Rabies surveillance in Senegal 2001 to 2015 uncovers first infection of a honey-badger
Abstract
Despite the establishment of Rabies surveillance in animals and humans since 2008, there is a lack of data on its circulation, dynamic of transmission and real burden in Senegal. To better understand the molecular epidemiology of rabies virus in Senegal, we investigated the genetic diversity of 18 new characterized Senegalese rabies virus sequences collected over 14 years, including a honey-badger-related isolate. Phylogeographic analyses demonstrated that the Senegalese isolates belong to a monophyletic cluster into the Africa-2 clade and supported two RABV introductions in Senegal from West-African neighbour countries, 36–40 years ago. Our study is noteworthy for reporting on the first characterization of an African honey-badger-related rabies virus that did not have the N-glycosylation site NKT at position 338-G of the glycoprotein. The identified amino acid polymorphisms found in the Senegalese rabies virus sequences are worthy of further investigation. Although a strong multidisciplinary stepwise cooperation is important for the successful elimination of Rabies in dog populations in Senegal by 2030, the establishment of surveillance in wildlife could be necessary to avoid future re-introductions into domestic hosts.
1 INTRODUCTION
Rabies is a neglected, acute as well as progressive neurological disease that is invariantly fatal, once symptoms onset, without prompt post-exposure prophylaxis (PEP) (World Health Organization, 2013a). Rabies is one of the most important zoonotic diseases and occurs in over 150 countries and territories around the world, causing between 50,000 and 70,000 human deaths annually (Hampson et al., 2015). Most human rabies deaths are associated with bites or scratches from an infected dog, mainly in developing countries in Africa and Asia, where Rabies remains a major public health concern (World Health Organization, 2013a, 2013b).
Rabies is caused by the rabies virus (RABV), which is a neurotropic and enveloped virus belonging to the Lyssavirus genus (order Mononegavirales, family Rhabdoviridae). In countries where RABV is endemic, enzootic transmission is maintained in the orders of wild Carnivora and Chiroptera, which is peculiar in the Americas (Rupprecht et al., 2002). The RABV's genome is a negative single-stranded RNA of approximately 12 kb in size, which encodes for five structural proteins (N, P, M, G, and L). The nucleoprotein (N), the phosphoprotein (P), and the RNA-dependent RNA polymerase form the ribonucleoprotein (RNP), which plays a crucial role in RABV transcription and replication (Riedel et al., 2020). The N and L proteins harbour the most conserved regions among the RABV genome (Riedel et al., 2020). Known as the key protein involved in virus assembly, budding and virions release, the matrix protein (M) is also a regulator of viral transcription and replication (Luo et al., 2019). The glycoprotein (G) is the only surface protein and plays a major role during the initial steps of the infectious cycle. It is also essential for induction of host's innate and adaptive immunity (Li et al., 2012; Zhang et al., 2013).
The considerable advances in understanding RABV biology (Rupprecht et al., 2002), pre-and post-exposure treatment on exposed people (Li et al., 2012; Zhang et al., 2013), and massive immunization campaigns in dog populations conducted to the success of elimination and control programs in Europe (Rupprecht et al., 2008) and the Americas (Freire de Carvalho et al., 2018). Nevertheless, the objective of no Rabies death by 2030 is difficult to achieve in developing countries despite some RABV-targeted control efforts (World Health Organization, 2013a). Although the elimination of dog-mediated Rabies is an achievable objective in developing countries through a strong One health cooperation, the elimination of Rabies in the wildlife seems more difficult considering that rabies viruses continue to be discovered in new hosts such as various species belonging to the genera Melogale, Meles, and Mellivora in the weasel family Mustelidae (Barnard, 1979).
Sequenced RABV genomes demonstrate expansive intra-species genetic diversification with seven major phylogenetic groups circulating throughout the world (Troupin et al., 2016). Three of these groups are circulating in Africa including the Africa-2, Africa-3, and Cosmopolitan clades (Davis et al., 2007; Talbi et al., 2009; Troupin et al., 2016). Previous phylogenetic studies based on the N gene sequences have shown that Africa-2 clade circulates in Central and West African countries including in Senegal (Talbi et al., 2009). Prior to this work, only two complete viral genomes from Senegal were available on GenBank (KX148238-39) and were generated from strains isolated in 1991 and 1992, respectively (Troupin et al., 2016).
Here, we investigated the genomic and phylodynamic characteristics of the RABV circulating in Senegal and West Africa, using complete genomes from 18 isolates collected over a 14-year period to gain a better understanding of virus spread patterns and timing. We also analyzed the disease burden and addressed public health considerations of Rabies within Senegal over the past two decades.
2 MATERIALS AND METHODS
2.1 Data collection
The Rabies Treatment Center at the Institut Pasteur de Dakar (IPD) provided us with data on pre-exposure and post-exposure prophylaxis in Senegal while data on human Rabies and animal Rabies were retrieved from annual reports of the Senegalese Ministry of Health and Social Actions (MoHSAS) and the Office of veterinary services, Senegalese ministry of livestock and animal production, respectively.
2.2 Sample collection
In Senegal, intra-vitam and post-mortem clinical samples are collected by the clinicians while the original brain tissues from animals are provided by the veterinarians. Clinical specimens from suspected patients and brain tissues from animals observed in private veterinary clinics are sent to the national reference centre for RABV at the Institut Pasteur de Dakar, Senegal (NRC-Rabies IPD) and the brain tissues from animals observed in private veterinary clinics are transported to the National laboratory of livestock and veterinary research (LNERV) located at the Senegalese Institute of Agricultural Research (ISRA), according to the WHO guidelines (OIE, 2021). At IPD, clinical specimens and brain tissues from animals are screened using direct fluorescent antibody test (DFAT), real-time reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR), and virus isolation in mice. Positive samples are sent for sequencing. At ISRA, the specimens from animals are tested DFAT and end-point RT-PCR (OIE, 2021).
All 18 virus isolates analyzed in this study were derived from experimentally infected suckling mice brain tissues preserved in the collection of the NRC-Rabies IPD (Table 1). One of these strains was isolated in 2011 from a honey-badger or ratel (Mellivora capensis) at Dahra and this is the first time in Senegal. Dahra is a city of up to 30,000 people, located in the Linguère department, Louga region in Senegal (15°21′N; 15°36′W), at approximately 264 km from the capital Dakar. The main activities in Dahra are agriculture and animal breeding.
| Isolate ID | Collection year | Origin | Host | GenBank accession number |
|---|---|---|---|---|
| SH155966 | 2001 | Dakar | Homo sapiens | MH514968 |
| SH177846 | 2005 | Dakar | Homo sapiens | MH514969 |
| SA217694 | 2011 | Linguère | Canis lupus familiaris | MH514970 |
| SA206776 | 2010 | Dakar | Canis lupus familiaris | MH514971 |
| SA194858 | 2008 | Dakar | Canis lupus familiaris | MH514972 |
| SA212203 | 2011 | Dahra | Mellivora capensis | MH514973 |
| SA217750 | 2011 | Dahra | Canis lupus familiaris | MH514974 |
| SA252913 | 2013 | Diamniadio | Canis lupus familiaris | MH514975 |
| SA204014 | 2010 | Dakar | Canis lupus familiaris | MH514976 |
| SA262037 | 2013 | Dakar | Canis lupus familiaris | MH514977 |
| SA262503 | 2014 | Dakar | Canis lupus familiaris | MH514978 |
| SA262518 | 2014 | Dakar | Canis lupus familiaris | MH514979 |
| SA252888 | 2013 | Dakar | Canis lupus familiaris | MH514980 |
| SA267115 | 2014 | Dakar | Canis lupus familiaris | MH514981 |
| SH189343 | 2007 | Dakar | Homo sapiens | MH514982 |
| SA217695 | 2011 | Dahra | Canis lupus familiaris | MH514983 |
| SH218152 | 2011 | Dakar | Homo sapiens | MH514984 |
| SA272282 | 2015 | Dakar | Canis lupus familiaris | MH514985 |
RABV infection was confirmed by RT-qPCR and DFAT as previously described (OIE 2021; World Health Organization, 2013b). Extraction of viral RNA from 140 μl of supernatants from suckling mice brain tissues was performed with the QIAamp viral RNA mini kit (Qiagen, Heiden, Germany) according to manufacturer's instructions and eluted in a final volume of 60 μL. Extracted RNA was stored at –80°C prior to downstream applications.
2.3 Complete genome sequencing
Reverse-transcription was performed using the Avian Myeloblastosis Virus (AMV) kit (Promega, Madison, WI, USA) with random primers, following the manufacturer's instructions. The complete polyprotein was assembled using overlapping polymerase chain reactions (PCR) with each primer set and the GoTaq® DNA polymerase kit (Promega, Madison, WI, USA) in a final volume of 50 μl, according to manufacturer's instructions. Primers details are indicated in Table A1. Briefly, 5 μl (around 10 μg) of cDNA was added to 45 μl of an RT-PCR mix and PCR was carried out as previously described (Faye et al., 2018). Subsequently, 5 μl of each PCR product was analyzed by gel electrophoresis on 1% agarose gels stained with ethidium bromide using a DNA molecular weight marker (HyperLadder™ 1 kb; Bioline, Taunton, MA, USA) to check the size of amplified fragments. The DNA amplicons were purified (QIAquick Gel Extraction Kit, Qiagen, Heiden, Germany) and sequenced from both ends for each positive sample using Sanger (Beckmann Coulter, High Wycombe, UK). Sequencing of the 3′ leader and 5′ trailer of the viral genome was performed using a 5′ RACE kit (Invitrogen, Carlsbad, CA, USA) and a 3′ RACE kit (Roche, Basel, Switzerland) following the manufacturer's protocols.
2.4 Sequence analysis
Overlapping genomic sequences were assembled using the Unipro UGENE software (http://ugene.net/download.html) (Okonechnikov et al., 2012) and obtained full genomes were aligned using Muscle algorithm (http://www.drive5.com/muscle/) (Edgar, 2004) within Unipro UGENE software. Based on these alignments, the genetic properties such as pairwise nucleotide and amino acid distances at gene level were analyzed among the newly characterized Senegalese sequences and between them and the previously available complete Africa-2 clade genomes from Nigeria (GenBank accession: KC196743) and Central African Republic (GenBank accession: KF977826).
In addition, amino acid substitutions were also assessed on the most divergent sequences from the previously available complete RABV genomes from Senegal (KX148238-39) and compared to specific mutations previously described for mongoose-related RABV (Africa-3 clade) and ferret-badger-related RABV (SEA5 sub-clade and the SEA2b lineage) (Troupin et al., 2016).
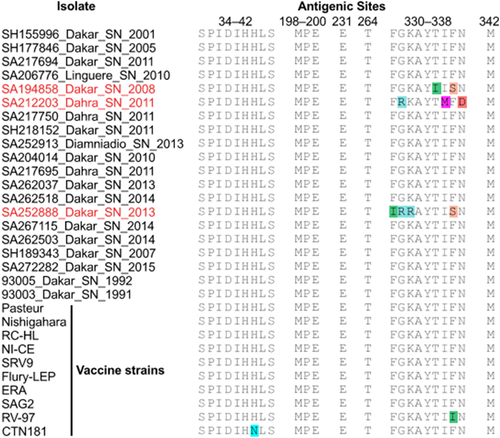
As it represents the major contributor to RABV pathogenicity (Faber et al., 2005), the glycoprotein (G) sequences from the new characterized isolates were screened for conservation of amino acid residues of virulence previously described (Dietzschold et al., 1983; Marston et al., 2007; Tomar et al., 2017). A motive was considered as non-conserved when it presents more than two non-conservative mutations in more than two RABV isolates. In addition, polymorphisms on antigenic sites of 10 vaccine strain G protein sequences were assessed on the newly characterized Senegalese rabies virus sequences.
2.5 Recombination and positive selection detection
The presence of recombination events was assessed in 37 RABV complete genome sequences from Western and Central Africa using seven methods (RDP, GENECONV, MaxChi, BootScan, Chimaera, SiScan, and 3Seq) implemented in the Recombination Detection Program (RDP4.97) (http://web.cbio.uct.ac.za/~darren/rdp.html) (Martin et al., 2015) and the Genetic Algorithms for Recombination Detection (GARD) method implemented in Datamonkey web server (http://datamonkey.org) (Pond et al., 2005). The settings were kept at their default values. In addition, episodes of positive diversifying selection were analyzed applying four different methods (SLAC, MEME, FUBAR, aBSREL) implemented in the HyPhy package from Datamonkey web server (Pond et al., 2005). An episode of positive selection was considered if it was detected by at least two different methods.
2.6 Molecular evolution and phylodynamics
Additional RABV sequences were downloaded from GenBank; long stretches of ambiguous bases or sequences derived from experimental infections or constructs were removed. Our initial dataset included 19 sequences from Western and Central Africa (Table A2). Alignments were done in Geneious, version 11.1.4, using MAFFT, version 7.388. For each RABV alignment, maximum-likelihood phylogenies were generated using PhyML version 3.3 with the GTR+ Γ model with 1000 bootstrap replicates. The maximum-likelihood phylogenies were used for estimates of root-to-tip distances, regression slopes, and correlations using TempEst with the best-fitting root option (Rambaut et al., 2016). Tip divergence data were exported and mapped with linear regression (95% prediction interval) in Rstudio, version 3.2.3, with the R Stats and ggplot2 packages (Wickham, 2016).
Bayesian analysis using BEAST (versions 1.10.0) was used to generate maximum-clade credibility (MCC) trees. Phylogenetic relationships and evolutionary rates were modelled according to the Hasegawa-Kishino-Yano 85 (HKY85) model with gamma-distributed rate variation and invariant rate distribution among sites (HKY85 + Γ4 + I) (Hasegawa et al., 1985). Analyses shared a strict molecular clock, a nonparametric coalescent “SkyGrid” model (number of grid points + 1 = 50) as a prior density across trees, and an approximate continuous time Markov chain rate reference prior (Gill et al., 2013; Ferreira & Suchard, 2008). Tracer v.1.6 was used to ensure run convergence (effective sample size > 200) (Rambaut et al., 2018). Each chain consisted of up to 1.0 × 108 Markov chain Monte Carlo steps (1.0 × 107 were discarded as burn-in). Parameters and trees were sampled every 100,000 generations. TreeAnnotator1.8.4 was used to calculate an MCC tree using a posterior probability limit of 0.7, which removes summary statistics from nodes with low support (Rambaut & Drummond, 2010).
To infer the historical movement of RABV across Western Africa, we implemented an asymmetric continuous-time Markov chain (CTMC) approach using country as a discrete trait. Bayesian stochastic search variable selection (BSSVS) and SpreadD3 (https://rega.kuleuvwn.be/cev/ecv/software/SpreaD3_tutorial) were used to identify strongly supported migrations (i.e., any with a Bayes factor ≥3) (Table A3).
3 RESULTS
3.1 Rabies burden and dynamics in Senegal
From 2009 to 2017, a total of 7359 human exposures to Rabies have been reported in Senegal with an average number of 17 exposures/week while steadily increasing annually. In 2017, the highest Rabies exposition rates on 10,000 people were found in the Senegalese administrative regions of Fatick, Kédougou, and Ziguinchor. However, only 43 human Rabies cases have been reported during this time period (unpublished data from the MoHSAS, 2018).
To date, there are only three rabies vaccination and treatment centres in Senegal including two in Dakar and one in the Fatick administrative region. From 2013–2019, an average of 1180 individuals consulted the RCT annually for rabies vaccination of which, around 90% received post-exposure prophylaxis (PEP) treatment. An overall rate of 17% (n = 220) received pre-exposure prophylaxis (PreP) and they are usually professionals. In 2020, this number decreased drastically with only 658 consultations and 591 PEP.
In 2017, the annual vaccine dose consumption at the IPD's Rabies vaccination and treatment centre represented in average 325 vaccine doses and 43 rabies immunoglobulin doses per month (unpublished data from the Rabies Treatment Center at IPD, 2018).
From 2000 to 2011, over 46 people have been exposed and died from Rabies in Senegal and 90% of rabies cases were associated with roaming dog bites and scratches. An overall mortality rate of 43.4% among these confirmed Rabies cases was documented in children and adolescents under 18 years of age (male–female sex ratio of 2.5) and 81% of them included agrarian people and students (unpublished data from the MoHSAS, 2018).
In Senegal, 90% of the human spillover of Rabies involves the dog population which represents between 72% and 75% of the annually reported rabies cases in animals. From 2014 to 2018, only 29 confirmed rabies cases in dogs have been reported with an average of six cases per year and the most affected Senegalese administrative regions were Ziguinchor with an overall rate of 31% (n = 9) and Kaolack with a rate of 21% (n = 6) from 2014 to 2018 (unpublished data from the Office of veterinary services, Senegalese ministry of livestock and animal production, 2018). A total of 43 and 69 rabies cases in animals were notified in 2019 and 2020, respectively. These confirmed cases have been mainly reported from dogs, but some cases from goats, donkeys and horses and notified by nine out of the 14 Senegalese administrative regions. As of September 30, 2019, a total of 11,534 domestic dogs were vaccinated while 27,869 stray dogs were euthanized (unpublished data from the Office of veterinary services, Senegalese ministry of livestock and animal production, 2020).
3.2 Genomic characterization
From 2001 to 2015, 43 RABV suspected samples were sent to the Institut Pasteur de Dakar for confirmation. An overall rate of 58.13% (n = 25) of them was found positive for RABV and submitted to further genomic characterization. A total of 18 RABV complete genome sequences were generated from four humans, 13 canines, and one honey-badger (Mellivora capensis) isolates (Table 1). As the previously available complete sequences from Senegal (Troupin et al., 2016), the newly characterized RABV isolates showed a genome length of 11,923 nt with similar size at protein and non-coding region levels.
3.3 Genetic distances
Pairwise nucleotide and amino acid distances of coding sequences were evaluated between isolates characterized in this study and in comparison, with previously available Senegalese RABV sequences and non-Senegalese sequences. Nucleotide percent (%) dissimilarity of RABV sequences from Senegal comparatively ranged from 0.2% to 1.7% while a nucleotide distance of 4% was found between the new characterized sequences from Senegal and previously described isolates from Nigeria (DRV-NG11, GenBank accession: KC196743) and Central African Republic (CAR_11_001h, GenBank accession: KF977826) belonging to the Africa-2 clade (Tricou et al., 2014; Zhou et al., 2013). However, the assessment of amino acid distances at protein level between the Senegalese RABV sequences showed a dissimilarity ranging between 0% and 2% for the N, P, M, and L proteins while the G protein exhibited more variability with an amino acid distance ranging from 0% to 7%. The highest amino acid distances in the G protein were observed between the isolate SA267115 (Dakar, SN 2014) from dog and the isolates SA212203 (Dahra, SN 2011) from honey-badger (7%) and SA252888 (Dakar, SN 2013) isolated from dog (7%). These three isolates presented also the highest pairwise amino acid distances from the other Senegalese sequences with ranges between 3%–4%, 3%–6%, and 5%–6% for SA267115, SA212203, and SA252888, respectively. The amino acid distances between the G protein sequences of the other Senegalese RABV isolates analyzed in this study ranged from 1% to 2%.
3.4 Amino acid substitutions on proteins of the most divergent Senegalese RABV sequences
The most divergent Senegalese RABV sequences from two dog-related RABV isolates (SA267115 and SA252888) and the honey-badger isolate (SA212203) were screened for mutations at genes level in comparison to the other Senegalese RABV sequences. No mutation was found in sequences of these three RABV isolates for the N, P, and M proteins. However, a total of three and nine substitutions were identified for the dog-related isolate SA267115, in the L protein and the G protein, respectively. The second dog-related RABV isolate SA252888 exhibited 100% homology in the polymerase L, while a total of eight amino acid substitutions were found in its G protein sequence. The RABV sequence from honey-badger showed eight and seven amino acid mutations for the polymerase L and the G protein, respectively. Previously described specific mongoose-related and ferret-badger-related RABV substitutions (Troupin et al., 2016) were assessed among the mutations identified in the glycoprotein and the polymerase of these three Senegalese isolates. The dog-related RABV isolate SA252888 have shown a mutation (Pro-386G-Leu) quietly similar to the Pro-386G-Ser mutation previously described as specific to mongoose-related RABV (Africa-3 clade) (Table 2).
| Protein and codon position | Honey-bagder-related RABVSA 212203_Dahra_SN_2011 | Dog-related RABVSA 252888_Dakar_SN_2013 | Dog-related RABVSA 267115_Dakar_SN_2014 |
|---|---|---|---|
| Glycoprotein | |||
| 25 | Thr = > Lys | ||
| 118 | Asn = > Asp | ||
| 146 | Thr = > Ile | ||
| 150 | Ser = > Cys | ||
| 197 | Thr = > Cys | ||
| 233 | Gly = > Ser | ||
| 248 | Gly = > Ala | ||
| 317 | Phe = > Ser | ||
| 321 | Ser = > Asp | ||
| 338 | Asn = > Asp | ||
| 339 | Lys = > Glu | ||
| 347 | His = > Trp | ||
| 352 | Arg = > Gln | ||
| 356 | Glu = > Ala | ||
| 362 | Gly = > Ala | ||
| 367 | Gly = > Asp | ||
| 368 | Gly = > Arg | ||
| 370 | Cys = > Arg | ||
| 373 | His = > Tyr | ||
| 386 | Pro = > Leua | ||
| 395 | |||
| 415 | Met = > Gly | Met = > Thr | |
| 417 | Pro = > Ser | ||
| 496 | Thr = > Ala | ||
| Polymerase | |||
| 622 | Trp = > Arg | ||
| 625 | His = > Gln | ||
| 1220 | Leu = > Pro | ||
| 1380 | Gly = > Trp | ||
| 1391 | Leu = > Ser | ||
| 1423 | Arg = > Pro | ||
| 1427 | Ser = > Leu | ||
| 1444 | Asp = > His | ||
| 1600 | Ser = > Arg | ||
| 1633 | Ser = > Leu |
- Note: Mutations were assessed with reference to the previously available complete genome sequences from Senegal (KX148238-39).
- a Substitution of the amino acid mutation Pro-386G-Ser previously identified as specific to mongoose-related RABV (Africa-3 clade) by 44).
3.5 Amino acid polymorphisms in the G protein
The presence of 13 highly conserved amino acid residues of virulence was assessed across the G protein of the new Senegalese RABV sequences using multiple alignments. The identified conservative mutations were highlighted in black while the non-conservative mutations were coloured in black and underlined (Table 3). A total of 10 amino acid residues of virulence among 13 were conserved in the G protein of Senegalese RABV isolates with sometimes non-conservatives amino acid changes. However, non-conservative mutations Arg333Ala and Asn194Tyr were identified in all Senegalese RABV sequences. Highly conserved in the Lyssavirus genus, the N-glycosylation site N319 was identified at amino acid position 338 in G protein of the Senegalese RABV isolates and the previously described motif of glycosylation NKT (Wickham, 2016) showed polymorphisms in some of the new characterized Senegalese sequences. Indeed, the isolate SA194858 (Dakar, SN 2008) from the dog exhibited a mutation Phe340Ala at the glycosylation site (NKA) while the isolate SA252888 from the dog had two non-conservative mutations (Lys339Glu and Phe340Ala) at the same site (NEA). In addition, the isolate SA212203 from honey-badger did not present this N-glycosylation site and exhibited an amino acid mutation Asn338Asp (DKT) (Table 3).
| Amino acid residues | Position | Motif's conservation | Replaced by this consensus sequence on these RABV isolates |
|---|---|---|---|
| Arginine (R) | 333 | NO | Alanine (A) on all the Senegalese isolates |
| Asparagine (N) | 194 | NO | Tyrosine (Y) on all the Senegalese isolates |
| Glycosylation site NKF | 319 | NO |
located at position 338 NKA on the isolate SA194858_Dakar_SN_2008; NEA on the isolate SA252888_Dakar_SN_2013; DKT on the isolate SA212203_Dahra_SN_2011 |
| EMQSSLLQQH | 394–403 | YES |
ETQSSLLQQL on the isolate SA267115_Dakar_SN_2014; EMQSSLLQQL on the isolate SA252888_Dakar_SN_2013 |
| NHDYTIWMPE | 191–200 | YES | NHDYTICMPE on the isolate SA267115SEN_Dakar_SN_2014 |
| HNPYPDYHWL | 132–141 | YES | HNSYPDYHWL on the isolate SA252913_Diamniadio_SN_2013 |
| AETYTNFVGY | 87–96 | YES | Similar on all the Senegalese isolates |
| TTFKRKHFRP | 99–108 | YES | TTFRRKHFRP on all the Senegalese isolates |
| DIHHLSCPNN | 37–46 | YES | Similar on all the Senegalese isolates |
| KWCSPDQLVN | 269–278 | YES |
KWCPPDQLVN on the isolates SH155996_Dakar_SN_2001 and SH177846_Dakar_SN_2005; RWCPPDQLVN on the isolate SA252888_Dakar_SN_2013; KWCPSDQLVN on all the other Senegalese isolates |
| WKMAGDPRYE | 119–128 | YES | WKMAGGPRYE on the isolate SA212203_Dahra_SN_2011 |
| CGFVDERGLY | 226–235 | YES | CGFVDERSLY on the isolate SA267115_Dakar_SN_2014 |
| TLMEADAHYK | 341–350 | YES |
ALMEAHAHYK on the isolates SA252888_Dakar_SN_2013 and SA194858_Dakar_SN_2008; TLMEADAWYK on the isolate SA212203_Dahra_SN_2011 |
- Note: The residues Arg333 and Asn194 were previously described by Dietzschold et al. (1983). The N-glycosylation motif NKT was previously described by Marston et al. (2007). The ten remaining RABV amino acid motifs were analyzed as described by Tomar et al. (2017). Positions with amino acid different on the glycoprotein of Senegalese RABV are highlighted in bold and black for conservative mutations and in bold and black and underlined for non-conservative mutations. A motif was considered as non-conserved when it presents more than three non-conservative mutations in more than three RABV isolates.
G protein antigenic sites (Tomar et al., 2017) were relatively conserved with a few exceptions. Isolates SA194858, SA212203, and SA252888 had multiple changes in antigenic site III (residues 330–338). Comparison with 10 vaccine strain G protein sequences also presents disagreements (Figure 1). G protein percent identity between SA252888 and vaccine strains ranged from 84.5% to 87.6%. On average, the percent identity of Senegalese G proteins was ∼91.0% when compared to the selected vaccine strains.
3.6 Recombination and episodic selection
There was no evidence for recombination detected in any RABV sequences used in this study. Several sites under strong negative selection were found with the SLAC (p value ≤ 0.1) and FUBAR (posterior probability [PP] ≥ 0.9) models and most of them were located in major proteins such as the L, N, and G protein, respectively. However, significant episodic positive selection was obtained for all the proteins, except for the polymerase L (Table 4). All positively selected sites estimated by the FUBAR model (PP ≥ 0.9) were also identified by the MEME method (p value < 0.1) and the majority of such sites were located in the G protein. The N, P, and M proteins of the West African RABV sequences only exhibited one amino acid site (N270, P131, and M60, respectively) under positive selection while the G protein showed a total of six amino acid sites (G144, G236, G318, G321, G347, G458) (p value ≤ 0.1 and PP ≥ 0.9). Two among these sites harboured mutations from the honey-badger-related RABV isolate (Ser-321-Asp and His-347-Trp). Branch-site analysis also showed one branch evaluated under positive selection (p value < 0.05) in the glycoprotein and this episodic selection event was identified in the sequence of the honey-badger-related isolate SA212203. The eight sites detected under positive selection in the polymerase protein have been found in a previously characterized strain from Guinea (Edgar, 2004) and were identified only with the MEME method; then considered as non-evident (Table 4).
| Number of sites detected by method | |||||
|---|---|---|---|---|---|
| Protein | SLAC (p value ≤ 0.1) | FUBAR (PP ≥ 0.9) | MEME (p value ≤ 0.1) | aBSREL (p value < 0.05) | Positive selection |
| N | 58/540 | 269/540 | 0/540 | 0/102 | YES |
| 0/540 | 1/540 | 1/540 | 0/102 | ||
| P | 23/297 | 43/297 | 0/297 | 0/55 | YES |
| 0/297 | 1/297 | 1/297 | 0/55 | ||
| M | 12/202 | 39/202 | 0/202 | 0/55 | YES |
| 0/202 | 1/202 | 1/202 | 0/55 | ||
| G | 55/524 | 269/524 | 0/524 | 0/75 | YES |
| 0/524 | 0 /524 | 6/524 | 1/75 | ||
| L | 110/2127 | 813/2127 | 0/2127 | 0/67 | NO |
| 0/2127 | 0/2127 | 8/2127 | 0/67 | ||
- Note: For each protein, the top row represents sites under negative selection (dN/dS < 1), and the bottom row represents site that are under positive selection (dN/dS > 1). Pervasive diversifying selection at posterior probability (PP) ≥ 0.9 with FUBAR model. Episodic diversifying selection at 0.1 significance level with SLAC and MEME models (p value p ≤ 0.1). Episodic diversifying selection at p ≤ 0.05 with aBSREL model.
3.7 Phylodynamics of RABV in Senegal and Western Africa
A maximum-likelihood phylogenetic tree (Figure 2) and Bayesian-inferred MCC tree (Figure 3) of Senegalese RABV sequences formed a strongly supported monophyletic clade. Additionally, Senegalese RABV sequences share a most recent common ancestor with sequences isolated from Cote d'Ivoire, Burkina Faso, and Mauritania. Due to the weak temporal signal in only Senegalese RABV genomes (r2 = 0.20), phylogeographic predictions included additional African RABV isolates. Altogether, related African RABV sequences showed a stronger temporal signal (r2 = 0.64, Figure 2a).
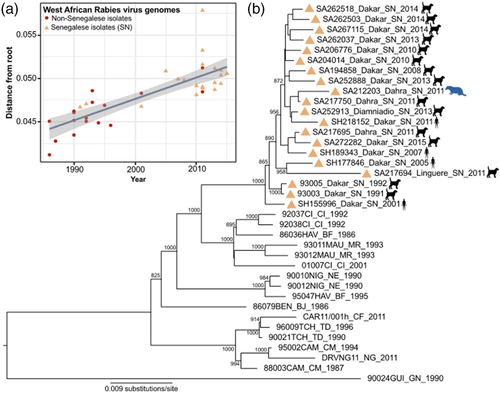
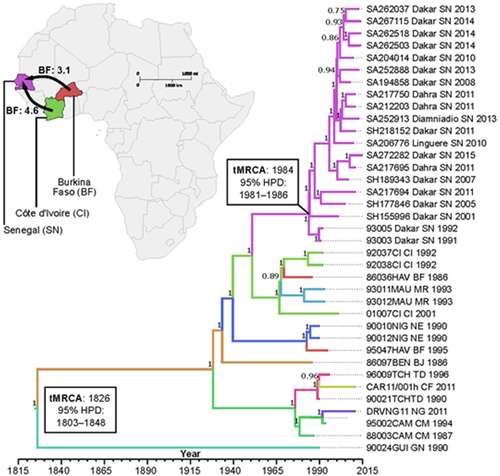
Time to the most recent common ancestor (tMRCA) for our RABV dataset was estimated to be in the early 19th century (95% HPD: 1803–1848, Figure 3). Based on currently available sequences, the tMRCA for the emergence of RABV in Senegal was in the 1980s (95% HPD: 1981–1986). Patterns of virus evolution were used to infer molecular epidemiology that supports two RABV introductions in Senegal from West-African neighbour countries. Moderate evidence points to two migration events: one from Cote d'Ivoire (Bayes factor: 4.6, Table A3) and the other from Burkina Faso (Bayes factor: 3.1, Table A3). Evolutionary rates of included RABV sequences had a mean substitution rate of 2.8 × 10–4 (95% HPD: 2.5–3.2 × 10–4).
4 DISCUSSION
The burden of RABV in Africa continues to hold steady around 20,000 deaths a year (World Health Organization, 2013b), with limited information on zoonotic circulation in West African countries (Mauti et al., 2019). Despite the establishment of a national surveillance program of Rabies infection in humans and animals since 2008, by the Institut Pasteur de Dakar and the Senegalese MoHSAS, only a few complete genomic data were available from Senegal prior to this study (Troupin et al., 2016). As usually reported in most of the developing countries, especially in Africa such as South Africa, Chad and Cameroon, officially reported human Rabies cases severely underestimated the actual incidence of rabies cases in Senegal (Tenzin and Ward, 2012). Here, we sequenced 18 RABV complete genome sequences isolated from humans, dogs and a honey-badger over a 14 year period. Notably, this is the first documented whole genome sequence of honey-badger-related RABV in West Africa. We assessed the genetic diversity of RABV from Senegal by evaluating amino acid distances and identifying mismatches in motives of virulence and antigenic sites located in the glycoprotein. In addition, we estimated the episodic selection pressures at the protein level and the phylodynamic analysis of RABV circulation in West Africa.
Overall, the new Senegalese RABV sequences are genetically closer to previously available genomes from Senegal (Troupin et al., 2016). The low pairwise nucleotide distances (<2%) observed between the Senegalese RABV sequences have resulted in a more apparent diversity at the amino acid level (<3%). The low amino acid sequence diversity observed in major proteins of the Senegalese RABV isolates such as the Nucleoprotein and the polymerase L, confirmed the description of these two regions as the most conserved among Lyssaviruses (Riedel et al., 2020). However, three isolates showed the highest amino acid distances between them and from the other Senegalese sequences of the G protein. The G protein-specific diversity exhibited in the Senegalese sequences could have served as a factor for virus spillover in a new host such as honey-badger (Talbi et al., 2009). In addition, the higher numbers of amino acid mutations from the three most divergent isolates were found in the Glycoprotein. Interestingly, the identification of a mutation closed to a mongoose-specific RABV's substitution from a dog-related RABV sequence in this study (Pro-386G-Leu), shows that the Pro-386G-Ser could be not only present in mongoose-related Africa-3 RABV (Bingham et al., 1994) but also shared between mongoose and dog-related isolates. Future studies could be conducted to investigate the possibility of RABV circulation in a wide variety of natural and incidental animal host species in Senegal. Furthermore, the substitutions observed in the polymerase protein could have participated in the RABV shift to new host species in Senegal such as honey-badger. Despite several isolations of RABV from honey-badger in Southern Africa (Bingham et al., 2001), no associated sequence was available. Our study is then noteworthy not only for generating the first whole-genome sequence of a honey-badger-related RABV but also for identifying the specific amino acid substitutions characterizing RABV isolated from the African honey-badger.
The G protein plays a crucial role in the biology and pathogenesis of neurotropic RABV infection (Li et al., 2012; Zhang et al., 2013). The amino acid mutation Arg-333-Ala has been shown to abolish virulence while the substitution Asn-194-Tyr is implicated in the ability of RABV to infect neurons (Faber et al., 2005; Schnell et al., 2010). These two substitutions in the Glycoprotein were highly associated with RABV neuropathogenesis in furious and paralytic forms of Rabies (Hemachudha et al., 2013). Previously described as one of the two most conserved N-glycosylation sites among sequences of the street RABV strains and the most stable site in the expression of the glycoprotein (Marston et al., 2007), the glycosylation site at position 319 (Asn319) was located at amino acid position 338 in the Senegalese sequences. Although the amino acid substitution at position 338 of the G protein plays no major role in viral pathogenicity, the identification of a glycosylation site at this position could be a new insight in the G-protein specific evolution of RABV in Senegal (Seif et al., 1985; Yamada et al., 2013). More noteworthy, our findings highlighted also the absence of this N-glycosylation motive (NKT) in sequence of the RABV isolate from honey-badger. Thus, further studies using recombinant technology could be needed to investigate the effect of this motive in the pathogenicity of the Senegalese isolates (Ding et al., 2017).
Although low levels of recombination have not been reported for RABV (Deviatkin & Lukashev, 2018), natural selection is believed to be one of the drivers of their evolution (Troupin et al., 2016). RABV is subject to strong purifying selection and the lowest nucleotide substitution rates were described in the following order: N, L, G, M and P (Troupin et al., 2016). As expected, our analysis also showed that the L and N proteins often described as more conserved for RABV, exhibited the strongest purifying selection. These two proteins are functionally important in RABV pathogenicity (Riedel et al., 2020). In addition, the absence of positive selection in the polymerase L suggests frequent purging of deleterious polymorphisms in this region during RABV transmission, since neither of the motives identified in the polymerase was found to be subject to positive selection using the methods employed in our study. Besides the other proteins, there is substantial evidence that episodic selection events in the G protein sequences are a result of the evolution of RABV in West Africa. The amino acid substitution 318G (Phe) has been previously associated with furious and paralytic Rabies and involved in p75 neurotrophin receptor binding, viral maturation and transport into the cell (Bingham et al., 1994). Although the other sites subjected to positive selection in the G protein were not documented in RABV pathogenicity, almost all functional sites directly related to host tropism and infectivity are located from positions 181 to 431 in the of the G protein derived from the carnivore RABV (Ding et al., 2017). The presence of specific mutations under positive diversifying selection in the sequence from the honey-badger-related RABV exhibited also the nature of selection pressures associated with host switching which could be considered as a pattern driving RABV evolution in West Africa, particularly in Senegal (Mollentze et al., 2014).
Phylogenetic analysis shows that the Senegalese RABV sequences are grouped into a monophyletic cluster (Africa-2 clade) and are not region (intra-Senegal) specific. The ML tree topology in this paper is in agreement with previously published data including sequences from Senegal (Talbi et al., 2009). However, the spillover of RABV into a novel ecological niche (host shift to the honey-badger) could lead to the rapid spreading and future diversification of the virus in Senegal. Spillover infections are important public health events that establish new reservoirs for human exposures and can affect the conservation of threatened or endangered wildlife species when they occur into those species (Randall et al., 2004). In addition, combinations of positively selected changes have been shown to be associated with genetic polymorphism during each RABV host shift (Streicker et al., 2012). Therefore, it is becoming increasingly clear that the adaptive changes necessary for RABV host shift are determined in part by viral factors that differ between strains, by the interaction between the viral factors and the identity of the donor and recipient host species involved (Katz et al., 2016). Data from the Senegalese RABV have been previously included in genome-wide phylogenetic analyses (Troupin et al., 2016). Nevertheless, our results represented the most comprehensive molecular analysis of RABV from Senegal, as it includes more whole-genome sequences than previously documented. The generated complete sequences could be useful not only for large-scale analysis of RABV dynamics in Africa but also in update of the existing pan-Lyssavirus or RABV-specific diagnostics.
Despite the host switching experienced since 2011 by the Africa-2 clade circulating in West Africa through spillover to honey-badger in Senegal, it showed a mean nucleotide substitution rate (2.8 × 10–4 (95% HPD: 2.5–3.2 × 10–4)) which is in agreement with findings reported in previous studies of RABV evolution (Chiou et al., 2014; Troupin et al., 2016). All the RABV sequences generated in this study formed a homogenous cluster in the MCC tree (nucleotide dissimilarity ranging from 0.2% to 1.7%), suggesting a close-related population of RABV in Senegal as previously described in a detailed analysis of the phylogeographical structure of the Africa-2 clade between Central to Western African regions (Talbi et al., 2009). Our data suggest two recent RABV introductions into Senegal during the 1980s (95% HPD: 1981–1986) from Cote d'Ivoire and Burkina Faso. Instead of a transboundary animal transmission of Rabies from these countries (Bourhy et al., 2008; Talbi et al., 2009), the phylogenetic evidence gathered in this study points more in the direction of long-distance transmission of Rabies facilitated by human-mediated animal movements (Bourhy et al., 2008; Fèvre et al., 2005). However, further phylogenetic studies including more complete data sequences from these two latest countries could be necessary to provide more consistency to these results as there are only 3 and 2 complete genome RABV sequences available from Cote d'Ivoire and Burkina Faso, respectively (Troupin et al., 2016).
Regarding the key genetic elements from honey-badger-related RABV, this spillover could have led to a less significant evolution of the Africa-2 clade RABV circulating in Senegal. Through selection mechanisms, a dog-related RABV isolate may have acquired the key genetic information to induce host switching, adaptation and viral spillover into honey-badgers as the honey-badger-related RABV clustered with a dog-related isolate collected from the same place during the same year. However, further characterization of other viruses from the honey badger would be crucial to identify genetic clustering of RABV in this specie and thus could help discriminate if this animal is just an accidental host or might be considered as a new reservoir.
In addition, through the identification of a quietly similar mongoose-related mutation in a dog-related RABV sequence (Pro-386G-Leu), our data could be worth considering that the Pro-386G-Ser mutation previously identified as specific to mongoose-related RABV (Africa-3 clade) could be also present in dog-related RABV isolates. Nevertheless, our phylogenetic data have exhibited the impact of purifying selection in the molecular evolutionary dynamic of the Africa-2 clade RABV circulating in Senegal.
Our study could constitute the starting point exhibiting the importance to establish more appropriate public health measures for rabies control. However, the limited genomic characterization and epidemiological data availability on Rabies enzootic cycle hosts and human disease burden collected from passive surveillance programs may hamper such efforts to retrospectively analyze and estimate the true extent of disease burden in Senegal. The current structure for coordination and the collection of all information relating to rabies, the lack of awareness by the population, the under-estimation of the number of cases and the little or complete absence of epidemiological studies in Senegal are all additional considerations health ministries must consider.
Coupled with data on dog populations at the country level, the future findings on genomic sequences, host reservoir clinical signs, and zoonotic transmission are crucial to define the true burden of rabies in Senegal. These data could help also strengthen the existing integrated surveillance program in humans and animals through the mobilization of local resources for mass dog vaccination, the improvement of access to PEP, especially for the poorest, the improvement of disease reporting and surveillance tools, and community and healthcare workers education and outreach. As rabid dogs constitute the main source of transmission to humans, rabies control in dog populations through a strong one health cooperation is essential for successful elimination in Senegal by 2030. However, the rabies spillover in honey-badger exhibits the importance of establishing a surveillance program in wildlife to improve control measures and avoid possible future re-introductions into domestic hosts.
ACKNOWLEDGEMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors and was only supported by the Institut Pasteur de Dakar. We acknowledge colleagues of the virology department at Institut Pasteur de Dakar, Senegal for sharing supportive information necessary for establishment and accomplishment of this study.
DISCLAIMER
No other person has any role in the study concept, study design, data collection, experimental work, data analysis, data interpretation or the decision to publish. Any use of products or commercial services mentioned in this publication is for descriptive purposes only and does not imply endorsement by the funding agencies. Any opinions, findings and conclusion or recommendations expressed in this material are those of the authors and do not necessarily reflect the view of the the U.S. Department of Defense, U.S. Department of Health and Human Services, U.S. Department of the Army, or the institutions and companies affiliated with the authors.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required.
APPENDIX
| Primer name | 3′–5′ Sequence | Direction | Location in viral genome | Melting temperature (°C) |
|---|---|---|---|---|
| SN3race243R | AGCTTGGCAGCATTCATGCCTG | Reverse | Nucleoprotein | 53 |
| SN3race298R | TGCTGCTGCCAAGTAGGAACAT | Reverse | Nucleoprotein | 53 |
| N127 | ATGTAACACCTCTACAATGG | Forward | Nucleoprotein | 56 |
| N8m | CAGTCTCYTCNGCCATCTC | Reverse | Nucleoprotein | 56 |
| SNRV1342F | TATGTKTCAGTCAGTTCC | Forward | Nucleoprotein | 55 |
| SNRV2939R | TCRTCCCAAGTGATCTCY | Reverse | Matrix | 55 |
| SNRV2745F | ATATTCYGGAAAYCACAGRAT | Forward | Matrix | 53 |
| SNRV4280R | TGAGACGTCTRAAACTCACTG | Reverse | Glycoprotein | 53 |
| M220 | TGGTGTATCAACATGRAYTC | Forward | Matrix | 53 |
| SNG5444R | GGTCATCATAGACCTCYC | Reverse | Glycoprotein | 53 |
| G4836-S3 | GGRARRGTYATATCTTCNTGGGA | Forward | Glycoprotein | 54 |
| PV08 | GGTCTGATCTRTCWGARYAATA | Reverse | Polymerase L | 54 |
| L7386-AS3 | CTRTCBGARTARTADAYCCANGACTT | Reverse | Polymerase L | 54 |
| SNRV5318F | GGGCTGGATCATCTATGCTT | Forward | Glycoprotein | 53 |
| SNRV6304R | TTGATGACCTCGTAGCCTGA | Reverse | Polymerase L | 53 |
| SNRV6281F | TGYGGAAAYTCCGGCTAT | Forward | Polymerase L | 54 |
| SNRV8504R | GCTCRCTGAGAAATCGRG | Reverse | Polymerase L | 54 |
| Taq3long | ATGAGAAGTGGAAYAAYCATCA | Forward | Polymerase L | 54 |
| L9633-AS3 | TTGCYRTATATGTTGACAGG | Reverse | Polymerase L | 54 |
| SNRV8401F | TTCAGAGTTYAGAGAGGCRAT | Forward | Polymerase L | 53 |
| SNRV9648R | ATGTTAACAGGGAAGATTGTT | Reverse | Polymerase L | 53 |
| SNRV9263F | ATGTTYCAGCCATTGATGCTT | Forward | Polymerase L | 54 |
| SNRV10590R | TGAAYACAAGCTTRGCATCYG | Reverse | Polymerase L | 54 |
| SNRV10331F | TGCTCTGCTCAACAGGTT | Forward | Polymerase L | 53 |
| SNRV11932R | ACGCTTAACAAATAAACAACA | Reverse | 5’ Trailer | 53 |
| SN5race11566F | GGTCTGGTGACACCCCGGTCTTCA | Forward | Polymerase L | 55 |
| SN5race11615F | GAGTCTGTCATCTCACTGGATCA | Forward | Polymerase L | 55 |
| Isolate | Collection year | Isolation place | Host | GenBank accession |
|---|---|---|---|---|
| 93005 | 1992 | SN | Canis lupus familiaris | KX148239 |
| 93003 | 1991 | SN | Canis lupus familiaris | KX148238 |
| 92037CI | 1992 | CI | Canis lupus familiaris | KX148232 |
| 92038CI | 1992 | CI | Canis lupus familiaris | KX148233 |
| 01007CI | 2001 | CI | Canis lupus familiaris | KX148235 |
| 86036HAV | 1986 | BF | Canis lupus familiaris | KX148234 |
| 95047HAV | 1995 | BF | Canis lupus familiaris | KX148230 |
| 93011MAU | 1993 | MAU | Canis lupus familiaris | KX148236 |
| 93012MAU | 1993 | MAU | Canis lupus familiaris | KX148237 |
| 90010NIG | 1990 | NE | Canis lupus familiaris | KX148231 |
| 90012NIG | 1990 | NE | Canis lupus familiaris | KX148229 |
| DRV_NG11 | 2011 | NG | Canis lupus familiaris | KC196743 |
| 86097BEN | 1986 | BJ | Felis catus | KX148107 |
| 90021TCH | 1990 | TD | Canis lupus familiaris | KX148240 |
| 96009TCH | 1996 | TD | Canis lupus familiaris | KX148241 |
| CAR_11_001 | 2011 | CF | Homo sapiens | KF977826 |
| 88003CAM | 1987 | CM | Canis lupus familiaris | KX148243 |
| 95002CAM | 1994 | CM | Canis lupus familiaris | KX148242 |
| 90024GUI | 1990 | GN | Canis lupus familiaris | KX148244 |
- Abbreviations: BF, Burkina Faso; BJ, Benin; CF, Central African Republic; CI, Côte d'Ivoire; CM, Cameroon; GN, Guinea; MAU, Mauritania; NE, Niger; NG, Nigeria; SN, Senegal; TD, Chad.
| From | To | Bayes factor | Posterior probability |
|---|---|---|---|
| TD | CF | 158.0 | 0.94 |
| CM | NG | 123.5 | 0.93 |
| CM | TD | 26.9 | 0.74 |
| NE | BF | 22.1 | 0.70 |
| CI | MR | 11.3 | 0.55 |
| CI | BF | 10.3 | 0.53 |
| BF | NE | 4.7 | 0.34 |
| CI | SN | 4.6 | 0.33 |
| BF | CI | 4.5 | 0.33 |
| BF | MR | 4.4 | 0.32 |
| BN | CM | 3.9 | 0.29 |
| MR | CI | 3.6 | 0.28 |
| BN | NE | 3.5 | 0.27 |
| BF | SN | 3.1 | 0.25 |
- Abbreviations: BF, Burkina Faso; BJ, Benin; CF, Central African Republic; CI, Côte d'Ivoire; CM, Cameroon; GN, Guinea; MAU, Mauritania; NE, Niger; NG, Nigeria; SN, Senegal; TD, Chad.
Open Research
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the Supporting Information of this article.




