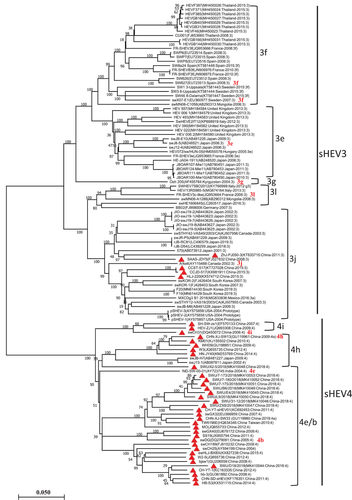
Previously, Smith et al. (2020) have updated the names of HEV subgenotypes based on the selected coding sequence of the viral genome. HEV3, in the Smith et al. (2020) report, has been classified into multiple subgenotypes, including 3a, 3b, 3c, 3d, 3e, 3f, 3g, 3h, 3i, 3j, 3k, 3l and 3m, while subgenotypes 4a, 4b, 4c, 4d, 4e, 4f, 4g, 4h and 4i were assigned to HEV4. According to that genotype classification system, herein, based on full-length genomes and genetic distances, the sHEV strains included in this study were still suggested to fall into corresponding subgenotypes. For example, swCH31 (GenBank ID: DQ450072), swDQ (GenBank ID: DQ279091) and CHN-XJ-SW13 (GenBank ID: GU119961) assigned previously as 4i, 4b and 4h, respectively (Smith et al., 2020), the other sHEV isolates that are genetically closer to those reference strains are genotyped as 4i, 4e/b and 4h, respectively (Figure 1, Figure S1). Moreover, SW627 (GenBank ID: EU723513) and swX07-E1 (GenBank ID: EU360977) assigned as 3f (Smith et al., 2020), the other sHEV isolates that are genetically closer to them in our genotype analysis are under 3f. The sHEV isolates that are genetically closer to FR-SHEV3c-like (GenBank ID: JQ953664) grouped under the newly added subtype 3l are genotyped as 3l. Viruses genetically closer to Arkell (GenBank ID: AY115488) grouped under 3j (Smith et al., 2020) are genotyped as 3j, while viruses genetically closer to swJ8-5 (GenBank ID: AB248521) under 3e are genotyped as 3e. Viral isolate Osh 205 (GenBank ID: AF455784) under 3g (Smith et al., 2020) is still shown as an independent lineage 3g in the full-length genome-based phylogenetic analysis (Figure 1 and Figure S1).
3.2 Genomic similarity
To further investigate the genetic characteristics of China sHEVs, we conducted a genomic similarity analysis based on ten representative full-length genomes, including five strains from each group: sHEV3 and sHEV4 (Figure 2). The complete genome sequence of SH-SW-zs1 (GenBank ID: EF570133) was included as a Query. The ORF1 coding region exhibited the lowest genetic similarity at the HVR fragment, while the OFR3/ORF2 region is shown the most stable. Consistent with phylogenetic analysis results, sHEV3 and sHEV4 strains isolated in China obviously fell into distinct groups in the genomic similarities (Figure 2).
3.3 Recombination analysis
Genetic recombination is the driving force of viral evolution that impacts the pathogenicity and geographical distribution of viruses. Despite decades of HEV discovery, the presence of recombination within HEV genomes has been understudied. So far, one report published in 2010 revealed the occurrence of recombination events among HEV strains (H. Wang et al., 2010). Thus, in this report, we analysed a total of 112 sHEV complete genome sequences to investigate the recombination patterns and their burden on the genetic diversity and evolution of sHEV. We detected eight potential recombination events between 2002 and 2019, seven of which were intragenotype (Events 1–6, and 8) and only one intergenotype recombination event (Event 7, Table 1). Four recombination events occurred between viruses isolated in China (Events 2, 4, 6 and 7); meanwhile, others occurred between sHEV3 strains from different countries, including Event 1 (Thailand vs. Canada), Event 3 (South Korea), Event 5 (Spain vs. France) and Event 8 (Thailand vs. Spain) (Table 1). These results suggest the domestic genetic exchanges and indigenous characteristics of sHEV in China with a clear global genetic interaction and substantial correlation between the geographic origins and the genetic variation of the worldwide sHEVs.
| Recombinant | Minor parent | Major parent | Note | Detection methods | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Recombination Event serial number | Virus name (GenBank ID: Country-Year: genotype) | Virus name (GenBank ID: Country-Year: genotype) | Virus name (GenBank ID: Country-Year: genotype) | Country | Genotype | R | G | B | M | C | S | T |
| 1 | HEVF46(MH450023:Thailand-2015:3) | swSTHY42-VAS49/2003/CA(KJ507956:Canada-2003:3) | HEVGB40(MH450029:Thailand-2015:3) | TH vs. CA | 3 | + | + | + | + | + | + | + |
| 2 | SWU/L9/2018(MK410050:China-2018:4) | swGX40(EU676172:China-2008:4) | SWU/7-173/2018(MK410053:China-2018:4) | CN | 4 | + | + | + | + | + | + | + |
| 3 | swKOR-1(FJ426403:South_Korea-2007:3)a | swKOR-2(FJ426404:South_Korea-2007:3) | F19(MN614429:South_Korea-2019):3 | SK | 3 | + | + | + | + | + | + | + |
| 4 | swCH31(DQ450072:China-2006):4 | WH09(GU188851:China-2009):4 | HEV-ZJ1(JQ993308:China-2009:4) | CN | 4 | + | + | + | + | + | + | – |
| 5 | SW626(EU723512:Spain-2008:3)a | FR-SHEV3f(JQ953666:France-2008:3) | SW627(EU723513:Spain-2008:3) | ES vs. FR | 3 | + | + | + | + | + | – | + |
| 6 | W3(JQ655735:China-2014:4) | MO(JQ655733:China-2012:4) | WH09(GU188851:China-2009):4 | CN | 4 | + | + | + | + | + | + | + |
| 7 | swCH31(DQ450072:China-2006):4 | SAAS-JDY5(FJ527832:China-2008:3) | HEV-ZJ1(JQ993308:China-2009:4) | CN | 3 vs. 4 | + | + | + | + | + | + | + |
| 8 | SW627(EU723513:Spain-2008:3)a | HEVGB144(MH450030:Thailand-2015:3) | SW626(EU723512:Spain-2008:3) | TH vs ES | 3 | + | – | + | + | + | – | + |
- Note: The potential recombination events were identified by each of the seven algorithms (RDP, GENECONV, Bootscan, MaxChi, Chimaera, SiScan, and 3Seq) embedded in the RDP4 package.
- Abbreviations: B, bootScan; C, Chimaera; CA, Canada; CN, China; ES, Spain; FR, France; G, GENECONV; M, MaxChi; R, RDP; S, SiScan; SK, South Korea; T, 3Seq; TH, Thailand; +, verified; −, not verified.
- a The major or minor parent may be the actual recombinant due to the possibility of misidentification.
The recombinant swCH31 (GenBank ID: DQ450072) was previously reported as resulting from both intragenotype recombination (sHEV4) between a sHEV virus (CHN-XJ-SW13) isolated in China and a hHEV virus (E067-SIJ05C) isolated from a Japanese patient who travelled to Shanghai and intergenotype recombination between E067-SIJ05C and JJT-Kan (sHEV3, isolated in Japan from a patient with acute hepatitis) (H. Wang et al., 2010). In our report, swCH31 recombinant (GenBank ID: DQ450072) was identified resulting from inter- and intragenotype recombinations among the pigs' population in China. Therefore, our data provide extra insight into sHEV behaviour, suggesting that the swCH31 virus may result from different viral recombinations.
We mapped the breakpoints of putative recombination events and observed that three of them occurred within ORF2, including Event 2, Event 5 and Event 8 (Figure 3; Table 1), three within the ORF1 (Event 1, Event 3 and Event 4) and two within ORF1 and ORF3/ORF2 overlapping genomic regions (Event 6 and Event 7) (Figure 3; Table 1), indicating that genomic recombination may occur all over the sHEV genome.
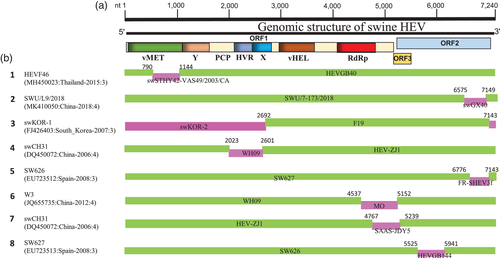
Out of 40 China sHEV isolates, a vast number of viruses (at least nine viruses) got involved in the recombination indicating that the intragenotype recombination could be the major factor driving the genetic diversity of sHEV in China during the past two decades. To provide evidence of the recombination authenticity, we further analysed all putative recombination events by constructing phylogenetic trees based on different genomic fragments (Figure S2). Recombination events are indicated with different symbols, in which the recombinants, minor parents and major parents are indicated by a red colour, a blue colour and yellow colour, respectively. As shown in Figure S2, the phylogenetic trees based on different individual fragments of the genome are not superimposed on each other. In Event 1, the recombinant strain HEVF46 in the fragment nt 700–1200-based phylogenetic tree is genetically closer to the minor parent swSTHY42-VAS49/2003/CA (Figure S2A). However, in the fragment nt 2000–2600-based phylogenetic tree, the recombinant HEVF46 is genetically closer to the major parent HEVGB40 (Figure S2B). The analyses of other recombination events showed similar results. These findings are in line with the recombination analysis conclusion (Figure 3; Table 1), confirming that our detected recombination events are real.
Swine serves as the major natural reservoir of HEV3 and HEV4 (Kamar et al., 2013). The zoonotic transmission of sHEV3 and sHEV4 is the primary cause of human HEV infection (Meng, 2013). The establishment of chronic HEV infection in immunocompetent and immunocompromised patients due to the zoonotic HEV cross-species goes beyond the hepatic manifestation to engender kidney failure and neurological diseases (Grewal et al., 2014; Meng, 2013). The emergence of swine HEV is a global health concern since it is endemic worldwide and not limited to developing countries, as was thought before (Izopet et al., 2019).
China is the largest swine-producing country worldwide (Chen et al., 2021). The seroprevalence of HEV in swine across multiple provinces in China is revealed high (Chen et al., 2021; Zhou et al., 2019), and the intraepidemic analysis of HEV genomes from different hosts suggested a potential swine–human HEV transmission (Zhou et al., 2019). The high prevalence of sHEV in China seems associated with economically developed regions (Chen et al., 2021). The pig trade movement worsens the regional characteristics of sHEV and increases its complexity (Y. Y. Li et al., 2018). While sHEV4 variants were most often detected in China (Figure 1) (X. Shu et al., 2014; Y. Shu et al., 2019; L. Wang et al., 2016), sHEV3 has also been identified in different areas since 2007 (Zhang et al., 2010). Genetic recombination is a pervasive phenomenon that significantly shapes the evolution of viruses. Since 2010, multiple recombination events of intra- and intergenotype drove the expansion of genetic diversity of sHEV in China (Figure 3). Si et al. (2012) demonstrated through analysis of swine faecal samples obtained from 39 pig farms during 2009−2010 in the Shanghai metropolitan area, a high coexistence level of sHEV3 and sHEV4, which increases the risk of coinfection. Si et al. (2012) also showed an inverse incidence rate between the two genotypes, suggesting their competition for existence within the host, which might increase the cross-species incidence rate. Furthermore, Liu et al. (2012) compared the partial nucleotide sequences of sHEV strains (sw-H04, sw-J02, sw-L05, CHN-XJ-SW61, SH-SW-zs1, SAAS-JDY5, bjsw1 and swGX40) with that of isolates obtained from patients with hepatitis E, living in the same regions as pigs, and reported a strong nucleotide identity, suggesting the pathogen cross-species and the ubiquity of zoonotic transmission. Herein, a total of eight potential recombination events were detected along the entire swine HEV genome, four of which occurred in China and were mainly between sHEV4, indicating that sHEV4 strains might remain the principal sHEV infection source that threatens the farming industry of China. However, the identified China intergenotype recombination predicts a risk of new emerging sHEV lineage which should be taken into consideration.
In summary, based on our results, genomic recombination is a major and substantial source of variability for sHEV in China, which may impact the evolution of zoonotic human HEV infection. The distribution of sHEV genotypes in China is obviously distinct from the global picture.
ACKNOWLEDGEMENTS
We thank all contributors to the collection and generation of swine HEV genomic sequences in GenBank. The Programme of Introducing Talents of Discipline to Universities (No. D21004).
CONFLICT OF INTEREST
The author declares that there is no conflict of interest.
AUTHOR CONTRIBUTIONS
Amina Nawal Bahoussi and Li Xing: conceptualization. Amina Nawal Bahoussi, Yan-Yan Guo, Pei-Hua Wang and Amina Dahdouh: data analysis. Amina Nawal Bahoussi and Amina Dahdouh: visualization and writing. Changxin Wu and Li Xing: administration. Amina Nawal Bahoussi and Li Xing: manuscript revision. All authors contributed to the article and approved the submitted version.
ETHICAL APPROVAL
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as the data analysed in this article have been collected.
Open Research
DATA AVAILABILITY STATEMENT
The data is available on the National Center for Biotechnology Information (NCBI) website.




