Intraspecific chemical variation of Tanacetum vulgare affects plant growth and reproductive traits in field plant communities
Abstract
- The study investigated the impact of intraspecific plant chemodiversity on plant growth and reproductive traits at both the plant and plot levels. It also aimed to understand how chemodiversity at stand level affects ecosystem functioning and plant–plant interactions.
- We describe a biodiversity experiment in which we manipulated intraspecific plant chemodiversity at the plot level using six different chemotypes of common tansy (Tanacetum vulgare L., Asteraceae). We tested the effects of chemotype identity and plot-level chemotype richness on plant growth and reproductive traits and plot-level headspace emissions.
- The study found that plant chemotypes differed in growth and reproductive traits and that traits were affected by the chemotype richness of the plots. Although morphological differences among chemotypes became less pronounced over time, reproductive phenology patterns persisted. Plot-level trait means were also affected by the presence or absence of certain chemotypes in a plot, and the direction of the effect depended on the specific chemotype. However, chemotype richness did not lead to overyielding effects. Lastly, chemotype blends released from plant communities were neither richer nor more diverse with increasing plot-level chemotype richness, but became more dissimilar as they became more dissimilar in their leaf terpenoid profiles.
- We found that intraspecific plant chemodiversity is crucial in plant–plant interactions. We also found that the effects of chemodiversity on plant growth and reproductive traits were complex and varied depending on the chemotype richness of the plots. This long-term field experiment will allow further investigation into plant–insect interactions and insect community assembly in response to intraspecific chemodiversity.
INTRODUCTION
Individuals of the same plant species can exhibit varying phenotypes, which reflect variation in growth, reproductive, physiological, and other traits (Raffard et al., 2019; Siefert et al. 2015; De Bello et al. 2011; Fridley & Grime 2010; Bolnick et al. 2003). This variation within plant species is a major element in individual performance and population-, community- and ecosystem-scale processes (Violle et al. 2012; Siefert et al. 2015; Guisan et al. 2019; Westerband et al. 2021). Functional traits are attributes of species that can be measured at the individual level, and are related to the individual's response to environmental conditions and which impact ecosystem properties and ecosystem functioning (Isbell et al. 2011). In addition to visible morphology-related functional traits, plants can vary intraspecifically in less apparent traits, such as chemical composition (Wetzel & Whitehead 2020). For instance, many plant species show pronounced intraspecific variation in specialized metabolites along environmental gradients (Moore et al. 2014; Bakhtiari et al. 2019), or even at finer spatial scales, such as plant patches located in areas of less than a few square kilometers (Kleine & Müller 2011). Variation in chemodiversity has recently gained increased attention in ecology as it might structure community assembly or community composition, as well as species interactions, and fulfil ecosystem functions, including structuring plant-associated food webs and biodiversity (Bálint et al. 2016; Erb & Kliebenstein 2020; Müller et al. 2020; Wetzel & Whitehead 2020).
Individuals of a plant species can be clustered into distinct groups – also called chemotypes – by their dominant compounds, the composition of volatile and non-volatile compound blends, or by comparisons of specific specialized metabolites produced by individuals (Eilers 2021). The consequences of specialized metabolites for intra- and interspecific interactions have been studied in various herbaceous plant model systems (Moore et al. 2014), including Tanacetum vulgare (Kleine & Müller 2011; Clancy et al. 2016; Eilers et al. 2021; Neuhaus-Harr et al. 2023), Jacobaea vulgaris (Macel 2011; Kostenko & Bezemer 2013; Carvalho et al. 2014), Plantago lanceolata (Harvey et al. 2005; Wurst et al. 2008), Senecio inaequidens (Cano et al. 2009; Macel & Klinkhamer 2010), and Brassica oleracea (Gols et al. 2008; Gols et al. 2009; Poelman et al. 2009; Kabouw et al. 2010; Kos et al. 2011; Bustos-Segura et al. 2017). Specialized metabolites can be stored in specialized structures, such as glandular trichomes in T. vulgare, and in the headspace when released into the environment as volatiles. Conversely, some compounds are only produced de novo and released immediately, for instance, in plants without storage structures. These volatile organic compounds (VOCs) and other compounds found in leaves shape the assemblage and interaction within the plant-associated insect community (Ponzio et al. 2013). Although ecologists are beginning to understand the consequences of plant chemodiversity for plant–insect interactions, much less is known about how it affects plant–plant interactions (Thorpe et al. 2011).
Plants can display intraspecific chemodiversity at different levels. Through variations in chemical composition at a small scale, groups of plants in a natural stand may differ in their chemotypes (Senft et al. 2017; Clancy et al. 2018; Müller et al. 2020; Clancy 2021; Eilers 2021). In such scenarios, intraspecific chemodiversity can be described by the richness (i.e., the number of chemotypes) and relative abundance of different chemotypes at the stand level. So far, the ecological consequences of variation in chemodiversity at the stand level have rarely been investigated (but see Glassmire et al. 2020).
The effects of increasing plant diversity on processes at the ecosystem level have been intensively studied in the framework of biodiversity–ecosystem functioning research (Chapin III et al. 1992; Weisser et al. 2017). In most empirical work, plant diversity was manipulated by creating plant communities that differed in the number of plant species they contained. In contrast to the wealth of studies manipulating plant community diversity at the plant species level, there are far fewer studies where plant community diversity is manipulated at the within-species level, i.e., by creating plant communities with the same number of plant species, but with different extent of intraspecific variation. In a seminal study, Crutsinger et al. (2006) showed that in Solidago altissima communities that differed in the number of Solidago genotypes they contained, increasing genotypic diversity enlarged arthropod community species richness and increased plant community biomass. The number of studies manipulating the intraspecific diversity of plant communities has been increasing in the past years (Genung et al. 2012; Koricheva & Hayes 2018; Raffard et al. 2019). However, the detailed results differ among studies and taxa, even when only considering research conducted on the effects on herbivores (Barton et al. 2015; Bustos-Segura et al. 2017; Fernandez-Conradi et al. 2022; Hauri et al. 2022). A general trend is that increased intraspecific genotypic diversity of the plant community increases the diversity of the associated arthropod community and that these diversity effects on natural enemies of herbivores are generally higher than effects on the herbivores themselves, suggesting changes in top-down control (see meta-analysis in Koricheva & Hayes 2018; Wetzel et al. 2018; Hauri et al. 2021). The mechanisms underlying these effects are still being discussed.
Plants in intraspecifically diverse communities may compete with their neighbouring conspecifics, resulting in changes in trait expression, such as growth (Viola et al. 2010; Genung 2012; Bustos-Segura et al. 2017; Ziaja & Mueller 2023). On the other hand, plants may also experience reduced competition through niche partitioning because of a priori differences in functional traits, such as phenology (Kuppler et al. 2016; Gallien 2017). Little is known about how the chemotypes of plants differ in other traits and how intraspecific chemotype richness at the stand level affects individual plant performance.
Here, we designed a field experiment using Tanacetum vulgare L. (Asteraceae), in which we manipulated plot-level chemotype richness and the presence of specific plant chemotypes within the plots, to test the effects on plant growth and volatile emission. T. vulgare exhibits high intraspecific variation in specialized metabolites (Rohloff et al. 2004; Bálint et al. 2016), mainly mono- and sesquiterpenes (Keskitalo et al. 2001; Ziaja & Mueller 2023), and is easy to propagate clonally (Bálint et al. 2016). Moreover, variation in T. vulgare in terpenoid profiles has been documented to differ strongly among different geographic regions and even among individuals within populations, meaning that plant–plant interactions of distinct chemotypes often occur in close proximity in nature (Kleine & Müller 2011; Clancy et al. 2016).
- At the individual plant level, chemotypes will differ in growth and reproductive traits, which will be affected by the plot-level chemical richness of the plots in which they grow.
- At the plot level, higher plot-level chemotype richness will increase plant growth, in line with biodiversity's generally observed positive effects described in the ecological literature.
- Individual chemotypes will differ in their effect on plot-level effects.
- Plant communities with higher plot-level chemotype richness will emit a more chemically diverse headspace of VOCs than plots with lower plot-level chemotype richness.
MATERIAL AND METHODS
Chemotypic characterization of T. vulgare lines and biological replicates
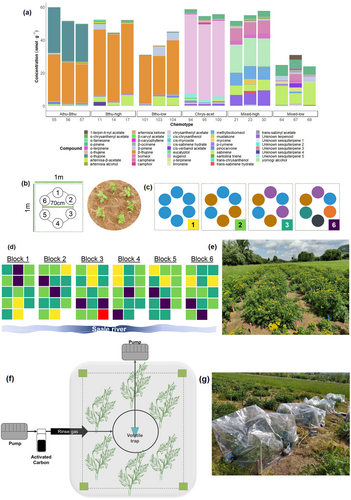
Plant chemotyping and chemotype selection are described in detail in Data S1 and in Neuhaus-Harr et al. (2023). The leaf terpenoid composition of the T. vulgare chemotypes used is shown in Fig. 1a and Table S1-1. We named the chemotypes according to their dominant compounds as follows. The ‘Athu-Bthu’ chemotype had both α- and β-thujone as prevalent compounds. The ‘Bthu-high’ and ‘Bthu-low’ chemotypes were dominated by β-thujone but had high or low relative levels of this terpenoid. There was a strong dominance of chrysanthenyl acetate in the ‘Chrys-acet’ chemotype. The ‘Mixed-high’ chemotype featured three terpenoids with a high relative total concentration, whereas the ‘Mixed-low’ chemotype featured several terpenoids evenly contributing to the total profile.
Propagation of plant material for the field experiment
Plants were propagated via stem cuttings. A detailed description can be found in Data S1.
Experimental design
The field experiment took place in the Jena Experiment site, located on the floodplain of the Saale River in Jena, Germany (50°55′ N, 11°35′ E, 130 m a.s.l.; Weisser et al. 2017). In May 2021, all vegetation was removed, and the soil was mechanically tilled. No fertilizer, chemical insecticides, or fungicides were used, and weeds were removed by hand every 2 weeks during the growing season.
We created plots of six tansy plants that were evenly distributed around a 70-cm diameter circle (84 plots × 6 plants = 504 plants total; Fig. 1b). The design followed suggestions of the design for biodiversity experiments: each plot type differed in the number and identity of tansy chemotypes assigned to it. The number of different chemotypes in one plot (hereafter: plot-level chemotype richness) ranged from 1, 2, 3, to 6 (Fig. 1c). A plot with a plot-level chemotype richness of 1 was assigned six plants of the same chemotype. Within the plot, we maximized the number of daughters per chemotype. For example, we used the three Athu-Bthu chemotype daughters (56, 57, and 58) for plot-level chemotype richness of 1 of Athu-Bthu. Daughters from each chemotype were equally distributed over the different treatment plots where possible or structurally assigned where an equal division was not possible (e.g., in the case of a shortage of plants of one daughter, they were replaced with a randomly picked daughter from the remaining two daughters of the same chemotype). So, a plot with plot-level chemotype richness 1 was assigned two clones from each of the three daughters of the plot chemotype. For plot-level chemotype richness 2, there was one clonal plant of each of the three daughters of each of the two chemotypes in the plot, etc. Lastly, a plot with plot-level chemotype richness 6 was assigned one clonal plant from one daughter of each of the six different chemotypes. Varying the daughter identities per chemotype in each plot allowed all level chemotype plots to be biologically replicated.
Prior to an in-depth analysis of the terpenoid composition, the composition of chemotype richness at the plot level was pre-assigned for each plot. Each chemotype combination was thus replicated twice, except for plot-level chemotype richness level 6, which was replicated 12 times but with different daughters. Hence, our field experiment contained 12, 30, 30, and 12 replicate plots for plot-level chemotype richness levels 1, 2, 3, and 6, respectively. Plots were distributed equally in six randomized blocks. The field was divided into 84 plots (1 × 1 m) separated from each other by 70-cm footpaths and distributed in six replicated blocks (Fig. 1d). Each block consisted of 14 plots: two plots of chemotype richness level 1, five plots of chemotype richness level 2 and 3, and two plots with chemotype richness level 6 (Fig. 1d, e). The exact assignment of plants to plots is given in Table S1-2.
Plant morphological trait measurements
We measured six morphological plant traits for individual plants, including four growth traits (number of stems per plant, height of the highest stem, and aboveground fresh and dry weight) and two reproductive traits (cumulative number of flower heads and flowering index). A more detailed description of each variable and time points used can be found in Data S1.
Headspace VOC collection
Headspace VOC emissions were collected at the plot level. A detailed description of the process is available in Data S1.
Statistical analysis
All analyses were performed in R version 4.2.2 and RStudio 2022.07.2 + 576 (R Core Team 2021). A description of all models is provided in Data S1.
To analyse plot-level chemotype richness, we calculated plot-level diversity metrics using the ‘chemodiv’ package (Petrén et al. 2023). Note that we calculated theoretical chemotype diversity metrics based on the cumulative leaf terpenoid profiles of each respected daughter present in a plot. Calculated realized volatile diversity metrics were based on our headspace VOC collection at the plot level. Data were visualized using the ‘ggplot2’, ‘grid’, ‘gridExtra’, and ‘ggpubr’ packages (Wickham 2016; Auguie 2017; Kassambara 2020; R Core Team 2021).
We distinguished between plant-level and plot-level analyses, whereby plant-level analyses focused on the performance of the individual plants growing in the different diversity plots, controlling for the plot in which they grow. Plot-level analyses focused on plot-level measures of plant performance calculated by averaging over all plants in a plot. All analyses of the effects of chemodiversity on plant traits were done with mixed-effect models using the ‘lme4’ package (Bates et al. 2015). All variables were analysed separately for each time point. For evaluating the effects on traits of individual plants, P-values were estimated by type II Wald-Chi-Square tests using the Anova() function in the ‘car’ package (Fox & Weisberg 2019) and by type I Wald-Chi-Square tests using the anova() function in base R for evaluating the effects of chemotype presence on plot-level measurements and overyielding indices. After fitting a model, post-hoc pair-wise comparisons among factor levels (i.e., chemotypes) were assessed with the ‘emmeans’ package with Tukey adjustment (Russell 2021). All plant- and plot-level model assumptions were met in all models, except for the model covering plant-level height on 28 October 2021, in which the residuals were not normally distributed, and transformations did not improve normality of model residuals. Despite the non-normal distribution of the data, we chose to keep all measurements untransformed, including outliers, as we found no ecological justification for their exclusion, and decided to maintain consistency with models at other time points to facilitate meaningful comparisons.
Effects of chemotype and plot-level chemotype richness on traits of individual plants
To test the effects of chemotype and plot-level chemotype richness on traits of individual plants growing in the different plots with six plants each, both factors (chemotype, plot-level chemotype richness) and their interaction were included as fixed factors in Generalized Linear Mixed-Effect Models (GLMM), with Poisson distribution for counting data (number of stems for both years and the cumulative number of flower heads), and with Linear Mixed-Effect Models (LMM) for the other individual plant traits (plant height, flowering index, and square root-transformed aboveground dry weight and square root-transformed aboveground fresh weight). We treated chemotype richness as a continuous variable with one degree of freedom. In the main analysis, random effects were Daughter ID (to account for technical replication of each daughter via cuttings) and Plot ID (84 in total) nested in Block ID (R scripts are provided as S3). In some variables, the random effect structure of the model led to singularity. We performed a second model excluding Plot ID since it had the lowest explanatory power of all random effects. We then chose the higher-quality statistical model using the estimator of prediction error AIC.
If the main effect of a factor was significant, post-hoc pair-wise comparisons with Tukey adjustment were conducted to assess significant differences between factor levels. While this is the most correct analysis from the point of daughter distribution within chemotypes, it results in a low degree of freedom. Hence, we carried out additional analyses to complement our understanding of within- and between-chemotype differences.
Effects of daughter and plot-level chemotype richness on traits of individual plants
We performed separate analyses per chemotype with Daughter, plot-level chemotype richness, and their interaction as fixed factors, and Plot ID nested in Block ID as a random factor. For count data, we used a GLMM with Poisson distribution and Nelder Mead optimizer and, for the other assessed variables, LMM models.
In a final analysis, we directly compared daughters' performance, using Daughter ID (with 18 levels) and plot-level chemotype richness as independent variables, and Plot ID and Block ID as random factors.
Effects of chemotype and plot-level chemotype richness on plot-level means
We first averaged each plant trait at the plot level to test the effects of chemotype and chemotype richness on plot-level plant traits. To test the effect of the presence/absence of a chemotype on plot-level trait values, we carried out a separate analysis per chemotype. This was done by running a linear model with Block ID, chemotype presence (indicating whether a specific chemotype is present in the plot), plot-level chemotype richness, and the interaction between chemotype presence and plot-level chemotype richness, separately for each chemotype.
Overyielding calculations
We assessed the plot-level performance (i.e., the plot-level mean yield of the measured trait) of plants growing in plots of different chemotype richness levels (plot-level chemotype richness = 2, 3, and 6) and compared this to their performance in a monoculture (i.e., the respective chemotype-specific plots in plot-level chemotype richness = 1). This was done by calculating the overyielding index (OI) according to Hector et al. (2002) and Hooper et al. (2004). Overyielding indices are positive when the yield for a given chemotype in a mixture is greater than expectations from monocultures and indicates overyielding (Loreau 1998). Overyielding indices for each plot-level chemotype richness (i = 1, 2, 3, or 6) were obtained for aboveground dry weight, aboveground fresh weight, the cumulative number of flower heads, and the flowering index for each plot. Mathematically, we calculated OIi = (Oci−Ec) × Ec−1, where Oci is the observed yield of a mixture plot I obtained by means of yields of the plants in the mixture, and Ec is the expected yield for the plot. Ec was calculated by averaging the yield of monoculture of each of the chemotypes present in the plot (Table S2-11).
In separate analyses per trait, we carried out LMM models with the calculated overyielding indices for all plot-level chemotype richness levels, with plot-level chemotype richness as a fixed factor and Block ID as a random factor.
Plot-level theoretical leaf and plot-level realized volatile chemodiversity metrics
We calculated chemodiversity metrics at the plot level based on the leaf terpenoid profiles of individual chemotypes before planting (plot-level theoretical leaf chemodiversity) and on the headspace VOC measurements in the field (plot-level realized volatile chemodiversity). Plot-level theoretical leaf chemodiversity metrics were calculated by summing the absolute leaf terpenoid concentrations (nmol·g−1) produced by each of the six specific daughters present in each plot (Fig. 1a). Those absolute leaf terpenoid profiles were obtained from the chemical analysis performed on leaves from greenhouse plants in 2020, before making the cuttings and planting them in the field. Based on the individual values of each plant present in a plot, we calculated plot-level theoretical leaf terpenoid richness, concentration, Hill Shannon index, and Hill evenness (Petrén et al. 2023). Plot-level realized volatile chemodiversity metrics were calculated based on the plot-level absolute terpenoid emissions (ng·h−1) detected by headspace VOC collection.
As plants were chemotyped a priori (in Neuhaus-Harr et al. 2023) before transplanting them to the field, and chemotyping was hence not affected by field conditions, plot-level theoretical leaf chemodiversity was analysed by linear models with plot-level chemotype richness as the independent variable, without putting block as a random factor into the model. Plot-level realized volatile chemodiversity metrics based on field-collected headspace VOCs were analysed by linear models with plot-level chemotype richness as the independent variable and Block ID (6 blocks) and Collection Day ID (3 days) as random factors.
We also analysed the correlation between leaf and headspace terpenoid profiles by calculating Bray–Curtis dissimilarity matrices for hierarchical analyses and performing a Mantel test by specifying 9999 permutations using the ‘vegan’ package (Oksanen et al. 2022).
RESULTS
Field establishment
The establishment of the field experiment was successful; all 504 plants survived until the first seasonal harvest date (28 October 2021). By early May of the second year (2 May 2022), 96.6% of the plants (487 of 504 plants) showed shoot regrowth. Of these 487 plants, 4 more plants, which had shown some aboveground growth, naturally died and were not present at the harvest date of the second year (05 October 2022). Missing data were excluded from analyses of plant traits at the plant level, with 17 plants missing for the first (May), 20 missing for the second (July), and 21 missing for the final measurements (October). Because of regrowth and mortality, the missing plants between time points only partially overlap. Plot-level averages were calculated according to the number of plants recorded in the plot at the measurement date, but we retained the original plot chemotype richness level for the statistical analysis as this was the treatment variable.
At the point of planting in May 2021, the plants were visually very similar. First trait measurements were conducted 2 weeks post-planting (Figure S2-2), and at various intervals leading up to the annual harvest. Notably, uniform height was observed among all plants during these assessments. Additionally, the number of stems per plant was recorded (see Figure S2-1). While all plants initially possessed a single main stem at planting, subsequent rhizome growth resulted in the rapid emergence of new stems.
Effects of chemotype and plot-level chemotype richness on traits of individual plants
To test hypothesis (i), we used analyses separated for each time point to assess the effect of chemotype and plot-level chemotype richness across and between seasons.
The number of stems of a plant was significantly affected by plant chemotype identity on 01 June (χ25 = 24.99, P < 0.001; Figure S2-1) and 22 June 2021 (χ25 = 21.49, P < 0.001; Fig. 2a), but not at the other time points (P > 0.05; Table S2-2). Post-hoc analyses revealed that in the 2021 season, the Mixed-high chemotype had the highest number of stems compared with all other five chemotypes. However, the effects of chemotype on the number of stems across chemotypes were not stable across nor between growing seasons (Figure S2-1, Table S2-2). Differences in the number of stems across chemotypes were less pronounced towards the end of the first season (Figure S2-1a–c) and even became indistinct for 2022 (χ25 = 3.28, P = 0.657; Fig. 2d).
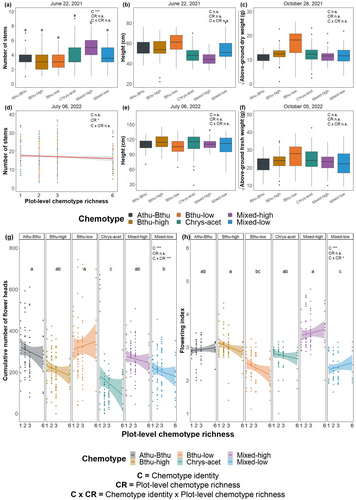
For plant height, chemotypes were marginally different on 22 June 2021 (χ25 = 10.54, P = 0.061; Fig. 2b) and strongly different when harvested on 28 October 2021 (χ25 = 12.26, P = 0.031; Figure S2-2c). At this time point, several outliers caused deviation from a normal distribution that could not be solved with transformation. Therefore, for this time point, the chemotype effect must be interpreted cautiously. For instance, even though the chemotype had a significant effect, a post-hoc test for 28 October 2021, did not reveal differences in height across chemotypes. Moreover, no effect of chemotype on height was retained in 2022 (Fig. 2e, Figure S2d–f, Table S2-3).
For plant biomass, the aboveground weight of the Bthu-low chemotype was slightly higher in both growing seasons, although the effect of the chemotype was not significant (aboveground dry weight 2021: χ25 = 9.35, P = 0.096; Fig. 2c; aboveground fresh weight 2022: χ25 = 2.65, P = 0.753; Fig. 2f, Table S2-4).
There was no significant effect of plot-level chemotype richness, nor the interaction between chemotype and plot-level chemotype richness, for any of the growth traits measured on individual plants, except for a slight negative effect of plot-level chemotype richness on the number of stems in 2022 (χ21 = 4.67, P = 0.031; Fig. 2d).
Both reproductive plant traits were significantly affected by plant chemotype: the cumulative number of flower heads (χ25 = 55.08, P < 0.001) and the flowering phenology (χ25 = 51.07, P < 0.001). Interestingly, reproductive plant traits depended on both chemotype and the plot-level chemotype richness, as indicated by an interaction between these two factors (cumulative number of flower heads: χ25 = 779.33, P < 0.001; flowering index: χ25 = 14.93, P = 0.011). For example, the Bthu-low chemotype produced more flower heads in plots with higher plot-level chemotype richness in the first growing season. The other five chemotypes produced fewer flower heads in plots with higher plot-level chemotype richness (Fig. 2g). The Mixed-high, Mixed-low, and Athu-Bthu chemotypes showed more advanced flowering phenology (i.e., higher flowering index) in plots with higher plot-level chemotype richness. In contrast, the Bthu-low, Chrys-acet, and Bthu-high chemotypes showed more advanced phenology in plots with lower plot-level chemotype richness (Fig. 2h).
We found strong variation among daughters – across all chemotypes and within individual chemotypes. More detailed results of the effect of daughter and plot-level chemotype richness on plant traits can be found in Data S2.
Chemotype presence and plot-level chemotype richness effects on plot-level measurements
Related to hypothesis (iii), the presence or absence of certain chemotypes in a plot affected plot-level variables related to plant growth: number of stems, plant height, aboveground dry weight, and reproduction: the cumulative number of flower heads and flowering phenology. However, the effects of specific chemotypes were variable and differed with time in the growing season. Effects on the number of stems, number of flower heads, and flowering phenology are described and presented below, and additional effects on plant height and biomass are described in Data S2 and Figures S2-8–S2-10.
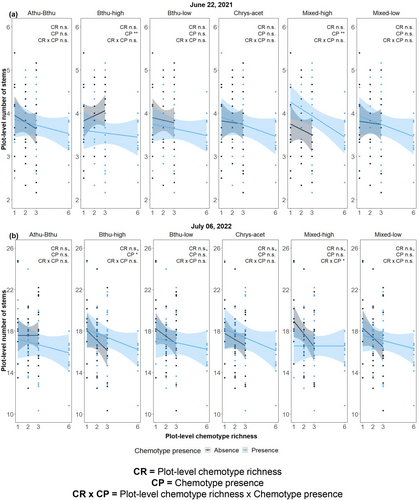
With respect to the mean number of stems per plot, three chemotypes significantly affected these in 2021, i.e., Bthu-low, Mixed-high, and Athu-Bthu on 01 July, and two on 22 July, i.e., Mixed-high and Bthu-high (Fig. 3a), but not on the other time point in 2022. The number of stems at the plot level significantly increased when Mixed-high was present in both time points (01 July: F1,75 = 12.85, P < 0.001; 22 July: F1,75 = 10.35, P = 0.002; Figure S2-7). The Mixed-high chemotype was also the one with the highest number of stems in 2021 for both dates. The presence of the Bthu-high chemotype lowered the number of stems at the plot level on 22 June 2021 (F1,75 = 8.10, P = 0.006; Fig. 3a). The effect of the presence/absence of the Mixed-high or Bthu-high chemotypes did not differ across chemotype richness levels, as indicated by the absence of interactions (Table S2-7). In 2022, chemotype presence/absence patterns on the number of stems differed slightly. Although we did not find a significant main effect of the plot-level chemotype richness of any of the chemotypes on the number of stems in any of the six separated models, we did observe an interaction between plot-level chemotype richness and the chemotype presence for the Mixed-high model (F1,75 = 4.29, P = 0.042; Fig. 3b). In plots where the Mixed-high chemotype was present, the number of stems did not differ, whereas in its absence, the number of stems decreased sharply with increasing plot-level chemotype richness. Across all six chemotype-specific models and at all time points in both years, plot-level chemotype richness had marginally significant negative effects on the mean number of stems per plot.
The number of flower heads per plot was affected positively by the presence of the Bthu-low chemotype in 2021 (F1,75 = 13.08, P < 0.001; Fig. 4a), probably because the Bthu-low chemotype produced the highest number of flower heads among the chemotypes (Fig. 2g). In contrast, the presence of the Chrys-acet chemotype negatively affected the number of flower heads (F1,75 = 12.12, P < 0.001). We also observed an interactive effect between the presence of the Bthu-low chemotype and plot-level chemotype richness (F1,75 = 5.25, P = 0.025), between the presence of the Mixed-high chemotype and plot-level chemotype richness (F1,75 = 4.98, P = 0.029), between the presence of the Bthu-high chemotype and plot-level chemotype richness (F1,75 = 7.08, P = 0.010), and between the presence of the Mixed-low chemotype and plot-level chemotype richness (F1,75 = 9.60, P = 0.003). In all the interactions, plot-level chemotype richness negatively affected flower number when the chemotypes were absent but not when they were present (Fig. 4a, Table S2-10).
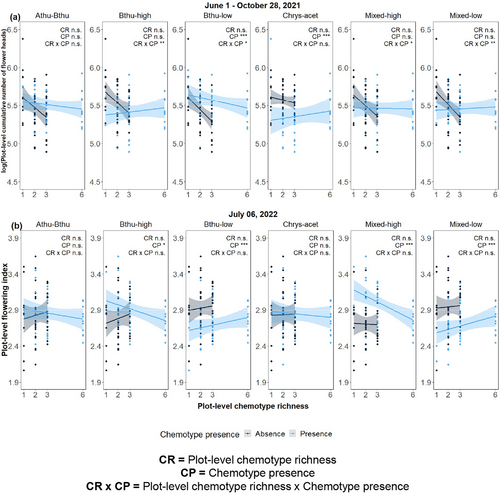
For the second growing season (2022), the presence/absence of certain chemotypes also strongly affected the flowering phenology of plots. This was true for the Bthu-high, Bthu-low, Mixed-low, and Mixed-high chemotypes (Fig. 4b, Table S2-10). The presence of the Bthu-high or Mixed-high chemotype resulted in a higher flowering index, i.e., advancing flowering phenology compared to the plots where they were not present (Bthu-high chemotype presence: F1,75 = 5.88, P = 0.018; Mixed-high chemotype presence: F1,75 = 37.90, P < 0.001). Conversely, plot-level flowering phenology was retarded (i.e., less advanced flowering phenology) when the Bthu-low or Mixed-low chemotype was present than when it was not (Bthu-low chemotype presence: F1,75 = 16.27, P < 0.001; Mixed-low chemotype presence: F1,75 = 22.42, P < 0.001). All other models of the presence/absence of other chemotypes were non-significant.
Overyielding calculations
Regarding our hypothesis (ii), we did not observe an overyielding effect in plot-level plant performance on plots with higher chemotype richness for any plot-level plant traits (plot-level chemotype richness always P > 0.05; Table S2-12). However, we observed a weak negative tendency towards plants producing fewer flower heads (χ2 = 2.57, P = 0.109) when they were associated with more chemotypes (chemotype richness 2, 3, 6) compared to their performance in monocultures (Figure S2-11).
Headspace VOC analysis
The T. vulgare headspace VOC collections led to the identification of 60 compounds (Table S2-1). Classification of VOCs and a comparison between plot-level headspace VOC profiles and plot-level chemotype richness are available in Data S2.
Plot-level theoretical leaf and plot-level realized volatile chemodiversity metrics
Our results confirmed that plots that had a higher plot-level chemotype richness (i.e., the number of different chemotypes present in the plot) also had higher theoretical leaf terpenoid richness (F1,82 = 20.48, R2 = 0.20, P < 0.001), diversity (F1,82 = 37.25, R2 = 0.31, P < 0.001), and evenness (F1,82 = 31.78, R2 = 0.28, P < 0.001) at the plot level (Fig. 5b–d). However, the theoretical leaf terpenoid concentration did not show any relationship with plot-level chemotype richness (F1,82 = 0.00, R2 = 0.00, P = 0.967; Fig. 5a). For the terpenoids in the headspace VOC profiles, plot-level chemotype richness had no significant effect on the abundance, richness, diversity, or evenness of terpenoids released as VOCs (Fig. 5e–h, Table S2-15). This rejects our hypothesis (iv) since plant communities with higher plot-level chemotype richness did not emit a more chemically diverse headspace of VOCs than plots with lower plot-level chemotype richness.
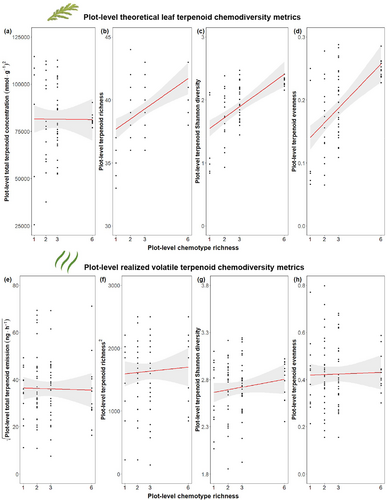
Plots that were more diverse in terms of their leaf terpenoids were not necessarily more diverse in their plot-level headspace terpenoid profile. However, there was a weak positive correlation between leaf and headspace terpenoid profiles (Mantel statistic R2 = 0.05, P < 0.001). As plots became more dissimilar in terms of their leaf terpenoids, they also became more dissimilar in terms of their headspace terpenoids.
DISCUSSION
Here, we designed a biodiversity field experiment in which we manipulated the number of chemotypes of Tanacetum vulgare plants at plot level to study how surrounding chemodiversity affects plant performance in the first 2 years after establishment. Our study showed that chemotypes initially differed in the studied morphological traits, confirming our first hypothesis (i). Concerning our second hypothesis (ii), the effects of plot-level chemodiversity on plot-level traits were only found for plot-level averages of reproductive traits, but not for growth-related traits. The third (iii) was also confirmed, i.e., the presence or absence of certain chemotypes and the plot-level chemotype richness influenced plot-level trait means. Importantly, the relationships between chemotypes, plot-level chemotype richness, and traits decreased over time in our 2-year study. Lastly, we found that the theoretical plot-level leaf terpenoid profiles significantly predicted the plot-level headspace terpenoid profiles, but the variance explained was low. However, plots that are chemically more diverse in terms of leaf terpenoids are not necessarily more diverse in their terpenoid headspace profile (hypothesis iv).
Our results show that tansy chemotypes vary in their chemical profiles and growth-related (number of stems and height) and reproductive traits (number of flower heads and flowering phenology) under field conditions. These findings broadly support the results of previous studies that used the same plant model system and found links between plant chemotypes and plant traits (Keskitalo et al. 2001; Neuhaus-Harr et al. 2023). For instance, in a previous study, tansy chemotypes with a high concentration of camphor were taller compared to other chemotypes rich in trans-thujone, artemisia ketone, 1,8-cineole, or davadone-D, and chemotypes rich in artemisia ketone or davadone-D produced more flower heads and flowered later compared with the other four chemotypes (Keskitalo et al. 2001). However, since Keskitalo et al. (2001) analysed the effect of chemical composition on certain traits at a broader geographic scale, and the chemotypes used by them and those used here are very different in their chemical composition, the current study's findings are – although conceptually similar – hard to directly compare. In an earlier study using the same chemotype lineages used here, the chemotypes showed similar growth patterns (Neuhaus-Harr et al. 2023). This is not entirely surprising but reveals that clonally-produced chemotypes show a high degree of consistency in phenotypes, at least in the early weeks of growth. As chemotypes in this study are maternally related, this seems a plausible reason why chemotypes may differ in some of their traits. However, daughter lines often differed more strongly within chemotypes than between chemotypes for many traits, making it unlikely that a maternal effect is the sole explanation (Figures S2-3–S2-6).
Variability in growth traits across chemotypes may enable individuals to partition local resources into different growth strategies and thus avoid intraspecific competition (Messier et al. 2010; Gallien 2017). For instance, the Bthu-low chemotype developed large biomass and typically few but taller and thicker stems compared with other chemotypes, such as Mixed-high, which developed more stems but these were shorter and thinner. Interestingly, however, the morphological differences between the chemotypes diminished over time. Only the Bthu-low chemotype remained quite distinct in both years, possibly because of its pronounced strategy of growth: tall and thick stems and high biomass. A likely explanation is the tendency for different daughters, even within chemotypes, to diverge in their trait expression over time, introducing enough variation to diminish chemotype-specific trait expression. This emphasizes the role of different growth strategies for individual survival, particularly during early establishment, and suggests that intraspecific chemodiversity might mediate niche realization processes (Müller & Junker 2022).
Contrary to our hypothesis, no differences were observed in plant growth parameters when plants grew in plots that differed in plot-level chemotype richness, indicating that the morphology of each chemotype is consistent across the environments in which they grow. Various possible explanations exist for why plant growth did not respond to plot-level chemotype richness. For instance, genotypes of T. vulgare can differ in their competitive ability in response to the presence of other plant species (Tse 2014), but little is known about the response to the presence of conspecifics. Although intra- or interspecific competition is expected to affect plant growth, differences between T. vulgare plants might be strongly determined by the genotype, especially in the early growth stage, so that intraspecific competition does not affect growth traits. It may very well be that growth responses to plot-level chemotype richness are not adaptive in T. vulgare, but as discussed below, responses do occur in other traits.
The measured T. vulgare reproductive traits also pronouncedly differed between chemotypes, but in contrast to what was observed for growth traits, plant-level reproductive traits responded to plot-level chemotype richness. Although Moreira et al. (2016) and Hughes et al. (2008) suggested that plants growing in plots that contained more chemotypes express higher individual plant fitness, in our study, all studied chemotypes, except for the Bthu-low chemotype, were inhibited (i.e., had lower flower head numbers) in plots with high plot-level chemotype richness compared to low-chemotype richness plots. This suggests that growth strategies that made the Bthu-low chemotype very dominant in height and weight in the first year might also bring some fitness advantages to this Bthu-low chemotype in highly diverse plant communities. It also suggests that most T. vulgare chemotypes might be negatively affected in more chemically diverse environments. This sharply contrasts with our expectations that chemically diverse environments would benefit plants. Our interpretation is based on the effect of chemodiversity on plant traits. Chemically diverse environments might still benefit plants by affecting plant interaction partners, as found by Ziaja & Müller (2023), who reported that some T. vulgare chemotypes benefit from neighbours that differ in chemotype in terms of lower herbivore load of Uroleucon tanaceti and Macrosiphoniella tanacetaria aphids. However, these authors did not report any plant performance parameters, and therefore, a direct comparison between their and our studies investigating plot-level chemotype richness effects is currently not possible.
Another key finding was that plot-level chemotype richness influenced flowering phenology in T. vulgare. In the case of the Bthu-low chemotype, the effect of plot-level chemotype richness was positive on the number of flower heads, but negative for the flowering index value, indicating a delayed onset of flowering. It appears that in different chemical environments, the flowering strategy differs across T. vulgare chemotypes. For instance, the Mixed-high, Athu-Bthu, and Mixed-low chemotypes had a more advanced flowering status at the time of assessment (high flowering index value) in plots with higher plot-level chemotype richness, while the Bthu-low, Chrys-acet, and Bthu-high chemotypes had more delayed phenology (low flowering index value) in such plots. We speculate that T. vulgare might be able to sense their neighbouring plants, either through direct competition (e.g., for space and nutrients), via the perception of volatiles (Heil & Karban 2010; Karban et al. 2014; Kessler & Kalske 2018; Ninkovic et al. 2019; Ninkovic et al. 2021), or absorption of semi-volatile compounds emitted by neighbouring plants (Himanen et al. 2010). As a result, T. vulgare may avoid competition by being reproductively active at different times, thereby optimizing their fitness. Variations in flowering phenology may also correlate with variations in interacting arthropod communities, which could affect reproductive success (Kuppler et al. 2016). Moreover, differences in flowering phenology across chemotypes might constitute a strategy to avoid cross-pollination between certain tansy chemotypes that could result in poor seed production (Keskitalo et al. 1998).
As predicted, plot-level average trait values were, in 2021, higher in mixture plots containing certain chemotypes that had either more stems, were taller, had larger aboveground dry weight, or produced more flower heads. In the same line, plot-level trait values decreased when chemotypes with fewer stems, smaller, lighter, or producing fewer flower heads were present. Several types of interaction can be observed, ranging from adverse effects related to competition, possibly for a limiting resource, to positive effects through facilitation, for instance, through increased resource availability or decreased herbivory (Roscher et al. 2005; Marquard et al. 2009; Ziaja & Müller 2023). In our study, the influence of a highly chemically diverse environment led to higher performance of the Bthu-low chemotype (i.e., a higher cumulative number of flower heads). At the plot level, individual chemotype contributions appear to become less pronounced over time, suggesting that the plots became more similar as they age, at least in terms of their morphological structure. Furthermore, we found no overyielding effects in our system, but instead, found a tendency towards lower plot-level plant trait values when increasing chemotype richness. This is important for future and ongoing work in this field experiment, which will investigate the role of chemodiversity on insect community assembly. A reduction of the plant- and plot-level differences in growth traits reduces the strength of potential confounding effects of plant growth on insect community assembly and will, therefore, help us to draw more robust conclusions and deepen our understanding of the consequences of chemodiversity. Moreover, chemotypes of tansy plants have been studied regionally in their distribution ranges (Wolf et al. 2012; Clancy 2021; Rahimova et al. 2023), and little attention has been paid to the implications of naturally co-occurring neighbouring chemotypes.
In line with our predictions, most diversity metrics based on theoretical plot-level leaf terpenoid profiles increased with increasing plot-level chemotype richness. We observed no differences between plot types for the total approximate terpenoid concentration. In contrast to what we hypothesized, we found no effect of plot-level chemotype richness on diversity metrics calculated based on headspace terpenoids. More diverse plots based on the theoretical leaf terpenoid of each chemotype were not necessarily more diverse in their plot-level headspace terpenoid profile. The headspace volatile terpenoid of a plant community is strongly dependent not only on the constitutive specialized metabolite profile of their individual plants, but also on the intrinsic chemical properties of their compounds (e.g., volatility and solubility), and on physiological and morphological plant features (such as the presence of trichomes; see Gershenzon & Dudareva (2007), He et al. (2011), Aschenbrenner et al. (2013)), and biological characteristics, such as the abundance and interacting organisms above- and belowground, plant age, and plant size (Takabayashi et al. 1994; McCormick et al. 2012; Kessler & Kalske 2018; Fabisch et al. 2019). Although volatile production is hard to accurately predict (Dudareva et al. 2006; Dicke et al. 2009), it is plausible that the volatile headspace profile will be related to the compounds found and stored in glandular trichomes in leaves of T. vulgare, since this plant has the capacity to store VOCs. However, while the hexane extraction was performed from the leaves of young plants before planting them in the field in 2021, the headspace terpenoids were obtained in the spring season 1 year after planting (2022) under field conditions (where the plants interacted with each other and other organisms) and analysed in a different lab. This could have led to changes in terpenoid diversity that may be reflected in our analysis (Eckert et al. 2023). Moreover, emitted volatile profiles might also differ from the stored profiles because some specific volatile organic compounds are produced only upon attack or under abiotic stress (e.g., green leaf volatiles, benzenoids, and terpenoids known as the herbivore-induced plant volatiles – HIPV; Rashid & Chung 2017; Unsicker et al. 2009). For example, Clancy et al. (2016) found that emitted T. vulgare headspace volatiles differed from compounds found in leaves at the individual level by 82%. However, the extent to which the chemotype richness and stored chemical profiles of a group of plants affect the volatiles in their communal headspace remains poorly understood. It will be interesting to see how the community-level volatile profile will develop over time when plants continue to grow and competition intensifies.
Further research should be undertaken to investigate the qualitative changes in plot-level volatile composition and how they correspond to plot-level cumulative leaf terpenoid profiles, but this was beyond the scope of the present study. Future investigations of leaf terpenoids and headspace terpenoids under field conditions over time, within and across growing seasons, upon induction, in response to environmental stresses, and including soil VOC from roots would help us to understand the temporal dynamics of plant terpenoid composition and volatile emission, and their effects on shaping plant–plant interactions. Furthermore, research on the mechanistic understanding of different defence strategies, such as the storage of defence metabolites and emission of volatiles, might help us understand these observed divergent patterns.
A mechanistic understanding of biodiversity–ecosystem functioning requires analysing the role of not only species number and functional groups, but also phylogenetic diversity and within-group variation in functional traits (Tilman et al. 1997). Our results provide insights into the underlying processes through which intraspecific chemodiversity acts on plant growth and reproductive traits. Although this study focuses on plant traits, this established field experiment also raises the possibility of further studying the role of intraspecific chemodiversity in interactions between plants and associated interaction partners, such as herbivorous insects, natural enemies, and pollinators. Insect diversity and abundance are typically positively correlated with plant species diversity and interspecific diversity of functional traits (Junker et al. 2015). However, the effect of intraspecific plant chemodiversity on shaping such interactions in the field has received limited attention to date (but see Bustos-Segura et al. 2017). The findings of this study suggest that intraspecific chemodiversity might influence ecosystem properties, such as primary productivity, resource use efficiency, ecosystem stability, and resilience. Given the degradation of morphological trait differences over time, this field experiment offers a unique opportunity to study the effect of chemodiversity by means of the constitutive plant terpenoid profiles in nature.
ACKNOWLEDGEMENTS
We thank the staff at TUM PTC Dürnast, particularly Sabine Zuber and Petra Scheuerer, for providing excellent care during the plant propagation and preparation phase. We thank Caroline Müller and Elisabeth Eilers for chemotyping the plants, and Anne Ebeling and Nico Eisenhauer for providing space within the Jena Experiment and allowing us to use core facilities under Gerlinde Kratzsch's guidance. We thank Nafiseh Mahdavi for her help in establishing the field experiment, Leonardo Moreno for his help in the maintenance of the plants and in collecting data in both years of the field experiment, and the Jena Experiment gardening staff for providing advice and help in the maintenance of this experiment. Many thanks to Beate Piecha, Annika Neuhaus-Harr, and other friends and colleagues who helped during the harvests. This study was funded by the German Research Foundation (DFG), project 415496540 (WE3081/40-1), as part of the Research Unit (RU) FOR 3000. We thank all members of the RU for their valuable discussions. Open Access funding enabled and organized by Projekt DEAL.
AUTHOR CONTRIBUTIONS
WWW conceived the original idea and designed the experiment. RH prepared the field experiment and propagated plants. LOP, RH, and WWW planted the field experiment. LOP collected all data and maintained the field experiment. LOP analysed ecological data with input from RH and WWW. PMvB, SBU, RH, and LOP prepared and executed volatile collections. PMvB chemically analysed volatiles. LOP wrote the manuscript with input from RH and WWW. All authors contributed to the final version and approved the final submission.
Open Research
DATA AVAILABILITY STATEMENT
Data are available on request.




