FAM46C controls antibody production by the polyadenylation of immunoglobulin mRNAs and inhibits cell migration in multiple myeloma
Abstract
FAM46C, frequently mutated in multiple myeloma (MM), has recently been shown to encode a non-canonical poly(A) polymerase (ncPAP). However, its target mRNAs and its role in MM pathogenesis remain mostly unknown. Using CRISPR-Cas9 technology and gene expression analysis, we found that the inactivation of FAM46C in MM down-regulates immunoglobulins (Igs) and several mRNAs encoding ER-resident proteins, including some involved in unfolded protein response and others that affect glycosylation. Interestingly, we show that FAM46C expression is induced during plasma cell (PC) differentiation and that Ig mRNAs encoding heavy and light chains are substrates of the ncPAP, as revealed by poly(A) tail-length determination assays. The absence of the ncPAP results in Ig mRNA poly(A) tail-shortening, leading to a reduction in mRNA and protein abundance. On the other hand, loss of FAM46C up-regulates metastasis-associated lncRNA MALAT1 and results in a sharp increase in the migration ability. This phenotype depends mainly on the activation of PI3K/Rac1 signalling, which might have significant therapeutic implications. In conclusion, our results identify Ig mRNAs as targets of FAM46C, reveal an important function of this protein during PC maturation to increase antibody production and suggest that its role as a tumour suppressor might be related to the inhibition of myeloma cell migration.
1 INTRODUCTION
Multiple myeloma (MM), the second most common haematological malignancy, arises from the abnormal proliferation of immunoglobulin-secreting clonal malignant plasma cells (PCs).1, 2 Genomic studies have demonstrated the genetic complexity of this cancer, which is characterized by marked clonal heterogeneity.3 Moreover, the role of the bone marrow (BM) microenvironment is crucial for the survival, migration and eventual dissemination of the disease.4-7 Recent progress in identifying somatic mutations using next-generation sequencing has revealed novel mutations in genes whose function in MM pathogenesis is largely unknown, such as FAM46C and DIS3.8
FAM46C is one of the most commonly mutated genes in MM, with somatic point mutations having been identified in about 10% of newly diagnosed MM cases.9, 10 The vast majority of these mutations are of an inactivating nature, frameshift or non-sense mutations, which indicates that FAM46C may function as a tumour suppressor.9 In addition, FAM46C is located in cytoband 1p12, which is known to be deleted in approximately 20% of MM patients. Loss of heterozygosity or mutations in FAM46C has been associated with shorter survival.11 Moreover, the acquisition of FAM46C mutations over time, as described in some longitudinal studies, suggests that loss of function of FAM46C might be a progression event in MM.12, 13
A recent study has demonstrated that FAM46C encodes an active non-canonical poly(A) polymerase14 and therefore may increase gene expression by extending the poly(A) tails of some mRNAs in the cytoplasm.15 Its authors showed that overexpression of FAM46C in human myeloma cell lines (HMCLs) carrying the mutated gene led to polyadenylation and stabilization of several mRNAs, and induced cell death.14 Another recent study found that overexpression of FAM46C induced substantial cytotoxicity in MM cells, up-regulated genes involved in unfolded protein response (UPR) and increased Ig light chain production.16 However, there are still many functional aspects of the impact of FAM46C loss on MM pathogenesis that remain to be elucidated. Here, we used CRISPR-Cas9 technology to delete endogenous FAM46C in different MM cell lines bearing the wild-type (WT) gene. The characterization of FAM46C KO clones revealed that the loss of FAM46C deregulated some migration-related factors and sharply increased the migratory ability of MM cells. These findings could explain the relationship between the presence of FAM46C mutations/deletions in patients with MM and the progression/poor prognosis of the disease. In addition, we revealed that both immunoglobulin light and heavy chain mRNAs are direct substrates of FAM46C. This finding demonstrates that the loss of polyadenylation activity is the mechanism by which antibody production is decreased in FAM46C KO clones.
2 MATERIALS AND METHODS
2.1 Cell lines
The human myeloma cell lines (HMCLs) JJN3 and RPMI-8226 were acquired from DMSZ, and U266 from ATCC. The cell lines were cultured as previously described.17 Cell line identity was confirmed within the last 3 years by STR analysis with PowerPlex 16 HS System kit (Promega) and online STR matching analysis. The presence of mycoplasma was routinely checked with MycoAlert kit (Lonza), and only, mycoplasma-free cells were used in the experiments.
2.2 CRISPR/Cas9-mediated generation of FAM46C knockout cells
FAM46C CRISPR-Cas9 knockout (KO) plasmids, consisting of a pool of three plasmids, each encoding the Cas9 nuclease and a target-specific 20 nt guide RNA (gRNA), were obtained from Santa Cruz Biotechnology (sc-407319). MM cells (1 × 106) were transfected with 5 μg of FAM46C CRISPR-Cas9 KO plasmids, or 5 μg of control CRISPR-Cas9 Plasmid (sc-418922), which contained a non-targeting 20 nt scramble guide gRNA. Transfections were carried out using the Amaxa Cell Line Nucleofector Kit V, the Amaxa Nucleofector device (Lonza), and programs T-016 for JJN3, X-005 for U266 and G-016 for RPMI-8226. Successful transfection of the CRISPR-Cas9 plasmids was confirmed by the detection of the plasmid encoded-green fluorescent protein (GFP). Single GFP + cells were sorted into 96-well plates 6 days after transfection using a Becton Dickinson FACSCalibur flow cytometer. To favour the growth of single cells, 50% filtered conditioned medium and 20% FBS were added to the culture medium. Isolated clones were expanded in culture over a period of 1 month, in the case of JJN3, or 2 months for U266 and RPMI-8226, and then genomic DNA was extracted. Clones were analysed by PCR using the primers FAM46C-FOR and FAM46C-REV (Table S1). Sanger sequencing was used to evaluate the alterations resulting from non-homologous end-joining (NHEJ) at the cut site.
2.3 Cell migration and invasion assays
MM cells were washed in serum-free culture medium and resuspended at a final concentration of 106 cells/mL. In the migration assays, 1.5 mL of cell suspension was seeded in the upper chamber of a 6-well, 8-µm pore Transwell plates (Corning-Costar) and 2.6 mL of RMPI-1640 with 20% serum was placed in the lower compartment. Invasion assays were performed using BioCoat Matrigel Invasion 24-well, 8.0-μm pore Transwell chambers (Corning-Costar). For these experiments, 500 µL of cells resuspended at 106 cells/mL in serum-free medium was seeded in the upper chamber, and 750 µL of medium containing 20% serum was used as the chemoattractant in the lower chamber. After 20 hours, cells migrating into the lower chambers were collected, resuspended in 400 μL PBS and counted using a BD Accuri C6 flow cytometer.
2.4 RNA extraction and microarray data analysis
Total RNA was extracted from three independent FAM46C WT or KO clones using an RNeasy mini kit (Qiagen). RNAs were then processed and used to hybridize Affymetrix PrimeView Human Gene Expression Arrays following the manufacturer's instructions. Raw data were background-adjusted, normalized and log2-transformed using the RMA algorithm18 available in the Affymetrix expression console (v.1.4.1). Microarray data were deposited in Gene Expression Omnibus (GEO) under accession number GSE114984. We compared the gene expression of the three FAM46C KO clones with each of the three control samples, resulting in nine comparisons. Genes with an absolute value of the fold change (FC) greater than 1.5 were selected for further analysis. A second approach using an absolute FC > 1.2 was carried out to assess the overrepresentation of gene ontology categories.
2.5 Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was reverse-transcribed to cDNA using a cDNA Reverse Transcription Kit from Applied Biosystems. qRT-PCR was performed using an iQ™ SYBR® Green Supermix kit (Bio-Rad), the iQ5 PCR detection system and the following gene-specific primers: FAM46C-FOR and FAM46C-REV2, GAPDH-FOR and GAPDH-REV (Table S1). Alternatively, and for other genes, expression was assessed using TaqMan qRT-PCR assays (Applied Biosystems). Relative gene expression was calculated by the 2−ΔCt method using GAPDH as the reference gene for normalization or 18S rRNA when indicated.
2.6 siRNA
HMCLs were transfected with 25 nmol/L of on-TARGET plus™ control pool or on-TARGET plus SMART pool Human FAM46C (Dharmacon). Transfections were carried out using the Amaxa Cell Line Nucleofector Kit V, the Amaxa Nucleofector device (Lonza) and program G-016.
2.7 Cell proliferation and cell viability assays
Cell proliferation and cell viability in the absence or presence of drugs was assessed by the MTT assay. Apoptosis was measured using annexin V-fluorescein isothiocyanate/propidium iodide (PI) double-staining (Immunostep) according to the manufacturer's procedure.
2.8 Poly (A) tail-length determination
mRNA poly(A) tail length was analysed using the USB® Poly(A) Tail-length assay kit from Affymetrix. Gene-specific forward primers were as follows: IGKC, SSR4, IGLC, BIP, IGHA1ns, IGHA1s, IGHEns and IGHEs (Table S1).
3 RESULTS
3.1 Knockout of FAM46C slightly affects the gene expression profile
To investigate the consequences of FAM46C inactivation for the pathogenesis of MM, we employed CRISPR-Cas9 technology to delete endogenous FAM46C in MM cells. The experiments were performed in the JJN3 cell line, which bears a wild-type (WT) FAM46C gene.10, 16 JJN3 cells were transfected with a pool of three plasmids each encoding a FAM46C-specific gRNA (Figure 1A) or with a control NT gRNA plasmid, and single GFP+ transfected cells were isolated by flow cytometry. The resulting clones were screened by PCR-Sanger sequencing (Figure 1B), qRT-PCR (Figure 1C) and Western blot (Figure 1D). Three clones knockout (KO) for FAM46C and three control WT clones were selected and used for further studies.
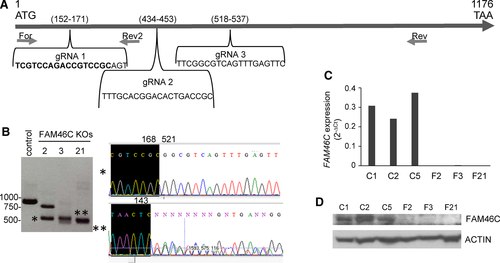
Recent studies have shown that FAM46C encodes a non-canonical mRNA poly(A) polymerase that may affect the stability of its target mRNAs.14, 19 In order to identify putative FAM46C substrates, total RNAs were extracted from FAM46C WT and KO clones and subsequently processed for microarray hybridization. Surprisingly, we found few genes whose levels of expression in all FAM46C KO clones changed relative to WT clones. Probe sets corresponding to eight genes (MAGED1, LRPAP1, RHOBTB1, GPX7, CKAP4, PCOLCE2, CD55 and HACD1) were underexpressed in the nine comparisons (three FAM46C KO cells compared with three controls) using a FC < −1.5 (Table S2), and 80 probe sets corresponding to 54 genes decreased their expression 1.2-fold (Table S3). On the other hand, no gene was up-regulated using a FC > 1.5, although 12 genes were overexpressed 1.2-fold in all FAM46C KO cells compared with controls (Table S4). When we considered at least six comparisons, nine genes were up-regulated 1.5-fold: EIF4E3, SKAP2, CCDC84, PAGE5, PROK2, SORBS2, MALAT1, PLSCR1 and TRA2 (Table S5).
3.2 Inactivation of FAM46C up-regulates oncogenic lncRNA MALAT1 and promotes cell migration and invasion in MM
Examining the functions of the genes deregulated in FAM46C KO clones, we found that some had previously been associated with cell migration and invasion: MAGED1 and RHOBTB1 inhibiting,20-22 and lnc RNA MALAT1, PROK2 and TRA2 promoting these activities.23-25 The deregulation of these five genes observed in the microarray analysis was validated by qRT-PCR (Figure 2A). Moreover, reduced levels of MAGED1 and RHOBTB1 proteins were confirmed by Western blot (Figure 2B). Then, we used the CRISPR-Cas9 technology to delete endogenous FAM46C in U266 and RPMI-8226, which also express wild-type FAM46C. In the same way as in JJN3, three WT and three FAM46C KO clones, confirmed by PCR, Sanger sequencing (data not shown) and Western blot (Figure S1), were selected and used in the experiments. Nevertheless, for some reason, U266 FAM46C KO clones stopped growing after several passages. RNA was obtained from the three U266 KO clones, but protein extracts only from clone F5. LncRNA MALAT1 was significantly up-regulated in FAM46C KO clones compared with WT clones in both U266 and RPMI-8226 (Figure 2C), whereas no statistically significant differences were detected in MAGED1, PROK2 and TRA2 expression between WT and KO clones (data not shown). An evident down-regulation of the tumour suppressor protein RHOBTB1 was observed in U266 FAM46C KO cells (Figure 2D), as previously found in JJN3 (Figure 2B). However, this effect was not detected in RPMI-8226 FAM46C KO clones.
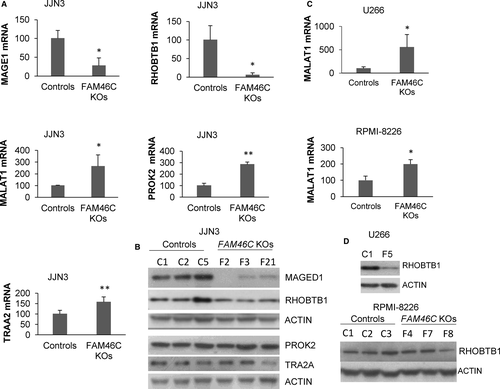
Deregulation of migration-related factors in FAM46C KO cells prompted us to carry out in vitro cell migration and invasion assays. Interestingly, we found that the number of migratory cells in JJN3 was almost 100 times greater in FAM46C KO clones than in WT controls (Figure 3A,B). Moreover, FAM46C KO cells also exhibited a greater invasive ability relative to WT cells (Figure 3C). To verify that the increase in cell mobility was independent of putative CRISPR-Cas9 off-target effects, we knocked down FAM46C expression by siRNA (Figure 3D) and performed cell migration assays. Down-regulation of FAM46C in JJN3 significantly increased the migratory ability of the cells (Figure 3E), confirming that loss of FAM46C promotes cell migration. Next, the in vitro cell migration assays were performed in the three RPMI-8226 FAM46C WT and KO clones. We found that the inactivation of the ncPAP induced a significant increase in migration, as previously demonstrated in JJN3 (Figure 3F).

3.3 Increased migration of FAM46C KO cells depends on PI3K activation
Next, we investigated molecular mechanisms that might underlie the increased migration and invasion of FAM46C KO cells. As matrix metalloproteinases (MMPs) are key mediators in cell invasion, we investigated whether the loss of FAM46C affected MMPs expression in MM cells. However, similar levels of MMP2 and MMP9 were found in FAM46C WT and KO cells (Figure 4A). In MM, an activation of the epithelial-mesenchymal transition (EMT) similar to the phenomenon observed in solid tumours, which is considered a key process for metastasis, has been described.26 EMT is characterized by the loss of E-cadherin, mediated by the up-regulation of its repressors such as Slug or Twist, and an increase in N-cadherin.27 Although MM cells are not epithelial cells, some MM cell lines express N and/or E-cadherin.28 We found that JJN3 cells, either FAM46C WT or KO, had no detectable level of N-cadherin, and a very low level of E-cadherin expression was observed in the different clones, with the exception of clone F2 (Figure 4A). Twist and Slug protein levels did not increase in FAM46C KO clones compared with WT cells. These results indicate that the increased rate of migration observed in FAM46C KO MM cells seems to be EMT-independent.
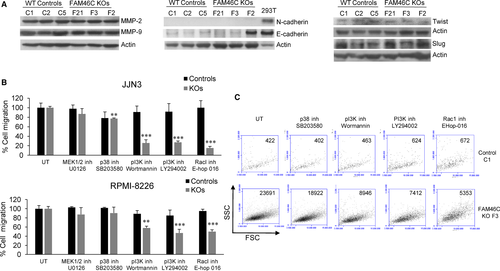
Previous studies have shown that MAPKs and PI3K can also regulate cell migration processes,29, 30 so we evaluated the influence of different kinase inhibitors on the migration ability of FAM46C WT and KO cells. We found that inhibition of MEK1/2 with U0126 did not affect migration of the cells and a small decrease in cell mobility was observed after inhibition of p-38 with SB203580 in JJN3 FAM46C KOs. However, a sharp reduction in cell migration occurred in FAM46C KO cells when PI3K was inhibited with Wortmannin or with the specific inhibitor LY294002 (Figure 4B). PI3K acts via diverse downstream signalling components, including the GTPase Rac1 and the kinase Akt (PKB), to promote cell mobility.31 We found that migration of FAM46C KO cells depended on Rac1 activation in these cells, as revealed by the fact that treatment with the specific inhibitor EHoP-016 also reduced cell mobility (Figure 4B). Representative dot-plots of JJN3 WT and FAM46C KO cells that migrated from the upper to the lower chamber in the absence or presence of the different inhibitors are shown in Figure 4C. The decrease in cell migration induced by the aforementioned inhibitors was not associated with any effect on cell proliferation, as WT and KO cells did not proliferate in the conditions assayed, neither on cell survival, as apoptosis was not observed at the doses employed (Figure S2).
3.4 Knockout of FAM46C does not increase proliferation rates or resistance to antimyeloma drugs
To determine whether loss of FAM46C affected cell growth rate, JJN3 and RPMI-8226 clones were cultured and proliferation was tested by the MTT assay. Growth rates did not increase in any of the FAM46C KO clones compared with controls. In fact, some of them, but not all, exhibited a slight decrease in growth ability (Figure 5A), suggesting that differences in growth rates may be clone-dependent and not FAM46C-dependent. On the other hand, silencing FAM46C in JJN3 and RPMI-8226 did not affect cell growth under the conditions tested (Figure 5B). We then evaluated the effect of common antimyeloma drugs in the different clones, and no significant differences were found in the sensitivity of FAM46C WT and KO clones to melphalan, bortezomib or dexamethasone (Figure 5C).
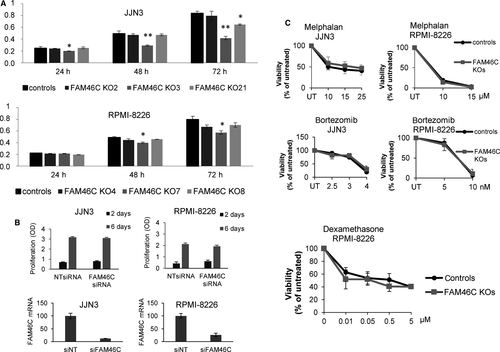
It has recently been reported that depletion of FAM46C in the XG1 MM cell line results in increased expression of IRF4, Bcl2 and ERK signalling activation.16 The authors proposed that these changes might explain the increased survival to dexamethasone and lenalidomide observed in this cell line. However, we observed similar amounts of these proteins in JJN3, U266 and RPMI-8226 cell lines in the presence or absence of FAM46C (Figure S3), indicating that the survival phenotypes could be cell line-dependent.
3.5 FAM46C is up-regulated during plasma cell differentiation and directly controls Ig production
Gene expression profiling of lymphomas from the Emu-myc transgenic mice identified FAM46C among the genes included in the B lymphocyte developmental signature32 using the GSEA platform33, 34 (MSigDB M1487 geneset, Table S6). These results suggested that FAM46C might be involved in PC differentiation, so we first quantified FAM46C mRNA levels by qRT-PCR in four BC populations, immature, naïve, memory B cells and PCs isolated from BM samples obtained from healthy donors.35 FAM46C expression was significantly higher in PCs than in the earlier stages of differentiation (Figure 6A, left panel) and was similar in PCs and in HMCLs (Figure 6A, right panel). However, when the expression of key genes involved in B-cell maturation was analysed in FAM46C WT and KO cells by qRT-PCR, no significant differences in their expression were observed between the samples (Figure 6B). These results indicate that loss of FAM46C does not revert the state of PC differentiation.
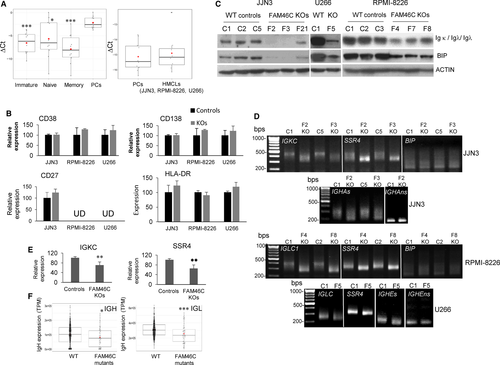
Zhu et al16 have recently described that FAM46C affects Ig light chain production. Consistent with this observation, we found in all FAM46C KO cells a clear reduction of Ig light chain and BIP protein, which is involved in the correct folding of the proteins in the ER36 (Figure 6C). Therefore, we wondered whether Ig and BIP mRNAs might be direct targets of the non-canonical poly(A)-polymerase activity of FAM46C. As shown in Figure 6D, poly(A) tails of mRNAs encoding Ig Kappa and Ig Lambda constant regions (IGKC, in JJN3 and IGLC1, in RPMI-8226 and U266) were clearly shorter in FAM46C KO cells than in the corresponding WT controls, revealing that Ig light chain mRNAs are direct substrates of the non-canonical poly(A) polymerase. In the case of heavy chains, there are two mRNAs, secretory and non-secretory, which are mainly expressed in PCs and B cells, respectively.37, 38 We found that IgH secretory mRNAs (IGHA1 in JJN3 and IGHE in U266) were also targets of FAM46C, but not the non-secretory IgH mRNAs, since in this case a band of similar size was amplified in both WT and FAM46C KO cells (RPMI-8226 only express Ig light chains). Conversely, we found that poly(A) tail length of BIP mRNA was similar in the different samples. SSR4, a known target of FAM46C,14 was used as a positive control. As expected, qRT-PCR showed that both SSR4 and IG mRNA were less abundant in FAM46C KO cells than in the WT clones (Figure 6E). The requirement of FAM46C for Ig production could therefore explain the increased expression of this ncPAP in PCs compared with previous stages of differentiation.
To validate the contribution of FAM46C to Ig production in the clinical setting, we used the RNAseq data corresponding to the MMRF CoMMpass trial (NCT01454297). We compared the levels of monoclonal Ig expression in patients carrying FAM46C non-sense mutations versus those expressing the FAM46C WT gene. A lower level of expression of monoclonal Ig, both Ig heavy and light chains, was observed in the patients with mutations of FAM46C (Figure 6F).
3.6 Inactivation of FAM46C decreases ER stress and down-regulates some genes involved in glycosylation
Functional enrichment analysis of the 54 genes down-regulated in JJN3 FAM46C KO cells (Table S3) failed to identify functional categories. However, the GO-term cellular component analysis showed that 17 genes encoded proteins located in the ER: AGA, DDOST, PDIA6, TMED10, CD55, ERP44, LMAN1, ERGIC3, MYDGF, GPX7, LRPAP1, TXNDC12, FKBP7, EDEM2, CANX, RPN1 and SSR4 (the known FAM46C mRNA substrate). Interestingly, four of them are involved in glycosylation: DDOST and RPN1, which both encode components of the N-oligosaccharyl transferase complex, LMAN1, involved in glycoprotein transport, AGA1, an aspartylglucosaminidase and SSR4, involved in translocating proteins across the ER membrane whose mutation causes a congenital disorder of glycosylation (www.genecards.org). Five of the genes, PDIA6, ERP44, TXNDC12, EDEM2 and CANX, have a role in tackling ER stress36 (www.genecards.org). These findings, together with the reduction in the level of BIP protein (Figure 6C), prompted us to consider the hypothesis that the decrease in Ig production, and probably of other proteins, in FAM46C KO cells could reduce the secretory cargo of the ER, leading to a diminished level of UPR. To test this, we monitored UPR activation in FAM46C WT and KO clones after treatment with the ER stress-inducer tunicamycin by means of RT-PCR analysis of spliced/unspliced XBP1 mRNA. We observed the appearance of spliced XBP1 4 hours after treatment with tunicamycin in all the tested clones, and its subsequent disappearance over time (Figure S4), as previously described.39 However, the XBP1 unspliced form remained more intense at 4 and 8 hours after treatment with tunicamycin in all JJN3 FAM46C KO clones compared with WT cells (Figure S4A). In RPMI-8226, the XBP1 unspliced forms reappeared after 30 hours of treatment and were also more intense in the FAM46C KO than in WT controls (Figure S4B). These results suggest a reduced level of ER stress in FAM46C KO cells.
Finally, we took advantage of the MMRF CoMMpass study (research.themmrf.org) and obtained a list of genes down-regulated in PCs from patients exhibiting FAM46C non-sense mutations. A total of 1681 genes were identified with an FDR < 0.05, of which 909 exhibited an FC < −2 (Table S7). The enrichment analysis using WebGestalt identified 13 functional categories (Table S8), and one of them was the ‘response to ER stress’ category, which included 33 genes. On the other hand, cellular component analysis found that 124 genes were located in the ER. When we cross-checked this list with that including the genes down-regulated in JJN3 FAM46C KO cells (Table S3), 11 genes emerged as being commonly down-regulated in all FAM46C KO clones and in MM patients carrying FAM46C mutations (Table S9). Interestingly, three of them participate in UPR (HSPA13, EDEM2 and PDIA6) and three are involved in glycosylation (DDOST, AGA and SSR4). These results suggest that inactivation of this non-canonical poly(A) polymerase may affect the glycosylation patterns of PCs.
4 DISCUSSION
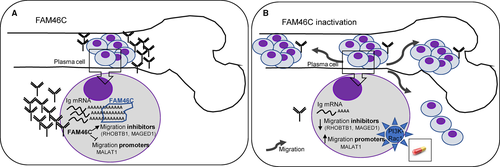
The poly(A) tail plays an important role in the post-transcriptional control of gene expression as it regulates mRNA transport, stability and translation. In addition to canonical nuclear poly(A) polymerase (PAP), which adds poly(A) tails to most eukaryotic mRNAs, seven non-canonical PAPs (ncPAPs) have been identified.15 FAM46C, which is frequently mutated in MM, has recently been described as a new ncPAP.14, 19 However, its function in myeloma cells remains to be elucidated. In this study, we showed that FAM46C is up-regulated during PC differentiation to increase antibody production by extending the poly(A) tail of Ig mRNAs. Moreover, we demonstrated that inactivation of FAM46C in MM sharply increased the migratory ability of PCs, which might explain the poor prognosis of MM patients with FAM46C abnormalities and the role of this gene as a tumour suppressor (Figure 7).
The terminal differentiation of B cells, which bear surface Ig, into antibody-secreting plasma cells is accompanied by a substantial increase in the abundance of the mRNAs of both Ig heavy and light chains.40 It has been reported that this increase is not due to a higher transcription rate but to an extended half-life of Ig mRNAs.40 In the case of IgH chains, two different mRNAs from a single primary transcript exist: B cells produce more mRNA encoding the membrane-associated protein, while plasma cells contain greater amounts of the Ig mRNA encoding the secreted protein. Regulation of this process requires competing splice and cleavage-polyadenylation reactions with balanced efficiencies.37, 38 In this study, we found that poly(A) tails of IgH secretory-specific mRNAs and Ig light chain mRNAs are controlled by FAM46C. Inactivation of the ncPAP reduced Ig poly(A) tail length and reduced the amount of Ig mRNAs and, consequently, the Ig protein levels. These results indicate that one of the functions of FAM46C in myeloma cells is to increase Ig production, which could explain the up-regulation of this ncPAP in PCs relative to that observed in earlier stages of B-cell differentiation. Importantly, a correlation between the presence of FAM46C mutations and significantly lower Ig mRNA levels was also observed in MM patients.
GEP of FAM46C WT and KO clones revealed that few genes were deregulated after FAM46C inactivation. These results indicate either that loss of FAM46C slightly affects mRNA stability under normal culture conditions and do not significantly alter RNA steady-state levels, or that FAM46C has few RNA targets. Our results contrast with those of Mroczek et al,14 who found 538 genes commonly up-regulated by FAM46C overexpression in two HMCLs carrying mutations in FAM46C. They also found hundreds of mRNAs that shifted towards longer poly(A) fractions in response to forced FAM46C expression. The most likely explanation for these apparently contradictory results is that ectopic overexpression of FAM46C may result in forced poly(A) addition to mRNAs. Another difference concerns the increased cell proliferation after FAM46C silencing previously reported,14, 16 which was not observed in our study, neither by silencing nor by complete inactivation. These discrepancies might depend on cell lines, clones or experimental conditions. In any case, the top hit FAM46C substrate described by Mroczek et al, SSR4, was also identified in our study, and several ER-resident proteins were found to be affected by FAM46C in both studies. SSR4 was down-regulated secondarily to the shortening of its mRNA poly(A) tail after FAM46C inactivation, which confirms this mRNA as a bona fide FAM46C substrate. It is of note that SSR4 was also down-regulated in MM patients from the CoMMpass study who carried FAM46C non-sense mutations. Several genes down-regulated by FAM46C inactivation have been associated with glycosylation. Down-regulation of these factors might affect glycosylation patterns of PCs, which may have implications for cell signalling or cell-matrix interactions.41 Alternatively, down-regulation of these genes may respond to the lowered glycosylation demand arising from decreased Ig production, as Igs are glycosylated proteins.42 This hypothesis is supported by the fact that some of them (DDOST, RPN1 and LMAN1) are up-regulated in PCs from mice (MSigDB M1487 geneset), suggesting a greater demand for these factors during PC differentiation to glycosylate Igs.
Inactivation of FAM46C is expected to induce pleiotropic effects in the cells, not only by deregulation of mRNAs, but also because changes in poly(A) tail length may affect mRNAs translatability giving rise to modifications in the proteome. Here, we found that FAM46C inactivation substantially increased the migratory ability of MM cells. We found that some of the few genes deregulated in the FAM46C KO clones, such as MAGED1, RHOBTB1 and MALAT1, had previously been associated with cell migration20, 22, 23, 43 MALAT1 was found to be up-regulated by FAM46C inactivation in the three HMCLs used in this study. This lncRNA was overexpressed in a wide variety of solid tumours and also in haematological malignancies, including MM.43-45 Moreover, high levels of expression of MALAT1 in MM have been associated with the onset of the disease, progression from normal PCs to MM and extramedullary dissemination.44-47 MAGED1 was down-regulated in JJN3 FAM46C KO clones and RHOBTB1 in two (JJN3 and U266) out of the three HMCLs analysed. The down-regulation of MAGED1 has also been reported in the XG1 HMCL knocked out for FAM46C.16 Although the down-regulation of these factors by FAM46C inactivation might contribute to the great migration ability observed in JJN3 FAM46C KO cells, this effect seems to be cell type-specific and genetic background-dependent. Consistent with our findings, a recent study has demonstrated that overexpression of FAM46C in hepatocellular carcinoma reduced cell migration and invasion.48 The authors showed that forced expression of the ncPAP suppressed EMT. However, we found here that the increased migratory ability of FAM46C KO MM cells seems to be EMT-independent. Conversely, we found that the increased rate of migration induced by FAM46C loss depended on PI3K-Rac1 activation in the KO cells, which may have important therapeutic implications, as patients with FAM46C mutations are likely to benefit from PI3K or Rac1 inhibitors49, 50 (Figure 7).
Future investigation should focus on the molecular mechanisms that explain the connections between FAM46C inactivation, the lowering of several ER-resident proteins and the UPR. Moreover, additional studies are needed to correlate all the findings obtained in MM cell lines with the phenotypes observed in patients, especially regarding migration of FAM46C-mutated myeloma cells and its relation with MALAT1 up-regulation.
ACKNOWLEDGEMENTS
This work was supported by the Samuel Solórzano Barruso foundation (FS/25-2015), the Spanish Association for Cancer Research (AECC, GCB120981SAN) and Institute of Health Carlos III/co-funding by FEDER (PI16/01074). The authors thank Isabel Isidro, Teresa Prieto, Vanesa Gutierrez and Laura San Segundo for their technical assistance, and Enrique Ocio and Mercedes Garayoa for helpful discussion. The authors thank the MMRF for access to the CoMMpass data set.
CONFLICT OF INTEREST
The authors declare no conflict interest.
AUTHOR CONTRIBUTIONS
ABH designed the study, performed most of the experiments and wrote the paper. DQ carried out the siRNA and qRT-PCR experiments. LAC performed all the bioinformatic and statistical analyses. RGS supervised the statistical analyses and contributed to the research tools. MVM revised the manuscript and contributed to the research tools. NCG supervised the experiments, corrected and approved the final version of the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. Additional methods are detailed in Appendix S1.




