Up-regulation of DDIT4 predicts poor prognosis in acute myeloid leukaemia
Abstract
The mammalian target of rapamycin (mTOR) inhibitor, DNA damage inducible transcript 4 (DDIT4), has inducible expression in response to various cellular stresses. In multiple malignancies, studies have shown that DDIT4 participates in tumorigenesis and impacts patient survival. We aimed to study the prognostic value of DDIT4 in acute myeloid leukaemia (AML), which is currently unclear. Firstly, The Cancer Genome Atlas was screened for AML patients with complete clinical characteristics and DDIT4 expression data. A total of 155 patients were included and stratified according to the treatment modality and the median DDIT4 expression levels. High DDIT4 expressers had shorter overall survival (OS) and event-free survival (EFS) than the low expressers among the chemotherapy-only group (all P < .001); EFS and OS were similar in the high and low DDIT4 expressers of the allogeneic haematopoietic stem cell transplantation (allo-HSCT) group. Furthermore, in the DDIT4high group, patients treated with allo-HSCT had longer EFS and OS than those who received chemotherapy alone (all P < .01). In the DDIT4low group, OS and EFS were similar in different treatment groups. Secondly, we analysed two other cytogenetically normal AML (CN-AML) cohorts derived from the Gene Expression Omnibus database, which confirmed that high DDIT4 expression was associated with poorer survival. Gene Ontology (GO) enrichment analysis showed that the genes related to DDIT4 expression were mainly concentrated in the acute and chronic myeloid leukaemia signalling pathways. Collectively, our study indicates that high DDIT4 expression may serve as a poor prognostic factor for AML, but its prognostic effects could be outweighed by allo-HSCT.
1 INTRODUCTION
One of the key features of acute myeloid leukaemia (AML), a group of very aggressive myeloid malignancies, is their strikingly heterogenous outcomes.1 Prognostication using clinical and molecular markers is crucial in designing treatment plans. The currently used risk stratification system still has significant intragroup heterogeneity, especially in the intermediate-risk group.2 Therefore, discovering appropriate biomarkers remains a research hotspot in AML. Over the years, it has been confirmed that NPM1 and double CEBPA mutations are favourable biomarkers, whereas FLT3-ITD mutation is associated with poor prognosis.3-5 In addition to genetic mutations, aberrant oncogene expressions have also been proposed as a tool for risk stratification. For example, high expressions of sFRP2 and DOK7 may suggest better prognosis,6, 7 but high expressions of FHL2 and iASPP may indicate poor survival in AML.8
DNA damage inducible transcript 4 (DDIT4), also known as REDD1 or RTP801, is induced by various cellular stress conditions, such as hypoxia, endoplasmic reticulum stress, oxidative stress, heat shock and starvation.9 It inhibits the activity of mammalian target of rapamycin complex 1 (mTORC1), a major player in cell growth, proliferation and survival. Abnormally elevated DDIT4 expression has been found in various malignant tumours.10, 11 Though rapamycin-derived mTOR inhibitors are powerful drugs in treating cancer, paradoxically, the naturally occurring DDIT4 seems to protect the cancer cells from apoptosis.10, 12, 13 Murine lymphocytes become more sensitive to dexamethasone-induced cell death after DDIT4 knockdown.12 Additionally, DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways.14
Recent studies indicated that high DDIT4 expression is also an adverse factor in AML.10, 15 However, the specific prognostic value of DDIT4 in AML is unknown. We aimed to evaluate the impact of DDIT4 expression on survival and its associated gene expression patterns in AML patients treated with chemotherapy or transplantation.
2 MATERIALS AND METHODS
2.1 Patients
The first cohort included 155 de novo AML patients with DDIT4 expression data, derived from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/).16 Eighty-four patients received chemotherapy alone, whereas 71 had allogeneic haematopoietic stem cell transplantation (allo-HSCT). The baseline clinical and molecular characteristics, follow-up and survival data were publicly available from TCGA website, including gender, age, white blood cell (WBC) count, bone marrow (BM) and peripheral blood (PB) blast percentages, French-American-British (FAB) subtype, karyotype, cytogenetic risk classification, RNA and microRNA sequencing data, and gene mutation spectrum.
The second cohort contained two Gene Expression Omnibus (GEO) datasets, GSE6891 and GSE12417, including 334 and 162 patients with cytogenetically normal AML (CN-AML), respectively. This cohort was mainly used to validate the findings of the first cohort. Affymetrix Human Genome 133 plus 2.0 and U133A gene chips were used to obtain gene expression profiles from the GSE6891 and GSE12417 datasets, and the entire process was fully compliant with the standard Affymetrix protocols. All patients’ clinical, molecular and microarray data were public accessible in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
2.2 Statistical analysis
Descriptive statistics were used to summarize the clinical and molecular characteristics of the patients. Datasets were described by median and/or range. Between-group comparisons of numerical and categorical data were performed by the Mann-Whitney U test and the chi-square test, respectively. Primary endpoints were event-free survival (EFS) and overall survival (OS). The former was defined as the time from diagnosis to the first event including relapse, death, failure to achieve complete remission, or was censored at the last follow-up. The latter was the time from diagnosis to death from any cause, or was censored at the last follow-up. Between-group comparisons of OS and EFS were performed by the Kaplan-Meier method and the log-rank test. Multivariate Cox proportional hazard models were constructed for OS and EFS using a limited backward elimination procedure. Spearman rank correlation was used to determine the associations between gene expression profile and DDIT4 expression. Multiple testing errors were assessed by false discovery rate (FDR). Gene Ontology (GO) enrichment analysis was conducted to assess enrichment of gene expression products associated with DDIT4. All tests were two-tailed. Statistical significance was defined as P < .05. All statistical analyses were performed by R software 3.5.0, SPSS software 24.0 and GraphPad Prism software 7.0.
3 RESULTS
3.1 Differences in clinical and molecular characteristics between different DDIT4 expression groups
In order to evaluate the prognostic significance of DDIT4 in AML, the first cohort was divided into the chemotherapy-only group and the allo-HSCT group. Within each group, the respective median DDIT4 expression level was used to divide the group into high and low expression subgroups, and the clinical and molecular characteristics of subgroups were compared (Table 1).
| Characteristics | Chemotherapy-only group | Allo-HSCT group | ||||
|---|---|---|---|---|---|---|
| High DDIT4 (n = 42) | Low DDIT4 (n = 42) | P | High DDIT4 (n = 35) | Low DDIT4 (n = 36) | P | |
| Age/years, median (range) | 70 (35-88) | 63 (22-82) | .003a | 53 (18-72) | 50 (21-65) | .411a |
| Age group/n (%) | ||||||
| <60 y | 8 (19.0) | 18 (42.9) | .018b | 23 (65.7) | 29 (80.6) | .158b |
| ≥60 y | 34 (81.0) | 24 (57.1) | 12 (34.3) | 7 (19.4) | ||
| Gender/n (%) | ||||||
| Male | 25 (59.5) | 20 (47.6) | .274b | 20 (57.1) | 21 (58.3) | .919b |
| Female | 17 (40.5) | 22 (52.4) | 15 (42.9) | 15 (41.7) | ||
| WBC/×109/L, median (range) | 13.3 (1.0-297.4) | 16.1 (0.7-171.9) | .522a | 34.2 (0.6-223.8) | 29.4 (0.9-115.4) | .761a |
| BM blast/%, median (range) | 77 (32-99) | 66 (30-95) | .041a | 72 (30-100) | 70 (34-99) | .913a |
| PB blast/%, median (range) | 22 (0-98) | 25 (0-91) | .449a | 53 (0-96) | 45 (0-94) | .366a |
| FAB subtypes/n (%) | ||||||
| M0 | 7 (16.7) | 0 (0.0) | .012b | 4 (11.4) | 5 (13.9) | .755b |
| M1 | 10 (23.8) | 10 (23.8) | 1.000b | 17 (48.6) | 6 (16.7) | .004b |
| M2 | 7 (16.7) | 14 (33.3) | .078b | 9 (25.7) | 9 (25.0) | 1.000b |
| M3 | 0 (0.0) | 0 (0.0) | 1.000b | 0 (0.0) | 1 (2.8) | .493b |
| M4 | 8 (19.0) | 12 (28.6) | .306b | 1 (2.9) | 12 (33.3) | .001b |
| M5 | 6 (14.3) | 6 (14.3) | 1.000b | 2 (5.7) | 2 (5.6) | 1.000b |
| M6 | 1 (2.4) | 0 (0.0) | 1.000b | 1 (2.9) | 0 (0.0) | .493b |
| M7 | 3 (7.1) | 0 (0.0) | .241b | 1 (2.9) | 1 (2.8) | 1.000b |
| Cytogenetics/n (%) | ||||||
| Normal | 19 (45.2) | 21 (50.0) | .662b | 18 (51.4) | 15 (41.7) | .410b |
| Complex karyotype | 9 (21.4) | 2 (4.8) | .048b | 9 (25.7) | 2 (5.6) | .019b |
| 8 Trisomy | 0 (0.0) | 0 (0.0) | 1.000b | 3 (8.6) | 3 (8.3) | 1.000b |
| inv(16)/CBFβ-MYH11 | 0 (0.0) | 6 (14.3) | .026b | 0 (0.0) | 5 (13.9) | .054b |
| 11q23/MLL | 1 (2.4) | 2 (4.8) | .614b | 1 (2.9) | 2 (5.6) | 1.000b |
| −7/7q- | 2 (4.8) | 1 (2.4) | .614b | 2 (5.7) | 1 (2.8) | .614b |
| t(15;17)/PML-RARA | 0 (0.0) | 0 (0.0) | 1.000b | 0 (0.0) | 1 (2.8) | 1.000b |
| t(9;22)/BCR-ABL1 | 1 (2.4) | 0 (0.0) | 1.000b | 1 (2.9) | 1 (2.8) | 1.000b |
| t(8;21)/RUNX1-RUNX1T1 | 0 (0.0) | 6 (14.3) | .026b | 0 (0.0) | 1 (2.8) | 1.000b |
| Others | 10 (23.8) | 4 (9.5) | .079b | 1 (2.9) | 5 (13.9) | .199b |
| Risk/n (%) | ||||||
| Good | 0 (0.0) | 12 (28.6) | .000b | 0 (0.0) | 7 (19.4) | .011b |
| Intermediate | 26 (61.9) | 20 (47.6) | .188b | 19 (54.3) | 21 (58.3) | .731b |
| Poor | 16 (38.1) | 10 (23.8) | .157b | 16 (45.7) | 8 (22.2) | .036b |
| FLT3-ITD/n (%) | ||||||
| Positive | 6 (14.3) | 9 (21.4) | .393b | 9 (25.7) | 8 (22.2) | .730b |
| Negative | 36 (85.7) | 33 (78.6) | 26 (74.3) | 28 (77.8) | ||
| NPM1/n (%) | ||||||
| Mutation | 13 (31.0) | 14 (13.3) | .815b | 9 (25.7) | 9 (25.0) | .945b |
| Wild type | 29 (69.0) | 28 (66.7) | 26 (74.3) | 27 (75.0) | ||
| DNMT3A/n (%) | ||||||
| Mutation | 12 (28.6) | 11 (26.2) | .807b | 10 (28.6) | 7 (19.4) | .368b |
| Wild type | 30 (71.4) | 31 (73.8) | 25 (71.4) | 29 (80.6) | ||
| IDH1/IDH2/n (%) | ||||||
| Mutation | 9 (21.4) | 6 (14.3) | .393b | 11 (31.4) | 6 (16.7) | .145b |
| Wild type | 33 (78.6) | 36 (85.7) | 24 (68.6) | 30 (83.3) | ||
| RUNX1/n (%) | ||||||
| Mutation | 12 (28.6) | 2 (4.8) | .003b | 3 (8.6) | 5 (13.9) | .710b |
| Wild type | 30 (71.4) | 42 (95.2) | 32 (91.4) | 31 (86.1) | ||
| NRAS/KRAS/n (%) | ||||||
| Mutation | 6 (14.3) | 6 (14.3) | 1.000b | 1 (2.9) | 6 (16.7) | .107b |
| Wild type | 36 (85.7) | 36 (85.7) | 34 (97.1) | 30 (83.3) | ||
| TET2/n (%) | ||||||
| Mutation | 4 (9.5) | 7 (16.7) | .332b | 0 (0.0) | 4 (11.1) | .115b |
| Wild type | 38 (90.5) | 35 (83.3) | 35 (100) | 32 (88.9) | ||
| TP53/n (%) | ||||||
| Mutation | 9 (21.4) | 2 (4.8) | .024b | 3 (8.6) | 1 (2.8) | .357b |
| Wild type | 33 (78.6) | 40 (95.2) | 32 (91.4) | 35 (97.2) | ||
- Abbreviations: BM, bone marrow; FAB, French-American-British; PB, peripheral blood; WBC, white blood cell.
- a Denotes Mann-Whitney U test
- b Denotes chi-square test.
In the chemotherapy-only group, compared with the DDIT4low subgroup, the DDIT4high subgroup had more patients ≥60 years old (P = .018), FAB-M0 (P = .012), with complex karyotype (P = .048), more frequent TP53 and RUNX1 mutations (P = .024, P = .003, respectively), and higher BM blast percentage (P = .041). It had fewer patients with CBFβ-MYH11 or RUNX1-RUNX1T1 (all P = .026) or good cytogenetic risk (P < .001). Gender distribution, WBC count, PB blast percentage and the frequencies of other recurrent genetic mutations (NPM1, FLT3, NRAS/KRAS, IDH1/IDH2, DNMT3A and TET2) were similar in the two subgroups.
In the allo-HSCT group, compared with the DDIT4low subgroup, the DDIT4high subgroup had more patients with FAB-M1 (P = .004), complex karyotype (P = .019) and poor cytogenetic risk (P = .036), yet fewer patients with FAB-M4 (P = .001) or good cytogenetic risk (P = .011). Age, gender distribution, WBC count, BM/PB blast percentage and the frequencies of recurrent gene mutations (NPM1, FLT3, RUNX1, DNMT3A, NRAS/KRAS, IDH1/IDH2, TP53 and TET2) were not statistically different between the two subgroups.
3.2 Prognostic value of DDIT4 expression in AML
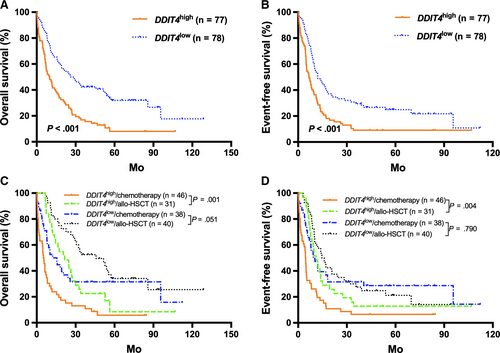
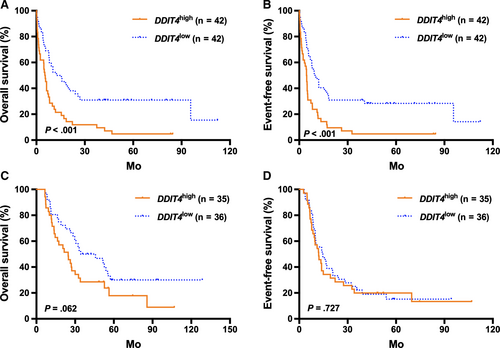
In the TCGA cohort, high DDIT4 expressers generally had significantly shorter OS and EFS than the low expressers (all P < .001; Figure 1A,B). Then, patients were further stratified according to the treatment modality and the median DDIT4 expression levels in the different treatment subgroups. For the high expressors (n = 77), those treated with allo-HSCT had significantly better survival than those who received chemotherapy alone (all P < .01, Figure 1C,D). For the low expressors (n = 78), treatment modality did not have outstanding influence on survival (all P > .05, Figure 1C,D). Kaplan-Meier analysis demonstrated that in the chemotherapy-only group, high DDIT4 expressers had significantly shorter OS and EFS than the low expressers (all P < .001, Figure 2A,B), whereas the survival was similar in the high and low expressors of the allo-HSCT group (all P > .05, Figure 2C,D).
3.3 Multivariate analysis of possible prognostic factors
To examine whether the impact of DDIT4 expression on AML survival was independent, we constructed multivariate Cox proportional hazard models using multiple variables, including DDIT4 expression (high vs low), WBC count (≥15 vs <15 × 109/L), age (≥60 vs <60 years), BM blasts (≥70 vs <70%), PB blasts (≥20 vs <20%), NPM1 (mutated vs wild), DNMT3A (mutated vs wild), TET2 (mutated vs wild), TP53 (mutated vs wild) and FLT3-ITD (positive vs negative) (Table 2).
| Variables | EFS | OS | ||
|---|---|---|---|---|
| HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Chemotherapy-only group | ||||
| DDIT4 (high vs Low) | 1.940 (1.158-3.251) | .012 | 1.878 (1.118-3.155) | .017 |
| Age (≥60 vs <60 y) | 3.051 (1.636-5.688) | .000 | 2.763 (1.478-5.167) | .001 |
| WBC count (≥15 vs <15 × 109/L) | 1.470 (0.816-2.646) | .199 | 1.407 (0.795-2.490) | .241 |
| BM blasts (≥70 vs <70%) | 1.863 (1.054-3.293) | .032 | 1.804 (1.025-3.176) | .041 |
| PB blasts (≥20 vs <20%) | 0.928 (0.548-1.571) | .781 | 0.868 (0.507-1.487) | .606 |
| FLT3-ITD (positive vs negative) | 1.245 (0.635-2.442) | .523 | 1.183 (0.582-2.408) | .642 |
| NPM1 (mutated vs wild) | 0.947 (0.491-1.826) | .871 | 0.772 (0.401-1.488) | .440 |
| DNMT3A (mutated vs wild) | 1.704 (0.979-2.965) | .059 | 1.772 (1.033-3.042) | .038 |
| TET2 (mutated vs wild) | 0.943 (0.429-2.074) | .885 | 0.672 (0.309-1.460) | .315 |
| TP53 (mutated vs wild) | 3.006 (1.372-6.587) | .006 | 2.444 (1.135-5.261) | .022 |
| Allo-HSCT | ||||
| DDIT4 (high vs Low) | 0.995 (0.557-1.775) | .985 | 1.622 (0.875-3.004) | .124 |
| Age (≥60 vs <60 y) | 1.097 (0.546-2.204) | .796 | 1.404 (0.710-2.777) | .330 |
| WBC count (≥15 vs <15 × 109/L) | 1.918 (1.032-3.563) | .039 | 1.537 (0.794-2.975) | .202 |
| BM blasts (≥70 vs <70%) | 0.777 (0.420-1.437) | .421 | 0.782 (0.391-1.566) | .488 |
| PB blasts (≥20 vs <20%) | 1.235 (0.633-2.408) | .536 | 1.445 (0.695-3.005) | .325 |
| FLT3-ITD (positive vs negative) | 2.462 (1.209-5.015) | .013 | 2.354 (1.048-5.291) | .038 |
| NPM1 (mutated vs wild) | 0.636 (0.312-1.298) | .213 | 0.625 (0.277-1.409) | .257 |
| DNMT3A (mutated vs wild) | 1.177 (0.605-2.292) | .631 | 1.377 (0.691-2.743) | .363 |
| TET2 (mutated vs wild) | 0.516 (0.145-1.838) | .307 | 0.961 (0.267-3.452) | .951 |
| TP53 (mutated vs wild) | 3.046 (0.884-10.495) | .078 | 7.196 (1.871-27.67) | .004 |
- Abbreviations: BM, bone marrow; CI, confidence interval; EFS, Event-free survival; HR, hazard ratio; OS, Overall survival; PB, peripheral blood; WBC, white blood cell.
In the chemotherapy-only group, high DDIT4 expression was an independent risk factor for both EFS and OS, along with age ≥ 60, BM blasts ≥ 70% and TP53 mutation (all P < .05). In addition, DNMT3A mutation was an independent risk factor for OS (P = .038). In the allo-HSCT group, FLT3-ITD was an independent risk factor for OS and EFS (all P < .05), and WBC count ≥ 15 × 109/L and TP53 mutation were independent risk factors for EFS (P = .039) and OS (P = .004), respectively, but DDIT4 expression was not an independent factor for survival.
3.4 Associations between genome-wide gene expression profile and DDIT4 expression
To elucidate the possible mechanism for the influence of DDIT4 in AML, DDIT4-associated gene expression profiles were summarized utilizing the high-throughput sequencing information from TCGA database. Three hundred and sixty-eight up-regulated and 171 down-regulated genes that were significantly associated with DDIT4 expression (P < .05, fold change = 1.5, Figure 3A) were screened. Eventually, with a more rigorous analysis (fold change = 2), 359 genes were excluded, and the remaining 180 genes were depicted in an aberrant expression heatmap (Figure 3B). Many leukaemia-associated genes were positively associated with DDIT4 expression, including WNT9A, NOTCH3, SOCS1, MCL1, HIF1A, ALOX5, CD47, CXCR4, CDK9, HRAS, PLK3 and ETS2. However, RPL5, a tumour suppressor in multiple cancers, was negatively correlated with DDIT4 expression. Furthermore, GO enrichment analysis suggested that the genes related to DDIT4 expression were mainly concentrated in "acute and chronic myeloid leukaemia," "bladder cancer," "hedgehog signalling pathway," "endometrial cancer," and "basal cell carcinoma" signalling pathways (Figure 3C).

3.5 Validation of the prognostic value of DDIT4 expression in AML
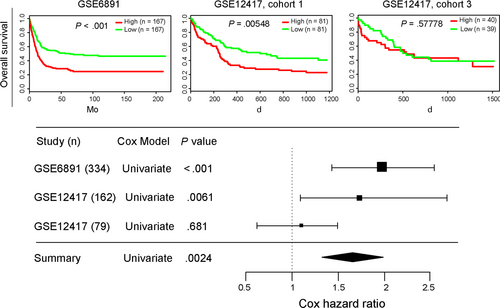
In the two other large CN-AML cohorts from the GEO database, high DDIT4 expression was also related to significantly shorter OS. Combining the data with the TCGA cohort, results consistently showed that up-regulated expression of DDIT4 had deleterious effect on survival of AML patient (all P < .01, Figure 4).
4 DISCUSSION
In this retrospective study, we demonstrated that up-regulated DDIT4 expression adversely affects the prognosis of AML patients who underwent chemotherapy alone, but not those who were treated with allo-HSCT, suggesting that allo-HSCT may neutralize its negative prognostic impact. Patients with low DDIT4 expression, on the other hand, had no survival benefit from allo-HSCT in the study.
Dysregulated DDIT4 expression is seen in various cancers and its role in tumorigenesis is likely tumour-dependent, based on previous studies.17-19 In breast cancer, DDIT4 is a tumour suppressor against miR-495-mediated oncogenesis and hypoxia resistance.20 In ovarian cancer, on the other hand, it is positively correlated with the oncogene p-AKT and predicts late FIGO stage and serous adenocarcinoma,13 indicating its role as a tumour promotor. DDIT4 is heavily involved in the PI3K-Akt-mTOR signalling pathway, a crucial pathway that regulates cell growth, motility, proliferation, apoptosis and one of the most commonly altered pathways in cancer.21 It is a downstream effector of PI3K-Akt-mTOR. By collaborating with other proteins, it is responsible for prostate cancer cells’ invasive behaviour.22 Moreover, DDIT4 also participates in the RAS signalling pathway and highlights the complex crosstalk among different cellular signalling pathways. Overexpression of DDIT4 after activation of RAS oncogene in RAS-transformed human ovarian epithelial cells can promote cell proliferation and colony formation, enhance the expression of anti-apoptotic proteins and reduce the expression of pro-apoptotic proteins.19, 23 In our study, high DDIT4 expression coexisted with other established poor prognostic factors, such as old age, complex karyotype and TP53 mutation, and did not coexist with other well-known favourable prognostic factors, such as CBFβ-MYH11 and RUNX1-RUNX1T. As an independent adverse prognostic factor in AML patients who received chemotherapy alone, DDIT4 up-regulation likely plays a positive role in leukaemogenesis.
Moreover, we found that enhanced DDIT4 expression was also an independent poor prognostic factor in CN-AML patients with a relatively consistent cytogenetic background. GO analysis demonstrated that genes (WNT9A, NOTCH3, SOCS1, MCL1, HIF1A, ALOX5, CD47, CXCR4, CDK9, HRAS, PLK3, ETS2 and RPL5) involved in “acute and chronic myeloid leukaemia,” “bladder cancer,” “hedgehog signalling pathway,” “endometrial cancer” and “basal cell carcinoma” signalling pathways were significantly correlated with the DDIT4 expression. Pinto et al observed a significant positive correlation between DDIT4 and NOTCH1 expression, and both of them tend to highly express in high-risk AML patients.24 These results indicate that DDIT4 expression may explain some of the aggressive features of AML by involving in the above pathways, though the exact role of DDIT4 in leukaemogenesis requires further study.
Multivariate analysis of the chemotherapy-only group was consistent with previous studies in that older age (≥60 years), more BM blasts (≥70%), mutations in TP53 and DNMT3A also independently contributed to shorter EFS and OS.25-28 The effect of high DDIT4 expression on survival was not reproduced in the allo-HSCT group, whereas WBC count ≥ 15 × 109/L, FLT3-ITD and TP53 mutation were associated with poor OS or EFS, suggesting that allo-HSCT could ameliorate the adverse prognostic effect of high DDIT4 expression in AML. Patients with high DDIT4 expression benefited more from allo-HSCT, whereas survival was not affected by treatment modality in the low expressers. Therefore, allo-HSCT may be a better option for patients with high DDIT4 expression, but may be not necessary for patients with low DDIT4 expression.
To summarize, our results indicated that enhanced DDIT4 expression could be a poor prognostic factor for AML patients treated with chemotherapy alone, and these patients might benefit from allo-HSCT. We were able to identify a unique gene expression pattern and cell signalling pathways associated with DDIT4 expression, which could shed lights on the role of DDIT4 in leukaemogenesis. It is reasonable to envision it as a marker for risk stratification and guidance for treatment in AML. Our study was limited by its small, retrospective nature, and the results would need to be verified by a larger prospective population.
ACKNOWLEDGEMENTS
This study was supported by grants from the National Natural Science Foundation of China (U190310003, 81500118, 61501519, 81600089) and the China Postdoctoral Science Foundation (2016M600443).
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Zhiheng Cheng and Yifeng Dai performed statistical analysis and drafted the manuscript. Yifan Pang and Yang Jiao critically revised the manuscript. Yan Liu, Longzhen Cui, Liang Quan, Tingting Qian, Tiansheng Zeng, Chaozeng Si, Wenhui Huang, Jinghong Chen and Ying Pang performed the acquisition and interpretation of the data. Lin Fu, Jinlong Shi and Xu Ye conceived this study and finally approved the version to be published. All authors approved the final manuscript.
Open Research
DATA AVAILABILITY STATEMENT
All data in this study were downloaded from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) and Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) databases. We did not involve direct interaction with patients. All analyses during this study were included in this article.




