Identification of diffusion routes of O/EA-3 topotype of foot-and-mouth disease virus in Africa and Western Asia between 1974 and 2019 – a phylogeographic analysis
Abstract
Foot-and-mouth disease (FMD) affects the livestock industry and socioeconomic sustainability of many African countries. The success of FMD control programs in Africa depends largely on understanding the dynamics of FMD virus (FMDV) spread. In light of the recent outbreaks of FMD that affected the North-Western African countries in 2018 and 2019, we investigated the evolutionary phylodynamics of the causative serotype O viral strains all belonging to the East-Africa 3 topotype (O/EA-3). We analyzed a total of 489 sequences encoding the FMDV VP1 genome region generated from samples collected from 25 African and Western Asian countries between 1974 and 2019. Using Bayesian evolutionary models on genomic and epidemiological data, we inferred the routes of introduction and migration of the FMDV O/EA-3 topotype at the inter-regional scale. We inferred a mean substitution rate of 6.64 × 10−3 nt/site/year and we predicted that the most recent common ancestor for our panel of samples circulated between February 1967 and November 1973 in Yemen, likely reflecting the epidemiological situation in under sampled cattle-exporting East African countries. Our study also reinforces the role previously described of Sudan and South Sudan as a frequent source of FMDVs spread. In particular, we identified two transboundary routes of O/EA-3 diffusion: the first from Sudan to North-East Africa, and from the latter into Israel and Palestine AT; a second from Sudan to Nigeria, Cameroon, and from there to further into West and North-West Africa. This study highlights the necessity to reinforce surveillance at an inter-regional scale in Africa and Western Asia, in particular along the identified migration routes for the implementation of efficient control measures in the fight against FMD.
1 INTRODUCTION
Foot-and-mouth disease (FMD) is a transboundary threat that affects livestock production of cloven-hoofed species, causing production losses, mortality of young stock and continued costs associated with FMD vaccination (FAO, Cameroon, 2015) and restriction of animal trade (Grubman & Baxt, 2004; Knight-Jones & Rushton, 2013; Perry & Rich, 2007; Tekleghiorghis et al., 2016). The disease is characterized by rapid diffusion and is present in parts of South America, Asia and across a wide geographical distribution in Africa, where pastoralism and unrestricted movement of livestock favour the dissemination of the causative agent, foot-and-mouth disease virus (FMDV) (Bertram et al., 2018; Brito et al., 2016; Di Nardo et al., 2011; Lazarus et al., 2012). FMDV, a member of the Picornaviridae family, has a single positive-strand RNA genome which is replicated by the RNA-dependent viral polymerase through a rapid and low-fidelity process (Morelli et al., 2013; Steinhauer & Holland, 1987). This results in a rapid viral evolution of seven immunologically distinct serotypes [O, A, C, Southern African Territories (SAT) 1, SAT 2, SAT 3 and Asia 1], most further branching into multiple genetic topotypes and lineages (Kitching, 1998; Knowles & Samuel, 2003; Knowles et al., 2016; Paton et al., 2009; Samuel & Knowles, 2001).
FMDV genomic RNA is particularly appropriate for phylogenetic analysis as this RNA virus of about 8500 nucleotides (nt) can change rapidly, accruing mutations of ∼1–8 nt per replication cycle (Domingo et al., 1995, 2002). The 1D FMDV genome region coding for an outer capsid protein (VP1) bears the most varied part of the genome, and is therefore frequently used for molecular epidemiological analyses (Brito et al., 2016; Ehizibolo et al., 2020; Pedersen et al., 2015; Sangula et al., 2010; Wekesa et al., 2014). Understanding the global dispersion of FMDV by tracking its viral evolution with phylogeographic models can help to reconstruct the disease spread from endemic regions and potentially predict the risks of incursion into FMD-free countries (Dellicour et al., 2018; Lemey et al., 2009).
Indeed, multiple FMDV lineages and topotypes have historically spread beyond their initial location to inter-regional and international destinations, even reaching a pandemic status for the FMDV O/ME-SA/Ind-2001 lineage that caused outbreaks in the Gulf States of the Western Asia, Southeast Asia, East Asia, the FMD-free islands of Mauritius and that reached FMD-free North Africa in 2013 to 2015 (Knowles et al., 2005; Bachanek-Bankowska et al., 2018). The geographical proximity as well as the numerous trans-Mediterranean connections between North Africa and Europe increase the risk of a potential introduction of the disease to FMD-free European countries
More recently, in the second part of 2018 and early 2019, FMD serotype O outbreaks have been reported to the OIE by Algeria, Morocco, Tunisia and Libya in hundreds of cattle and small ruminant farms. The causative agent for the 2018/2019 outbreaks was identified as belonging this time to the serotype O, topotype East-Africa-3 (O/EA-3) that caused earlier in 2018 an upsurge in FMD cases in a number of West African countries including Burkina Faso, the Gambia, Guinea, Mauritania, Senegal and Sierra Leone (EuFMD, 2019). The spread of this topotype parallels that of another FMDV lineage, A/AFRICA/G-IV, in West African Countries in 2015 and then in North Africa in 2017 (Pezzoni et al., 2019).
Through the activities of the OIE/FAO Network of FMD reference laboratories, clinical samples belonging to the O/EA-3 topotype were collected from many FMD outbreaks reported in West and North Africa over 2018 and 2019. We took the opportunity of this extensive collaborative collection to use phylogeographic approaches on new and publicly available sequences encoding the FMDV VP1 region (n = 489) to trace the global spread of the O/EA-3 topotype. We mapped and dated its initial recorded geographic origins and showed the establishment of viral migrations outside its first pool of distribution in East Africa.
This study incorporates samples from 25 different African and Western Asian countries where at least one O/EA-3-related FMDV outbreak occurred between 1974 and 2019, providing the most comprehensive investigation of this lineage spread so far. We further detail the inferred transmissions and trans-regional introductions, providing novel insights into the geographical origin but also into the spatial and temporal distribution of the FMDV O/EA-3 topotype.
2 MATERIAL AND METHODS
2.1 Sequence collection
Sequences from samples collected from FMD outbreaks investigations in 25 African countries between 1974 and 2019 were retrieved either online from the NCBI GenBank database or from OIE/FAO FMD reference laboratories, which included: Anses (Maisons-Alfort, France), The Pirbright Institute (Woking, UK), Sciensano (Bruxelles, Belgium), IZSLER (Brescia, Italy) and NCFAD (Winnipeg, Canada).
2.2 VP1 sequencing of FMD viruses
Virus RNAs were extracted either from clinical sample suspensions or from cell culture passaged virus through primary bovine thyroid cells (Snowdon, 1966), IB-RS-2 cell monolayers (De Castro, 1964) or ZZ-R 127 cells (Brehm et al., 2009). Sequences prior to 1993 were obtained by direct sequencing from vRNA templates (Knowles & Samuel, 1995). Sequences obtained after 1993 were obtained from PCR products sequenced according to the technical protocol previously described (Knowles et al., 2016). Nucleotide sequences covering the VP1-coding region were assembled from at least one forward and one reverse read for each sample using SeqMan Pro™ (Lasergene 15.0 software; DNAStar Inc., Madison, WI, USA). Sequences described here for the first time were obtained from samples collected in West and North African outbreaks that occurred in 2018–2019. Live virus could not be recovered from two poor quality samples. However, virus isolates O/SEL/13/2018 and O/SSD/6/2017 were rescued following the transfection of extracted RNA into LFBK cells using Lipofectamine. VP1 sequences were compared to the homologous genomic regions available in the NCBI GenBank database or in the World Reference Laboratory of Foot-and-Mouth Disease (WRLFMD) database. For VP1 sequences submitted to GenBank, accession numbers are reported in Table S1.
2.3 Sequence data and primary phylogeny
Considering the length of FMDV VP1 serotype O coding sequence (639 nt), we excluded sequences shorter than 630 nucleotides and for sequences with a length of 630–638 nucleotides, the terminal missing nucleotides were replaced with the N code rather than discarding the sequences. Unambiguous, unique VP1 sequences were collected from 25 countries including Algeria, Burkina Faso, Cameroon, Egypt, Eritrea, Ethiopia, Gambia, Ghana, Guinea-Conakry, Israel, Ivory Coast, Kenya, Libya, Mali, Mauritania, Morocco, Nigeria, Palestine, Senegal, Sierra Leone, Somalia, Sudan, South Sudan, Tunisia and Yemen. Multiple sequence alignments were conducted using CLUSTAL W (Edgar, 2004; Thompson et al., 1994) as implemented in the MEGA X software (Kumar et al., 2018).
2.4 Epidemiological data
For all sequences, we consulted the available metadata for each isolate and collected the following variables: date of collection, country and host species (Table S1). When the date of collection was incomplete, we excluded the sample. Geographical distribution of the samples, respective times of collection and frequency in host species are reported in Figure 1. Samples were collected from five different species: cattle, goats, pigs, sheep and water buffalo (Table S1).
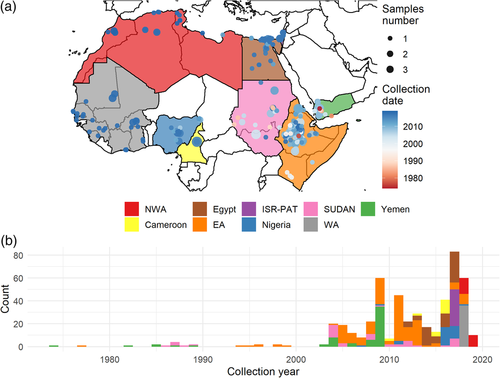
The number of sequences greatly differed between countries. Therefore we decided to group countries belonging to the same United Nations (UN) sub-region (UN standard country or area codes for statistical use, 1996, M49, https://unstats.un.org/unsd/methodology/m49/) and with less than ten samples with the neighbouring countries as follow: (i) we selected the country with the least number of samples, (ii) we grouped it with the neighbouring country with the least number of sample and assessed the total number of samples from this first group, (iii) we repeated steps (i) and (ii) until the total number of samples reached at least ten samples in each group.
2.5 Bayesian evolutionary model
We resolved the spatial history of the O/EA-3 FMDV topotype by reconstructing the virus migration network using an asymmetric continuous-time Markov chain model (Lemey et al., 2009) in BEAST2 version 2.6.4 (Drummond & Rambaut, 2007). We first set the population model to be a coalescent model with constant population size. We then selected the most appropriate substitution model using Bayesian model averaging as implemented in the package bModelTest (Bouckaert & Drummond, 2017). We then selected molecular clock and population models, based on their marginal likelihood estimated by nested sampling, as implemented in the NS package (Russel et al., 2019; Skilling, 2006). Constant population size and a relaxed uncorrelated molecular clock branch rate model were used to examine the temporal origin of each clade.
BEAST2 provides for each migration rate between two clusters an indicator whose mean is the posterior probability (pp) that this particular transition rate is positive. For each rate indicator, we computed the Bayes factor (BF) as the ratio of the posterior and prior odds that this indicator is positive. The level of support can be assessed by the BF as follow: no evidence for BF = 1, anecdotal support for 1 ≤ BF < 3, moderate support for 3 ≤ BF < 10, strong support for 10 ≤ BF < 30, very strong support for 30 ≤ BF < 100 and extreme support for BF ≥ 100.
We then counted the labelled transition for the migration rates with a BF > 3 in a subset of 101 posterior trees sampled after a burn-in of 10%.
We generated posterior distributions of parameters and posterior set of trees running a Markov Chain Monte Carlo (MCMC) of 500 million iterations, sampling every 50,000 and removing the 10% as burn-in.
We assessed the inference validity by inspecting each parameter trace to determine whether a stationary distribution was reached. We also inspected the effective sample sizes (ESS) of different parameters to check that the MCMC were well mixed and added iterations, if necessary, to attain ESS greater than 200.
We generated the maximum clade credibility (MCC) tree using TreeAnnotator version 2.6.3 (Drummond & Rambaut, 2007). From the MCC tree, we defined clades as the largest subtrees whose most recent common ancestor (MRCA) had a pp > .9.
We present thereafter each parameter range, consisting of the median estimate and its 95% highest posterior density (HPD) and ESS.
All phylogenetic trees were drawn with ggtree 2.4.1 package (Yu et al., 2017). Other graphics were drawn with the ggplot2 package 3.3.3 (Wickham, 2011).
3 RESULTS
3.1 Data cleaning and description
From the 559 available sequences obtained from samples collected between 1974 and 2019, two were excluded due to truncated sequence length (402 nt) and 12 were duplicates, leading to 545 unique sequences. In addition, 56 sequences were also excluded due to the missing sampling time. In total, 489 sequences were included in the analysis (Table S1).
The spatial and temporal distributions of the 489 unique sequences are shown in Figure 1. In order to have at least ten samples per group of countries, Algeria, Libya, Morocco and Tunisia were clustered in a region that we thereafter define as NWA (North-West Africa). Similarly, Burkina Faso, Ghana, Guinea, Ivory Coast, Mali, Mauritania, Senegal and Sierra Leone were grouped, with the resulting region defined as WA (West Africa). Sudan and South Sudan were grouped in the SUDAN region; Eritrea, Ethiopia, Kenya and Somalia were grouped in the EA (East Africa) region; and Israel and Palestine were grouped in the ISR-PAT region. Cameroon, Egypt, Nigeria and Yemen remained as individual countries (Figure 1(a)). The time window differed for each country and region (Figure 1).
3.2 O/EA-3 FMDV evolutionary model
The substitution model selected by model averaging was the General Time Reversible (GTR) model with Gamma distribution (with four categories) and a proportion of invariant sites (set to 0.4). According to the BF, an uncorrelated lognormal relaxed molecular clock was also preferred to the strict molecular clock (Table S2). Characteristics of parameter posterior distribution according to the subsamples are shown as an extract in Table 1.
| Variable | Median | 95%HPD | ESS |
|---|---|---|---|
| Tree height | 48.36 | 45.09; 67.17 | 3848 |
| Uncorrelated lognormal clock mean | 6.64 × 10−3 | 5.65 × 10−3; 7.70 × 10−3 | 1402 |
| Uncorrelated lognormal standard deviation | 0.63 | 0.51; 0.77 | 853 |
| Effective population size | 7.39 | 6.27; 8.57 | 878 |
- For each parameter the median, the 95% highest posterior density (95% HPD) and the effective sample size (ESS) are reported.
The molecular clock rate was estimated having a median of 6.64 × 10−3 nt/site/yr (IC) (95% HPD = [5.65 × 10−3 – 7.70 × 10−3]). The tree height, corresponding to the time when the MRCA of the included sequences circulated, was inferred as 48.36 years (95%HPD = [45.67; 52.37]), suggesting that the MRCA of the O/EA-3 topotype circulated between February 1967 and November 1973. The MRCA was predicted to have circulated early in Yemen with a strong probability (p = .89).
Figure 2 shows the MCC tree. The reconstructed topology is characterized by three clades, named A, B and C. Some strains were not included in any clade, according to our clade definition. Clade A contains the oldest sequences. Clade A's MRCA was predicted to circulate in Yemen (pp = .99), similarly to the overall tree MRCA. All sequences from this clade were collected either from Yemen or from the EA region between 1982 and 2015. Note that the two oldest strains (collected in Yemen in 1974 and in Ethiopia in 1979) were not included in clade A.
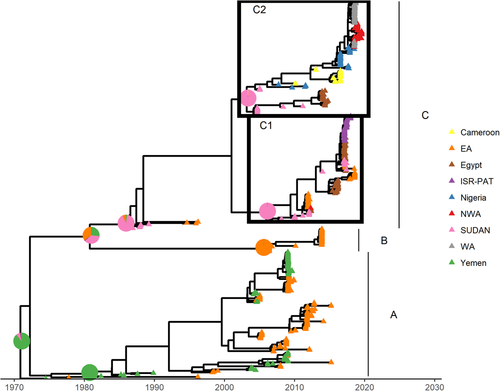
The location of the MRCA of both clades B and C could not be accurately predicted. Indeed, the probabilities for Yemen, SUDAN region and EA were .27, .37 and .35, respectively. Clade B was the smallest on the tree and contained only sequences collected from EA region between 2006 and 2013. As expected, its MRCA was predicted (p = 1.0) to have circulated in the EA region as well. Clade C was the largest and contained sequences collected from all countries or regions besides Yemen between 1986 and 2019. Its MRCA was predicted to be located in SUDAN region (p = .91). Within clade C, two sub-clades could be identified (C1 and C2) and for both sub-clades, the MRCA was also predicted in SUDAN region (p = 1.00 for both). C1 contained sequences collected from SUDAN region, EA, Egypt and ISR-PAT region collected between 2008 and 2018, whereas C2 contains sequences collected from SUDAN region, Egypt, Nigeria, Cameroon, WA and NWA collected between 2004 and 2019.
We then predicted the likelihood of migration routes which were identified by their BF (BF > 3) in Figure 3.
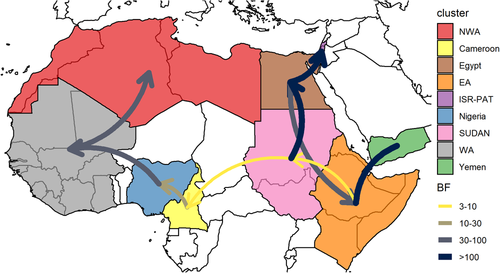
FMDV O/EA3 was predicted to have migrated between Yemen and the EA region (BF ≥ 100) and to have migrated north to SUDAN region (3 ≤ BF < 10)). O/EA-3 spread from SUDAN region to Egypt (BF ≥ 100) and from Egypt to ISR-PAT (BF > 100). Of note, migration between Egypt and the EA region was also well supported (30≤ < BF < 100). For the O/EA-3 spread from the Eastern to the Western regions of Africa, a migration from SUDAN region to Cameroon was found moderately well supported (3 ≤ BF < 10). Once in the Western part, FMDV O/EA3 spread first to the West from Cameroon towards Nigeria (10 ≤ BF < 30) and from Nigeria towards the WA region (30 ≤ BF < 100) then North from the WA region to the NWA region (30 ≤ BF < 100).
Figure 4 shows the proportion of each statistically-supported migration identified per year and per tree, computed from 101 trees sampled after the burn-in.
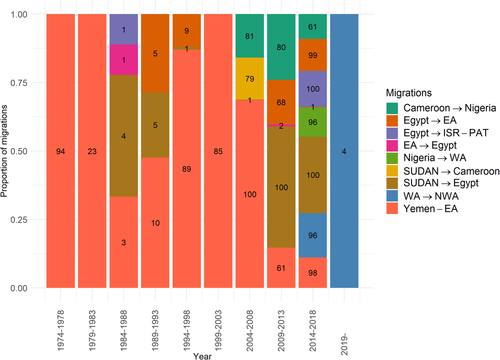
The migrations identified differed before and after 2004. Before 2004, the majority of predicted migrations were clustered to the Eastern part, mainly between Yemen and the EA region starting in 1974. In addition, migrations from EA to Egypt in 1984–1988, from SUDAN region to Egypt in 1984–1993 and from Egypt to EA between 1989–1993 and 1994–1998 were also predicted, but in less than 10 out of 101 posterior trees.
The transition from East to West was predicted to appear from SUDAN region to Cameroon in 2004–2008 in 80 out of 101 posterior trees and from Cameroon to Nigeria from 2004–2013 in 80 out of 100 posterior trees to 2014–2018 in 61 out of 101 posterior trees.
During the 2009–2013 and 2014–2018 5-year time intervals, the predicted migrations were more diverse and more frequent (Figure 4) on both the Eastern and Western parts of the African continent. Migrations from SUDAN region to Egypt were predicted in all posterior trees for both time intervals 2009–2013 and 2014–2018 on one hand and from Egypt to EA on the other hand in 68 out of 101 posterior trees in 2009–2013 and 99 out of 101 in 2014–2018. Migration from Egypt to the ISR-PAT region during the 2014–2018 time interval was predicted in all posterior trees. Finally, migrations between Yemen and EA, from Nigeria to WA and from WA to NWA in 2014–2018 were predicted in more than 97 out of 101 posterior trees.
4 DISCUSSION
In this work, we used combined genomic and epidemiological data to identify FMDV O/EA-3 topotype migration routes across Western Asia and the African continent. For this purpose, we gathered the most exhaustive dataset for this topotype to our knowledge, larger than similar ones previously collected for the entire serotype O (Duchatel et al., 2019; Tully & Fares, 2009). However, the number of samples collected before 2000 was limited and no sample bearing FMDV O/EA-3 topotype was collected for some countries central in our study area, such as Chad.
We reconstructed the phylogeographic history of the O/EA-3 FMDV topotype since its first detection in 1974 to its incursion in North-West Africa in 2019. Using Bayesian evolutionary models, we estimated a substitution rate for the O/EA-3 topotype of similar order of magnitude or slightly higher than previous maximum estimates for the entire serotype O in Africa and in Europe/Asia of 4.26 × 10−3 nt/site/yr (Tully & Fares, 2009), with 27 samples collected between 1951 and 2002 and 4.67 × 10−3 nt/site/yr with 192 samples collected in Africa between 1964 and 2016 (Duchatel, Bronsvoort & Lycett, 2019).
The topology of our MCC tree is reminiscent of the phylogeny found in Duchatel et al. (2019) for FMDV serotype O in Africa where the topotype EA-3 contained two clades: the first located in Ethiopia, Kenya, Somalia (similar to the strains collected in the EA region of our clade A) and the second clade (similar to our clade C) divided with one sub-clade mostly located in SUDAN region, and then in Nigeria/Cameroon (similar to our clade C2) and the second sub-clade mostly located in Egypt and Ethiopia (similar to our clade C1) (Duchatel, Bronsvoort & Lycett, 2019; Tully & Fares, 2009). With the inclusion of older strains from the Arabic Peninsula, we highlighted the intertwined history between Yemen and East Africa at the origin of the O/EA-3 FMDV topotype. However, due to the sparse number of early sequences, the direction of the migration should be considered with caution. Yemen has always mostly imported livestock from East Africa while exports are limited to null (Costagli et al., 2017; Samatar, 1988) which would favour migration from East Africa to Yemen rather than in the other direction. Our study also specified that the deployment in Africa happened first in the EA region before spreading to SUDAN region, which then acted as a spreading source for the O/EA-3 FMDV topotype. More specifically, our results strongly support virus transmission routes from SUDAN region to Israel and Palestine through Egypt and, while not as strongly supported, our results suggest that a second virus transmission route from SUDAN region via Cameroon and Nigeria reaches West Africa and then North West Africa. Our results therefore extend previous phylogenies to new territories North (Israel and Palestine) in clade C1 and West (Western African countries and the North-West Africa region) in clade C2. Finally, in 2021, outbreaks due to an O/EA-3 FMDV were reported in Bahrain. The field strain analyzed was most closely related to FMDV found in Ethiopia in 2015 and to strains detected in Yemen in 2006 and 2008 (WRLFMD, 2021) which belonged to the clade A from our results, suggesting the ongoing extension of this clade.
The routes of FMDV diffusion described in this study could result in part from the recently improved trans-continental connectivity, allowing for both livestock and livestock products to move easily, thus increasing the chance of FMD spread not only from East to Central Africa but also from Central Africa to West Africa and Maghreb (Di Nardo, Knowles & Paton, 2011). Indeed, the trans-Saharan Highway running from Lagos in Nigeria directly north to Algiers in Algeria might represent a possible entry gate for infected livestock, meat products and contaminated material. This specific 4600 km section has seen great improvements in the recent years in each country it crosses, more specifically with extensive funding and improvements in 2018–2019 for the Malian, Nigerian and Algerian transepts that should be achieved in 2021 (Trans-Sahara highway - OPEC Fund for International Development, 2021).
In addition to the existing transnational transhumance pastoralism livestock movements between Sudan and Central Africa, population displacement resulting from conflicts also favour FMD diffusion (Luizza, 2017). Our findings showing the pivotal role of the SUDAN region in the FMDV O/EA-3 spread at the continental level, especially after 2000, could relate to the Darfur war, which induced massive population displacements starting in 2003 (Olsson & Siba, 2010). The FMDV O/EA-3 migration events we reconstructed between sub-Saharan countries and North-African countries might also relate to climatic events [e.g., 2018 Mauritanian drought (FAO, 2019)] or political changes [e.g., 2010s Arab spring (Kandeil et al., 2013)], which altered pattern of animal movements and trading relationships (Hamoonga et al., 2014; VanderWaal et al., 2017).
Besides their potential impact on FMD diffusion, conflicts and political instability led to a heterogeneous monitoring and sampling of FMD outbreaks in Africa, both between countries and in time. For instance, the Lake Chad area was attractive for the nomadic pastoralists until the 2000s (Seignobos, 2015). However, the crisis caused by the presence of Boko Haram and the measures adopted by Nigeria (closure of borders and livestock markets) led to the fled of some population and change in transhumance routes (Magrin and Raimond, 2018). This resulted in missing information from countries located in the core of our study area (e.g., Niger, Chad and the Central African Republic). Although sampling bias can alter statistical inference when migrations are treated as continuous-time Markov chain, other models known to be less affected by these biases, such as structured coalescent models, are in their turn disturbed by unsampled deme (Beerli, 2004; Ewing & Rodrigo, 2006; Slatkin, 2005). According to the frequency of sampling and outbreaks’ occurrences in our study, these biases are probably more important before 2000 and in the Western part (Nigeria, Cameroon, WA, NWA) than in the Eastern part of our study region (EA, Yemen, Egypt, SUDAN region and ISR-PAT region). Although older samples cannot be retrieved, additional prospective samples from this Western part would improve the robustness of our results in this area.
In this study, we described the early migration routes of the O/EA-3 FMDV topotype between Yemen and East Africa. We showed that SUDAN region acted as a spreading source at the continent level. Other FMD virus topotypes circulating in Central and West Africa such as SAT2/VII or O/WA represent a potential threat and their spread should therefore be closely monitored as the epidemiological connectivity described here can ease the movement of these topotypes in the near future with similar devastating outcomes.
ACKNOWLEDGMENTS
We would like to acknowledge the veterinary services and laboratory teams from all participating institutes involved in the collection and processing of samples. In particular, for the sequences newly described in this paper: Samples from Algeria were provided by Dr Hafsa Madani Laboratoire Central Vétérinaire d'Alger, Institut National de Médecine Vétérinaire (INMV), Mohammadia, Algeria. Samples from Burkina Faso were provided by Dr. Moctar SIDI, Dr. Bruno Lalidia OUOBA and Dr. Habibata Lamouni Zerbo/Ouermi laboratoire national d’élevage of Ouagadougou. Samples from Guinea were provided by Dr. Mamadou Lamarana Souare Central Veterinary Diagnostic Laboratory in Conakry, Guinea. Samples from Ivory Coast were provided by Dr. Cyprien Yapi Boke, Laboratoire central vétérinaire de Bingerville (LCVB), Côte d'Ivoire. Samples from Libya were provided by Professor Ibrahim Eldaghayes and Professor Abdunaser Dayhum Faculty of Veterinary Medicine, University of Tripoli, Libya. Samples from Mauritania were provided by Dr Meina Hasni, Centre national d'élevage et de recherches vétérinaires, Nouakchott, Mauritanie. Samples from Morocco were provided by Dr Nabil Abouchoaib Office National de Sécurité Sanitaire des produits Alimentaires (ONSSA), Rabat, Morocco. Samples from Tunisia were provided by Dr. Soufien Sghaier Institute of Veterinary Research of Tunisia. Tunis, Tunisia. The work of the EURL for FMD was supported with funding provided from the European Union within the framework of the EURL 2019–2020 Work Program. V. M. is funded by the United Kingdom Department for Environment, Food and Rural Affairs (DEFRA; project SE1130).
CONFLICT OF INTEREST
The authors declare no conflict of interest; the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
AUTHOR CONTRIBUTIONS
Conceptualization: S. B., L. C., B. D., D. K. and L. B.-K.; methodology: L. C., S. B. and A. D.-N.; sample collection and sample analysis: J. W., A. E. S., A. Ro., A. Re., C. B.-C., A.-L. S. A. H., U. H. and D. L.; data curation: N. K., S. B. and L. C.; formal analysis: L. C.; investigation: L. C. and S. B.; resources, L. B.-K., D. K. and B. D.; writing—original draft preparation: S. B. and L. C.; writing—review and editing: S. B., L. C., A. D.-N., D. K., S. B.-B., C. B.-C., E. B., K. D., G. P., C. N. and L. B.-K.; visualization: L. C.; supervision: S. B.; project administration: S. B.; funding acquisition: L. B.-K., B. D. and D. K. All authors have read and agreed to the published version of the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




