Characterization of parainfluenza virus 5 from diarrheic piglet highlights its zoonotic potential
Abstract
Parainfluenza virus 5 (PIV5), a member of paramyxoviruses, causes respiratory and neurological infection in several animal species. Whereas information on PIV5 infection in digestive system is very scarce. Here, we successfully isolated one PIV5 strain from diarrheic piglets. After four times plaque purification and ultracentrifugation, the paramyxovirus-like particles were observed by electron microscopy. The genome-wide phylogenetic analysis showed that the isolated strain was closely related to the PIV5 strain from a lesser panda and pigs in China. Therefore, we characterized this isolated PIV5 and found that this virus could haemagglutinate red blood cells from both guinea pigs and chickens. Further, we observed that this PIV5 could infect cell lines from various host species including pig, human, monkey, bovine, dog, cat, rabbit, hamster and mouse, which was confirmed with the immunofluorescent assay. To evaluate the distribution of PIV5 in the field, we developed an indirect ELISA (iELISA) for the first time to detect the specific antibodies based on recombinant nucleocapsid protein. A total of 530 porcine serum samples were tested and the PIV5-positive rate was 75.7%. To our knowledge, this is the first report describing the full characterization of PIV5 strain isolated from a diarrheic piglet. The ability of this PIV5 strain to infect a wide range of mammalian cell types indicates that PIV5 can transmit across different species, providing a remarkable insight into potential zoonosis. The virus strain and iELISA developed in this study can be used to investigate the pathogenesis, epidemiology, and zoonotic potential of PIV5.
1 INTRODUCTION
Paramyxoviruses comprise a large family of negative-strand RNA viruses, which contain important respiratory and systemic pathogens in both animals and humans (Knipe & Hetsley, 2007; Philbey et al., 1998; Thibault et al., 2017; Zeltina et al., 2016). The family Paramyxoviridae possesses one of the highest rates of cross-species transmission among RNA viruses, leading to zoonotic infections (Kitchen et al., 2011). Over the last few decades, several paramyxoviruses have been emerging or re-emerging in humans and animal species (Thibault et al., 2017). Under the family Paramyxoviridae, parainfluenza viruses (PIV) are classified into five types, designated PIV 1−5.
Parainfluenza virus 5 (PIV5) was renamed to mammalian orthorubulavirus 5 in 2016 (Adams et al., 2017); however, the term PIV5 has been used in this article to provide a better connection to the scientific context. PIV5 belongs to genus Orthorubulavirus within subfamily Rubulavirinae and has the smallest genome among the family Paramyxoviridae with 15,246 nucleotides, and a pleomorphic-shaped virion of 50−200 nm in diameter (Chen, 2018; Rima et al., 2019; Terrier et al., 2009). The genome of PIV5 contains seven genes encoding eight proteins in the order of 3′-N (nucleocapsid protein)-V (V protein)-P (phosphoprotein)-M (matrix protein)-F (fusion protein)-SH (small hydrophobic protein)-HN (haemagglutinin-neuraminidase)-L (large protein)−5′(Parks et al., 2011; Tompkins et al., 2007). During virus replication, HN and F proteins facilitate virus attachment, entry and the fusion between viral and cellular membrane (Samuel et al., 2010).
Recently, PIV5 infection has been increasingly reported and causes respiratory and neurological diseases in a variety of host species, including human, pig, dog, cattle, cat, hamster, guinea pig, pangolin, lesser panda and horse (Charoenkul et al., 2021; Lee & Lee, 2013; Liu et al., 2015; Wang et al., 2019; Xie et al., 2020; Zhai et al., 2017). The co-infection of PIV5 with porcine reproductive and respiratory syndrome virus in pigs was found to be associated with respiratory clinical signs (Heinen et al., 1998; Lee et al., 2013). However, the prevalence of PIV5 and its impact on swine health are largely unknown, especially the information regarding PIV5 infection in the porcine digestive system is scarce.
Currently, diagnosis of PIV5 infection is generally based on the detection of nucleic acid by PCR and confirmed by virus isolation (Charoenkul et al., 2021; Yang et al., 2014). Although direct detection of the presence of viral RNA by PCR is sensitive, ELISA test can be more simple, less expensive, and applicable to wider field-based applications. To our knowledge serology assays to diagnose PIV5 infection are not yet available. Based on these facts, highly sensitive and effective serodiagnosis assays are crucial to investigate the epidemiology of PIV5. This study aimed to characterize the PIV5 strain associated with porcine diarrhoea and develop an indirect ELISA to assess the prevalence of PIV5 in swine farms.
2 MATERIALS AND METHODS
2.1 Sample collection
Faecal samples from diarrheic piglets in Heilongjiang province of China were collected and submitted to our lab for routine diagnostic purposes. In addition, a total of 530 serum samples were collected from randomly selected pigs from different age groups in Heilongjiang province, China. Serum samples and stool suspensions were stored at −80°C until use.
2.2 Cell culture
Twenty-one cell lines derived from different species were used, including, porcine (PK15, ST, IPEC-J2), human (293T, Huh7, HepG2, A549, Hela, CaCo2), primate (Marc145, Vero E6), bovine (MDBK), canine (MDCK), feline (CRFK), rabbit (RK13), hamster (BHK21, CHO), mouse (NIH/3T3, RAW 264.7), duck (DEF) and chicken (DF-1) cell lines. Marc145 cell was cultured in RPMI-1640 medium (Gibco), DEF and DF-1 cells in Eagle's Minimum Essential Medium (Multicell) and the remaining cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco). All media were supplemented with 10% (v/v) foetal bovine serum (Biological Industries), 100 U/ml penicillin and 100 U/ml streptomycin.
2.3 Isolation of PIV5 strain
To identify the causative agent of diarrheic piglets, faecal samples were examined by RT-PCR for common porcine enteric viruses, including transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhoea virus (PEDV), porcine deltacoronavirus (PDCoV), genogroup A and C rotavirus (RVA and RVC), porcine astrovirus (PAstV), porcine sapovirus (PSaV), porcine norovirus (PNoV), mammalian orthoreovirus (MRV) and porcine parvovirus (PPV), using specific primers (Table S1) as previously described (Ding et al., 2020; He et al., 2021; Kim et al., 2006; Kim et al., 2010; Lelli et al., 2013; Reuter et al., 2011; Wang et al., 2006). Faecal sample tested negative for above enteric viruses was subjected to virus isolation. Briefly, faecal sample was diluted to prepare a 10% suspension in PBS. The suspension was vigorously homogenized by vortex for 5 min, clarified by centrifugation at 10,000 g for 30 min, and filtered through a 0.22 μm filter (Merck Millipore Ltd. United States). One millilitre of filtered faecal suspension was diluted 1:1 in DMEM containing 1% penicillin and streptomycin, inoculated onto confluent PK15 cells and incubated at 37°C and 5% CO2 for 1 h. The inoculum was discarded, and cells were then washed two times with PBS, replaced with DMEM without FBS, incubated at 37°C and 5% CO2 and observed daily for cytopathic effects (CPEs). After 4 days, cell lysates were collected and re-inoculated into PK15 cells for three blind passages under the same conditions.
2.4 Viral plaque assay
To purify the isolated virus, plaque assay was performed according to the previously described procedure with slight modifications (Guo et al., 2014). PK15 cells were inoculated with diluted viruses, overlaid with 1% agarose in DMEM containing 1% penicillin and streptomycin and 2% FBS, and incubated at 37°C and 5% CO2. After plaque development, uniform and clear plaques were picked using sterile pipette tips, and the agarose plug was placed into a 0.2 ml medium and re-inoculated into cell monolayer to harvest the positive clone. After four rounds of plaque purification, the virus clones with the highest titre were successfully obtained. The plates were also fixed with 4% paraformaldehyde and stained with 1% crystal violet for visualization the production of CPEs.
2.5 Transmission electron microscopy (TEM) of virus particles
The virus supernatant was passed through 0.22 μm filters and centrifuged at 35,000 rpm for 4 h in Beckman SW32Ti rotor. The resulting pellet was resuspended in DMEM and then centrifuged through 13 ml 20−50% (w/v) sucrose cushion (Beckman SW55Ti rotor, 4 h, 35,000 rpm, 4°C). The virus particles which formed a white opalescent band at the interface of the sucrose solutions were collected and resuspended in 0.5 ml DMEM and centrifuged for 3 h (Beckman TLA55 rotor, 35,000 rpm, 4°C). The purified virus was examined under TEM (Hitachi Model H-7650). To further determine the viral particles, cell supernatants were analysed for paramyxoviruses including parainfluenza viruses (PIV1, PIV2, PIV3, PIV4 and PIV5), porcine parainfluenza virus 1 (PPIV1), porcine rubulavirus LPMV (La Piedad Michoacan Mexico Virus), Nipah virus and paramyxoviruses under subfamily of Rubulavirinae using RT-PCR with the specific primers (Table S2) as previously reported (Aguilar et al., 2000; Dénes et al., 2021; Jiang et al., 2018; Tong et al., 2008; Wiman et al., 1998).
2.6 Haemagglutination assay (HA)
The protein spikes of parainfluenza viruses have haemagglutinating activity, thus a direct HA was used to determine whether the isolated PIV5 has haemagglutinating activity. In brief, chicken and guinea pig red blood cells were washed three times with PBS and resuspended as a 1% suspension. HA test was performed by preparing serial twofold dilutions of isolated PIV5 against chicken and guinea pig red blood cells. The HA titre was expressed as the reciprocal of the highest dilution of PIV5 showing HA.
2.7 Virus growth kinetic
To determine the growth curve of the isolated virus, PK15 cells were infected in triplicate with PIV5 at 0.01 multiplicity of infection (MOI). Cell supernatants were harvested at 1, 12, 24, 36, 48, 72, 96 and 120 h post-inoculation and stored at −80°C until use. Virus titres were measured by 10-fold serial dilutions in PK15 cells. The Reed-Muench method was used to calculate viral titres as 50% tissue culture infectious dose (TCID50) per ml (Reed & Muench, 1938).
2.8 Whole-genome sequencing and phylogenetic analysis
To better understand the molecular characteristics of the isolated PIV5, the whole genome was sequenced. In brief, total RNA was extracted from plaque-purified virus clones using the TIANamp virus RNA Kit (Tiangen Biotech, Beijing), cDNA was transcribed using the PrimeScript cDNA Synthesis Kit (Takara, Dalian, China) and cDNA libraries were prepared using the Agencourt AMPure XP-Medium kit (Beckman Coulter, A63881, USA) according to the manufacturer's instructions. The prepared libraries were then sequenced using BGISEQ-500 Sequencing System. After filtering out low-quality raw reads, the viral genome was assembled using CAP3, IDBA, SPAdes and Edena as described previously (Bankevich et al., 2012; Hernandez et al., 2014; Huang & Madan, 1999; Peng et al., 2012). The prediction of viral genes was performed using the software GeneMarks (http://topaz.gatech.edu/GeneMark/genemarks.cgi), and annotation of the predicted virus was carried out through the non-redundant protein database (NR) using BLASTn and BLASTx. Phylogenetic analysis of whole-genome, N, F and NH genes was performed by comparing nucleotide sequences of the isolated PIV5 with reference sequences from GenBank (Table 2). Nucleotide and amino acid sequences were aligned using Clustal W software. Phylogenetic analysis was constructed via the maximum-likelihood method using MEGA v.6.0 software with the neighbor-joining method with Kimura 2-parameter and 1000 bootstrap replicates (Hall, 2013; Tamura et al., 2013).
2.9 Cell susceptibility test to the PIV5 isolate
To determine the viral tropism, 21 cell lines derived from different species (Table 1) were subjected to PIV5 at MOI of 0.01 and observed for CPEs. To confirm PIV5 infection in different cell lines, an immunofluorescence assay (IFA) was performed to determine viral N protein production as described previously (Zhang et al., 2021). Briefly, 21 cell monolayers were inoculated with PIV5 at MOI of 0.01 and incubated for 24 h. After fixation with 4% paraformaldehyde, cells were permeabilized with 0.1% Triton X-100 and blocked with 2% bovine serum albumin. Mouse polyclonal antibody specific for PIV5 N protein at dilution of 1:200, or porcine PIV5 positive serum (this serum was collected from swine positive to PIV5 tested by RT-PCR and confirmed by WB) at 1:100 dilution was used as primary antibody and incubated at 37°C for 1 h. Then cells were incubated with fluorescein isothiocyanate (FITC) conjugated goat anti-mouse IgG (Thermo Fisher Scientific, 1:500) or rabbit anti-pig IgG (Sigma, catalogue #F1638, 1:50) for 1 h at 37°C. Cellular nuclei were stained with 10 μg/ml DAPI (Solarbio, China) for 10 min at 37°C, and cells were then visualized under an inverted fluorescence microscope equipped with a camera (Evos FL, USA).
| Cells | infection | |||||
|---|---|---|---|---|---|---|
| NO | Name | ATCC® Number | Tissue origin | Species | CPE | IFA |
| 1 | 293T | CRL-11268 | Embryonic kidney | Human | + | + |
| 2 | A549 | CCL-185EMT | Lung carcinoma | Human | + | + |
| 3 | Hela | CCL-2 | Cervix adenocarcinoma | Human | + | + |
| 4 | HepG2 | HB-8065 | Hepatocellular carcinoma | Human | + | + |
| 5 | Huh7 | N/A | Hepatocellular carcinoma | Human | + | + |
| 6 | Caco2 | HTB-37 | Colon adenocarcinoma | Human | + | + |
| 7 | PK15 | CCL-33 | Porcine Kidney | Swine | + | + |
| 8 | ST | CRL-1746 | Swine testicular | Swine | + | + |
| 9 | IPEC-J2 | N/A | Small intestinal epithelium | Swine | + | + |
| 10 | Marc145 | N/A | African green monkey kidney | Monkey | + | + |
| 11 | Vero E6 | CRL-1586 | African green monkey kidney | Monkey | + | + |
| 12 | RAW 264.7 | TIB-71 | Mouse monocyte /macrophage | Mouse | + | + |
| 13 | NIH-3T3 | CRL-1658 | Mouse embryonic fibroblasts | Mouse | + | + |
| 14 | MDBK | CCL-22 | Madin-darby bovine kidney | Bovine | + | + |
| 15 | MDCK | CCL-34 | Madin-darby canine kidney | Canine | + | + |
| 16 | BHK-21 | CCL-10 | Baby hamster kidney | Hamster | + | + |
| 17 | CHO | CCL-61 | Chinese hamster ovary | Hamster | + | + |
| 18 | CRFK | CCL-94 | Crandell Reese Feline Kidney | Feline | + | + |
| 19 | RK13 | CCL-37 | Rabbit kidney | Rabbit | + | + |
| 20 | DEF | CCL-141 | Duck embryo fibroblasts | Duck | – | – |
| 21 | DF-1 | CRL-12203 | Embryo fibroblasts | Chicken | – | – |
- N/A: not available; (+) infection; (–) no infection or obvious lesions.
2.10 Expression and purification of recombinant PIV5 N protein
The PIV5 N gene was amplified by RT-PCR, and the purified PCR product was inserted into prokaryotic expression vector pMAL-c5X. Recombinant N protein was expressed as maltose-binding protein (MBP)-tagged fusion protein in Escherichia coli ER2523. The fusion protein was expressed in the presence of 1 mM isopropyl-D-1-thiogalactoside (IPTG) and examined by SDS-PAGE. The recombinant N protein was purified using a pre-packed MBP Trap column (GE Healthcare Bioscience) by AKTA AVANT liquid chromatography system (Cytiva, USA), and its purity was evaluated by SDS-PAGE and Western blot. The purified fusion protein was used to immunize BALB/c mice for antibody production. Animal experiments were performed under the guidelines of the Animal Ethics Committee of the School of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (Approval Number: 210602-01).
2.11 Western blot analysis
Western blot analysis was conducted as previously described with a slight modification (Zhang et al., 2021). Lysates of PIV5-infected PK15 cells and recombinant N protein tagged to MBP were separated in 10% SDS-PAGE and transferred into a PVDF membrane. After blocking, the membrane was incubated with primary antibodies, either mouse polyclonal antibody against recombinant N protein (1:200), mouse anti-MBP mAb (1:10,000), or porcine serum (1:100). The IRDye 680 conjugated goat anti-mouse IgG (Li-Cor Biosciences, USA), or goat anti-pig IgG (Biodragon-immunotech, China) was used as secondary antibodies at 1:10,000 dilution. Lastly, the membrane was scanned using Odyssey infrared imaging system (Li-Cor Biosciences, USA).
2.12 Development of iELISA
The N protein-based indirect ELISA (iELISA) was established to detect anti-PIV5 antibodies as described previously with some modifications (Domańska-Blicharz et al., 2019). For iELISA optimization, IFA assay was used as a standard evaluating method for verifying truly positive and truly negative serum, and Western blot was used as a confirmatory assay. To optimize iELISA conditions, antigen concentration, serum dilution and secondary antibody dilution were determined by checkerboard titration. Briefly, 96 well plates were coated with purified recombinant N protein (0.1–0.5 mg/ml) at 4°C overnight. The plates were washed five times with PBST and blocked with 200 μl 5% skimmed milk for 2 h at 37°C. After washing five times, the positive and negative serum against PIV5 at dilution of 1:100 to 1:6400 was added at 50 μl/well and incubated for 2 h at 37°C. After washing, HRP conjugated goat anti-pig IgG (Sigma, catalogue #SAB3700434) was diluted 1:10,000 to 1:60,000, added at 50 μl/well and incubated at 37°C for 1 h. After washing, the substrate was added at 50 μl/well and incubated at room temperature for 15 min. Lastly, the reaction was stopped by adding 50 μl 2 M sulphuric acid per well. The results were read at 450 nm optical density (OD). The dilution with the highest OD 450 nm ratio between positive and negative serum (P/N value) was considered optimal. The cut-off value was determined as previously described (Zhang et al., 2020), by analysing 39 PIV5-seronegative samples, which were verified by IFA and confirmed by Western blot assay.
To validate the specificity of this developed iELISA, porcine positive sera against other pathogens such as porcine epidemic diarrhoea virus (PEDV), porcine circovirus type 2 (PCV2), classical swine fever virus (CSFV), African swine fever virus (ASFV) and Seneca Valley virus (SVV) were tested in triplicates and the mean OD 450 nm value was calculated based on the cut-off value. These sera were certified by Harbin Guosheng Biological Testing Technology Co., Ltd, Harbin, China. To evaluate the validity of the developed iELISA, 230 porcine sera were tested by the iELISA and IFA, respectively. The sensitivity was determined by the ratio of iELISA positive to IFA positive samples, whereas the specificity was determined by the ratio of iELISA negative to the IFA negative samples. Finally, the developed iELISA was used to test 530 sera randomly collected from swine of different age groups.
2.13 Statistical analysis
All statistical analysis was done using GraphPad Prism software (version 8.0.2) (GraphPad Software Inc. La Jolla, CA, USA). The experiments were carried out at least three times independently.
3 RESULTS
3.1 Isolation and identification of PIV5
PK15 cells were inoculated with diarrheic stool samples which were tested negative for other common enteric viruses such as TGEV, PEDV, PDCoV, RVA, RVC, PAstV, PSaV, PNoV and MRV by RT-PCR (data not shown). In the first two passages, no CPE was observed, but at passage three the cells started to show CPEs characterized by initial cell rounding, aggregation and detaching (Figure 1a–c). Again, RT-PCR of cell supernatants was negative for the enteric viruses mentioned above, suggesting that the CPEs were caused by other virus. After four rounds of plaque purification, the purified virus formed clear plaques with approximately 0.5–1 mm in diameter (Figure 1d). After ultracentrifugation, the plaque-purified virus was examined under TEM, and the enveloped paramyxovirus-like particles of approximately 80–200 nm in diameter were observed (Figure 1e). In addition, enveloped virus with burst envelope and typical herringbone appearance was also observed (Figure 1f). These results suggest that the isolated virus is associated with paramyxovirus. To further determine this unknown virus isolate, we utilized RT-PCR by using specific primers for paramyxoviruses. Interestingly, the RT-PCR results showed that the isolated virus was PIV5 (Figures 1g and Figure S1). The whole-genome sequencing confirmed the result (GenBank access No. OK505006).
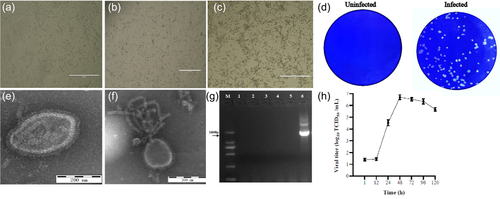
To identify whether the newly isolated PIV5 has haemagglutination activity, cell supernatants were subjected to haemagglutination assay, and the results revealed that the isolated virus was able to haemagglutinate red blood cells of chicken and guinea pig up to titre of 1:512 and 1:28, respectively. To determine the growth kinetics of the PIV5 isolate, PK15 cells were inoculated with the virus at MOI of 0.01 and the mean titres of three independent experiments at indicated time points were measured. The results showed that after a latent phase of approximately 12 h the virus reached a peak of 106.7 TCID50/ml at 48 h, suggesting that the cycle of PIV5 multiplication is completed within 48 h (Figure 1h).
3.2 Analysis of whole-genome sequence
To understand the molecular characteristics of the isolated PIV5, we performed whole-genome sequencing. The result revealed that isolated PIV5 had a genome size of 15,246 bp, containing seven genes in the order 3′-N-V/P-M-F-SH-HN-L-5′. Comparison of the whole-genome sequence of the isolated PIV5 to published reference strains showed that nucleotide and amino acid sequence similarity of this PIV5 to other strains were 98.4−99.9% and 96.7−99.8%, respectively. The PIV5 isolate shared the highest similarity to swine PIV5 strains SH/122 and SH/1202 identified in China, and the lowest similarity to human PIV5 strains from the United Kingdm. Whereas, the PIV5 isolate had lower nucleotide and amino acid sequence similarity to PIV1-4 36.3−61.1% and 19.8−40.5%. The pairwise comparison of N, F, HN and V proteins showed that the PIV5 isolate possessed 98.4−99.8%, 97.5−100%, 97.3−99.8% and 97.8−99.6% amino acid similarity to reference PIV5 strains from various hosts, and the PIV isolate was more closely related to swine strain SH/1202, pangolin strain GD18 and panda strain ZJQ-221, and overall nucleotide and amino acid similarity among PIV5 strains identified in China was 99.6–100% and 99.5–100%, respectively (Table 2).
| Nucleotide (nt) and amino acid (aa) sequence identity % | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WGS | NP | F | HN | ||||||||||
| No | Strain | Year | Host | Country | Accession no | nt | aa | nt | aa | nt | aa | nt | aa |
| 1 | SH/122 | 2015 | Swine | China | MT890698 | 99.9 | 99.8 | 99.9 | 99.8 | 99.9 | 99.8 | 99.8 | 99.8 |
| 2 | HLJ/DP2-1 | 2015 | Swine | China | MT890697 | 99.9 | 99.7 | 99.9 | 99.8 | 99.9 | 99.6 | 99.8 | 99.8 |
| 3 | SH/1202 | 2015 | Swine | China | MK028670 | 99.9 | 99.8 | 99.8 | 99.6 | 100 | 100 | 99.8 | 99.8 |
| 4 | HuB/YC | 2015 | Swine | China | MT890699 | 99.9 | 99.7 | 99.9 | 99.8 | 99.9 | 99.6 | 99.8 | 99.8 |
| 5 | JX/1221 | 2015 | Swine | China | MT890700 | 99.9 | 99.7 | 99.9 | 99.8 | 99.9 | 99.6 | 99.6 | 99.8 |
| 6 | HLJ/DP1-1 | 2015 | Swine | China | MT890696 | 99.9 | 99.7 | 99.9 | 99.8 | 99.9 | 99.6 | 99.8 | 99.8 |
| 7 | GD18 | 2017 | Pangolin | China | MG921602 | 99.8 | 99.7 | 99.9 | 99.8 | 100 | 100 | 99.6 | 99.5 |
| 8 | HMZ | 2018 | Tiger | China | MH370862 | 99.8 | 99.7 | 99.7 | 99.6 | 99.9 | 99.8 | 99.7 | 99.6 |
| 9 | ZJQ-221 | 2015 | Panda | China | KX100034 | 99.8 | 99.6 | 99.7 | 99.6 | 100 | 100 | 99.6 | 99.6 |
| 10 | 1168-1 | 2019 | Canine | Korea | KC237064 | 99.8 | 99.6 | 99.8 | 99.8 | 99.8 | 99.3 | 99.7 | 99.5 |
| 11 | CC-14 | 2000 | Canine | China | KP893891 | 99.2 | 98.4 | 99.4 | 99.4 | 99.2 | 98.6 | 99.2 | 98.6 |
| 12 | SER | 1998 | Swine | Germany | JQ743328 | 99.2 | 98.4 | 99.5 | 99.4 | 99.2 | 98.6 | 99.2 | 98.8 |
| 13 | HB01 | 2019 | Snake | China | MT124463 | 99.1 | 98.3 | 99.4 | 99.4 | 99.2 | 98.6 | 99.1 | 98.4 |
| 14 | BC14 | 2014 | Calf | China | KM067467 | 99.1 | 98.3 | 99.2 | 99.2 | 99.2 | 98.6 | 99.1 | 98.4 |
| 15 | W3A | 1964 | Macaque cells | USA | JQ743318 | 99.0 | 97.9 | 99.2 | 99.2 | 98.7 | 98.2 | 98.3 | 97.3 |
| 16 | KNU-11 | 2011 | Swine | Korea | KC852177 | 99.0 | 97.9 | 99.0 | 98.4 | 99.0 | 98.4 | 98.9 | 98.2 |
| 17 | H221 | 1998 | Dog | UK | JQ743323 | 98.9 | 97.7 | 98.6 | 99.0 | 98.2 | 98.4 | 98.8 | 98.8 |
| 18 | 78524 | 1980 | Dog | UK | JQ743319 | 98.9 | 97.6 | 98.5 | 99.0 | 98.2 | 98.4 | 98.8 | 98.8 |
| 19 | CPI+ | 1980 | Dog | USA | JQ743321 | 98.5 | 97.1 | 98.4 | 99.2 | 98.2 | 97.5 | 98.3 | 98.2 |
| 20 | CPI- | 1980 | Canine | USA | JQ743320 | 98.5 | 97.1 | 98.4 | 99.2 | 98.2 | 97.6 | 98.3 | 98.2 |
| 21 | DEN | 1980 | Human | UK | JQ743322 | 98.5 | 96.8 | 98.2 | 99.0 | 98.2 | 98.2 | 97.7 | 97.9 |
| 22 | MTL | 1980 | Human | UK | JQ743326 | 98.4 | 96.7 | 97.9 | 98.6 | 98.2 | 98.2 | 97.6 | 97.7 |
| 23 | MEL | 1980 | Human | UK | JQ743325 | 98.4 | 96.7 | 97.9 | 98.4 | 98.1 | 97.8 | 97.7 | 97.9 |
| 24 | RQ | 1976 | Human | UK | JQ743327 | 98.4 | 96.7 | 98.0 | 98.6 | 98.2 | 98.0 | 97.6 | 97.7 |
| 25 | LN | 1980 | Human | UK | JQ743324 | 98.4 | 96.7 | 98.0 | 98.6 | 98.2 | 98.0 | 97.6 | 97.7 |
| 26 | HPIV1/AUS/53 | 2007 | Human | Australia | KF530221 | 36.3 | 20.2 | – | – | – | – | – | – |
| 27 | S033N | 2009 | Swine | Hong Kong | JX857410 | 36.9 | 20.3 | – | – | – | – | – | – |
| 28 | HPIV-2 | 1991 | Human | Japan | NC003443 | 61.1 | 40.5 | – | – | – | – | – | – |
| 29 | HPIV3/AUS/3 | 2007 | Human | Australia | KF530243 | 36.3 | 19.8 | – | – | – | – | – | – |
| 30 | Texas-81 | 1981 | Swine | USA | EU439429 | 36.6 | 20.4 | – | – | – | – | – | – |
| 31 | HPIV4_DK (459) | 2000 | Human | Denmark | KF483663 | 48.3 | 28.7 | – | – | – | – | – | – |
3.3 Phylogenetic analysis
Phylogenetic tree constructed from nucleotide sequences of whole-genome showed that PIV5 strains identified in various hosts were grouped together and separated from PIV1-4 clusters, and PIV5 strains were further divided into three groups (I, II and III). Group I consisted of PIV5 strains identified in swine (SH/122, HLJ/DP2-1, SH/1202, JX/1221 and HLJ/DP1-1), pangolin (GD18), tiger (HMZ), panda (ZJQ-221) from China and dog (1168-1, H221, and 78524) from Korea and the United Kingdom. Group II was composed of PIV5 strains identified in dog (CPI+, CPI–) USA, bovine (BC-14), dog (CC-14), snake (HB01) from China, and swine (SER and KNU-11) from Germany and Korea. Group III mainly included PIV5 strains identified in human (DEN, LN, MIL, MEL and QR) from UK and simian (W3A) from USA. The PIV5 isolate was classified within group I and closely related with swine, pangolin, tiger and panda PIV5 strains identified in China, which may indicate the presence of various PIV5 genetic strains depending on geographic location (Figure 2a). Phylogenetic analysis based on the N, F and HN genes exhibited similar results of topological structure and branching of the evolutionary tree of PIV5, in which PIV5 strains were classified into two groups (A and B) and five clades (I, II, III, IV and V). Group A contains three clades I, II and III, and group B consists of clades IV and V. The isolated PIV5 was classified in group A, within clade I, and clustered with swine, pangolin, tiger and panda PIV5 strains identified in China and dog from Korea based on F, HN and N genes. Of interesting, swine strain from Germany was clustered within clade I only in case of N gene. Clade II consists of PIV5 strains identified in human (DEN, LN, MIL, MEL and QR) from the United Kingdom and simian (W3A) from the United States according to N, F and HN genes, in addition to PIV5 strains identified in dog (CPI+, CPI–) in the United States in case of HN gene (Figure 2b–d). While a distinct cluster of PIV5 from calf, dog, and snake from China as well as PIV5 from pig in South Korea were observed within clade III, implying that these viruses may originate from the common ancestor.
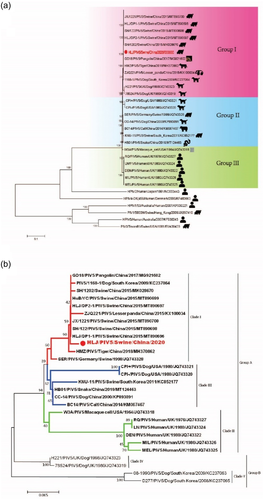
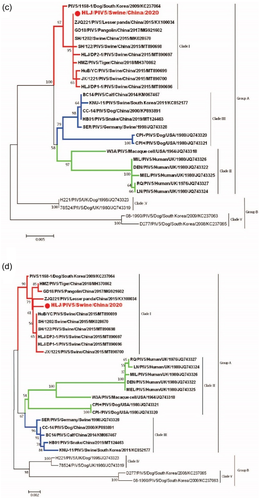
3.4 Cell susceptibility test to the PIV5 isolate
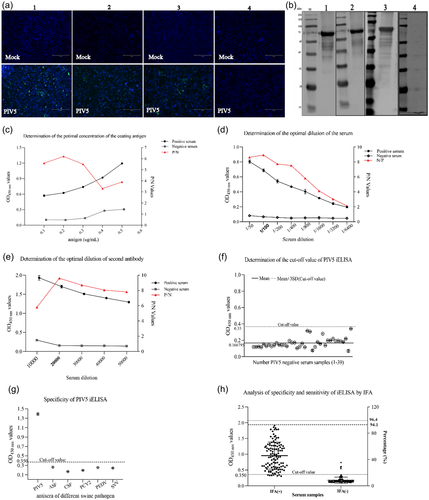
To examine the host tropism of the isolated PIV5 strain, 21 cell lines were tested. We found that 19 out of 21 cell lines, namely, the porcine (PK15, ST, IPEC-J2), human (293T, Huh7, HepG2, A549, Hela, CaCo2), primate (Marc145, Vero E6), bovine (MDBK), canine (MDCK), feline (CRFK), rabbit (RK13), hamster (BHK21, CHO) and mouse (NIH/3T3, RAW 264.7) cell lines showed CPEs at days 2–3 after infection, whereas the duck (DEF) and chicken (DF-1) cell lines did not show CPEs (Table 1). The evidence of productive infection of PIV5 in the susceptible cell lines was confirmed by the detection of viral N protein expression using IFA. As shown in Figure 3, the porcine, human, primate, bovine, canine, feline, rabbit, hamster and mouse cell lines had N protein expression, while the duck and chicken cell line did not, which was consistent with the CPEs results. These data indicate that this PIV5 strain isolated from diarrheic piglet may have the potential to infect mammalian hosts directly without adaptation.
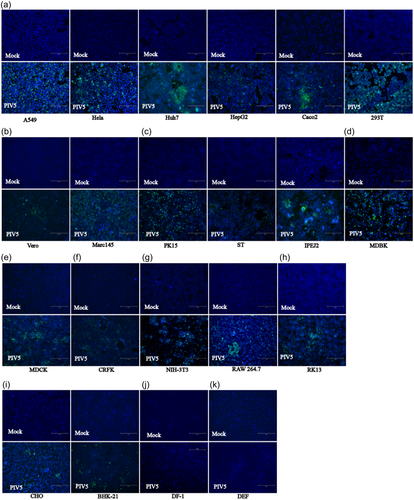
3.5 Establishment and optimization of iELISA
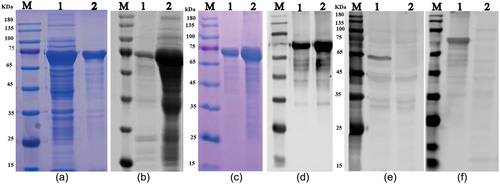
To further investigate porcine PIV5, we established a serologic assay. First, the PIV5 N gene was amplified, cloned into vector pMAL-c5X and expressed as MBP-tagged fusion protein in Escherichia coli ER2523, as shown in SDS-PAGE result (Figure 4a). The recombinant N protein at ∼75 KDa was confirmed by Western blot using MBP specific antibody (Figure 4b). The recombinant N protein was purified by AKTA liquid affinity chromatography system using MBP trap column. The purified N protein was evaluated by SDS-PAGE and Western blot, and the result showed that the recombinant N protein was efficiently purified (Figure 4c and d). The purified N protein was used to immunize mice, and the specificity of mouse anti-N protein polyclonal antibody was tested by Western blot. The result showed that the resulting antibody recognized one specific band at about 65 kDa in PIV5-infected cells and 75 KDa in the purified recombinant protein (Figure 4e and f), indicating that the mouse polyclonal antibody can specifically identify N protein of porcine PIV5.
Second, an iELISA was developed based on the purified recombinant N protein. Typically, the purified N protein was diluted in buffer and used to coat 96-well polystyrene plates overnight at 4°C. Plates were blocked with skimmed milk, and sera from PIV5 positive and negative pigs were serially diluted and placed into the wells of pre-coated plates. The presence of IgG reacting with the PIV5 N protein was revealed by incubating with goat anti-porcine IgG-HRP followed by chromogenic substrate TMB. IFA was used as a standard evaluating method, whereas Western blot was used as confirmatory method for verifying seropositive and seronegative samples for iELISA optimization (Figure 5a and b).
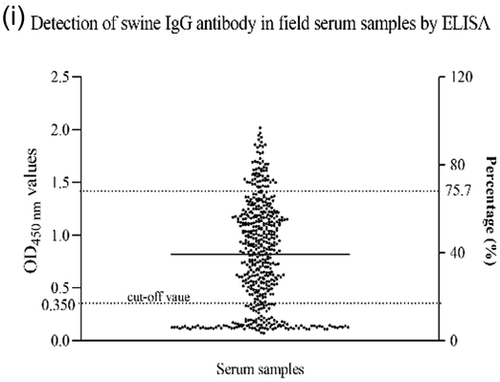
Third, the checkerboard titration was performed to determine the optimal concentration of the antigen and the positive or negative serum. The results showed that the optimal antigen coating concentration was 0.2 μg/ml, and the optical dilution for positive or negative serum was 1:100 (Figure 5c and d). Using this optimal dilution of coating protein and primary antibody, the optimal dilution of the HRP-conjugated anti-porcine IgG was 1:20,000, as shown in Figure 5e.
Fourth, 39 negative serum samples were used to determine the detection threshold of the assay. The cut-off point was based on the OD 450 nm of the 39 negative samples. Based on the mean OD 450 nm plus 3 × SD (0.166795 + 3 × 0.061079; n = 39; Max = 0.341; Min = 0.069), the cut-off value was determined to be 0.350 (Figure 5f). The negative–positive threshold was consequently set at 0.350, and serum samples with an absorbance >0.350 at OD 450 nm was considered PIV5 seropositive, and vice versa.
3.6 Evaluation of iELISA specificity and sensitivity
To determine the specificity of the developed iELISA, porcine positive sera against other viruses including PEDV, PCV2, CSFV, ASFV and SVV were examined. The results showed that there was no cross-reactivity between the N protein of PIV5 and antibodies against other porcine viruses, as shown in Figure 5g, and the average OD 450 nm of positive serum for PEDV, PCV2, CSFV, ASFV and SVV was 0.261, 0.168, 0.193, 0.254 and 0.240, respectively.
Furthermore, a total of 230 porcine sera collected from the field were tested in parallel using the iELISA developed in this study and IFA test. The results showed that positive and negative serum numbered 116 and 114 by IFA, but 119 and 111 by iELISA. In total, 112 samples were positive and 107 negatives in both cases, which represents a consensus for 95.22%. Seven samples that were positive in the iELISA test were negative in the IFA test, and four samples that were negative in the iELISA test were positive in the IFA. Hence, the sensitivity of iELISA was 94.12% among PIV5-seropositive samples, and the specificity was 96.4% among PIV5-seronegative samples using IFA as standard evaluation method (Table 3 and Figure 5h). These data suggest that the iELISA developed here can be used for the detection of antibodies in large numbers of samples.
| IFA results | ||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| iELISA results | Positive | 112 | 7 | 119 |
| Negative | 4 | 107 | 111 | |
| Total | 116 | 114 | 230 | |
| Data analysis | Sensitivity | 94.12% | ||
| Specificity | 96.4% | |||
| Coincidence rate | 95.22% | |||
3.7 PIV5 detection of field samples
The developed iELISA was applied to investigate the prevalence of PIV5 in the field, and a total of 530 clinical porcine sera were randomly collected from pig farms. As shown in Figure 5i, the positive rate was 75.7% (401/530), indicating that PIV5 infection is common among swine in China. Taken together, the prevalence of PIV5 among pigs may have pandemic potential.
4 DISCUSSION
The first report of porcine PIV5 is from Germany (Heinen et al., 1998); however, the prevalence of porcine PIV5 and its impact on swine health are largely unknown, especially the information on PIV5 infection in the porcine digestive system is scarce. Recently, Jiang et al. have identified PIV5 from diarrheic piglet samples positive for PEDV (Jiang et al., 2018), but it is not clear whether porcine PIV5 can cause enteric symptoms alone. Previous reports have demonstrated that co-infection with a diverse set of pathogens and environmental factors contributes to the severity and virulence of PIV5 infections in swine (Heinen et al., 1998; Lee et al., 2013). In this study, we have isolated a porcine PIV5 strain from a diarrheic piglet, which is confirmed by TEM, gene sequencing, and IFA. Our results suggest that the newly isolated porcine PIV5 is strongly associated with diarrhoea in piglet, implying the newly isolated PIV5 may be enteropathogenic, however, further research is required to confirm the pathogenicity.
Currently, the whole genome sequences of porcine PIV5 in GenBank database are limited, and the complete genome sequence of PIV5 isolated herein provides additional information for understanding the genetic diversity and pathobiology of PIV5 in general. Alignments of whole-genome sequences showed that the isolated PIV5 shared relatively high nucleotide and amino acid similarity with PIV5 strains from swine, pangolin, tiger, and panda identified in China. These data indicate that PIV5 strains identified from different hosts in China exhibit a close relationship and may be evolved from the same ancestor.
The phylogenetic analysis based on the whole genome sequence revealed that PIV5 strains were divided into three groups, and the PIV5 strain isolated in this study had higher similarity to PIV5 strains identified from pangolin and swine in China. Interestingly, PIV5 strains identified in swine intestine in China were grouped together and genetically different from the other PIV5 strains identified in swine lung in Germany and South Korea (Heinen et al., 1998; Lee & Lee, 2013). These indicate that PIV5 strains identified from the same host may be genetically different based on geographical location and tissue tropism. The phylogenetic analysis based on the N, F, and HN genes of PIV5 strains revealed that PIV5 strains divided into two groups and five clades, which is similar to the previous report from China (Jiang et al., 2018), Moreover, the PIV5 strain isolated in this study formed a clade with strains identified from China, Korea and Germany, suggesting a potential common ancestor of these viruses. Even though PIV5 strains were divided into different groups, the variation in nucleotide and amino acid among PIV5 strains from different hosts was relatively low. Consistent with our data, previous studies have reported that PIV5 strains from different hosts identified in different countries exhibit relatively high similarity (Chatziandreou et al., 2004; Rima et al., 2014).
Previous reports showed that PIV5 strains isolated from human could infect a wide range of mammalian cells (Arimilli et al., 2006; LAMB, 2001). In addition, Chen et al. (2012) reported that dog PIV5 neutralizing antibodies were detected in human serum samples and suggested that the tested individuals might have a previous exposure to PIV5 positive dogs. However, there is only one report related to the tropism of porcine PIV5, which showed that PIV5 isolated from pig lung could only infect cell lines of porcine origin (Lee et al., 2013). Interestingly, our results have demonstrated that a porcine PIV5 strain isolated from faeces could infect to a wide range of mammalian cell lines including swine, human, primate, bovine, canine, feline, rabbit, hamster, and mouse. Taken together, the ability of a porcine PIV5 strain isolated from faeces to infect and replicate in different mammalian cell lines might indicates interspecies transmission and zoonotic potential of this virus.
Serum neutralization test (NT) is one of serological assay for PIV5 test, which has been used for the detection of PIV5 specific antibodies in swine (Lee et al., 2013). However, NT is laborious and time-consuming. In contrast, ELISA test is simple, rapid, and can process multiple samples in parallel, which is the most commonly used classical serological test. The N protein possesses highly conserved amino acid regions among paramyxoviruses (Morgan, 1991; Lamb & Parks, 2013). Furthermore, the N protein is the most abundant and a highly immunogenic viral structural protein (Alayyoubi et al., 2015; Dortmans et al., 2011; Pearce et al., 2015). Furthermore, the N protein of isolated PIV5 contains highly homologous sequences with reference PIV5 strains from other species, sharing 98.4−99.8% amino acid identity (Table 2). Similarly, Rima et al. (2014) and Chatziandreou et al. (2004) reported more than 97% nucleotide similarity among PIV5 strains isolated from diverse hosts. Chen et al. (2012) and Johnson et al. (2018) also discovered antigenic cross-reactivity across PIV5 strains from various hosts. On the other hand, PIV5 showed no cross-reactivity with viruses that do not belong to the genus Orthorubulavirus (Johnson et al., 2018). As a result, we believe that the PIV5 N protein-based iELISA may not have cross-reactivity with sera from pigs infected with viruses other than PIV5.
Therefore, we selected PIV5 N protein as the coating protein to establish an ELISA test, and the detection conditions were optimized. IFA is another common serological method and widely used to detect viruses. Here, IFA assay was used as standard method to confirm the serum samples for the development of iELISA test. Our data have revealed that the developed iELISA has a sensitivity of 94.12% and a specificity of 96.4%, and the overall agreement between the iELISA and IFA is 95.22%. Moreover, this iELISA test has no cross-reactivity with antibodies against other porcine viruses. By using the iELISA developed here, we have investigated the distribution of PIV5 in swine and observed seropositivity of 75.7%, indicating a higher prevalence of PIV5 in Chinese swine farms. In contrast, a PIV5 seropositivity of 93.8 % was reported in swine farms in Korea (Lee et al., 2013). These findings indicate that PIV5 is widely epidemic not only in China but also in other pig-producing countries. Altogether, these results have showed that the iELISA established herein is specific and sensitive.
5 CONCLUSION
In summary, this report has described the isolation and characterization of PIV5 strain from a diarrheic piglet. The ability of this porcine PIV5 isolate to infect diverse types of cell lines from different species signals its zoonotic potential. The virus isolated here can be used as a reference strain to study the pathobiology of PIV5 in digestive system in swine. In addition, for the first time, viral N protein-based iELISA has been developed to effectively detect PIV5-specific antibodies from blood samples. This iELISA can be used to study the epidemiology of PIV5 in swine.
ACKNOWLEDGEMENTS
This research was supported by the National Key R&D Program of China (2021YFF0703100) and Key Research & Development Program of Heilongjiang province (GZ20210010).
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received. The US National Research Council's guidelines for the Care and Use of Laboratory Animals were followed.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.




