Recurrent GRHL fusions in a subset of sebaceoma: microscopic and molecular characterisation of eight cases
Abstract
Aims
Sebaceous neoplasms constitute a group of adnexal tumours, including sebaceous adenoma, sebaceoma and sebaceous carcinoma. Although mismatch repair deficiency may be observed, the nature of the genetic alterations contributing to the development of most of these tumours is still unknown. In the present study, we describe the clinical, microscopic, and molecular features of eight sebaceomas with GRHL gene rearrangement.
Methods and results
Among these sebaceomas, four occurred in women and four in men; the median age was 63 years (range = 29–89). The tumours were located in the head and neck area in all cases. Microscopic examination revealed a well-demarcated lesion located in the dermis with focal extension into the subcutaneous tissue (three cases). The neoplasms displayed macronodular (eight cases), cribriform (seven cases) and organoid (six cases) growth patterns, occurring in combination. The tumours were mainly composed of immature basophilic cells associated with scattered mature sebocytes. Numerous small infundibular cysts were present in seven cases. Mitotic activity was low (none/one to four mitoses/mm2). Immunohistochemistry showed positivity for androgen receptor and p63. Preserved expression of MLH1, PMS2, MSH2 and MSH6 was observed in all cases. RNA-sequencing revealed RCOR1::GRHL2 (three cases), BCL6::GRHL1 (two cases), a BCOR::GRHL2 (one case), RCOR1::GRHL1 (one case) and TLE1::GRHL1 (one case) fusion transcript. Methylation analysis demonstrated that GRHL-fused sebaceomas form an independent cluster and highlight the proximity of such tumours with poromas with folliculo-sebaceous differentiation.
Conclusions
In conclusion, we report recurrent fusions of the GRHL genes in a distinctive subset of sebaceomas harbouring infundibulocystic differentiation, a frequent organoid growth pattern and lack of mismatch repair deficiency.
Graphical Abstract
Abbreviations
-
- BCL6
-
- BCL6 transcription repressor
-
- BCOR
-
- BCL6 corepressor
-
- CK
-
- cytokeratin
-
- DNA
-
- deoxyribonucleic acid
-
- FFPE
-
- formalin-fixed paraffin-embedded
-
- FOXK1
-
- forkhead box K1
-
- GPS2
-
- G protein pathway suppressor 2
-
- GRHL
-
- grainyhead like transcription factor
-
- HPV
-
- human papillomavirus
-
- MAML2
-
- mastermind like transcriptional coactivator 2
-
- MMR
-
- mismatch repair
-
- MLH1
-
- mutL homolog 1
-
- MSH2
-
- mutS homolog 2
-
- MSH6
-
- mutS homolog 6
-
- Mm
-
- millimeter
-
- NCOR2
-
- nuclear receptor corepressor 2
-
- NGS
-
- next generation sequencing
-
- NUTM1
-
- NUT midline carcinoma family member 1
-
- PAK
-
- p21 (RAC1) activated kinase
-
- PCR
-
- polymerase chain reaction
-
- PMS2
-
- PMS1 homolog 2, mismatch repair system component
-
- RCOR1
-
- REST corepressor 1
-
- RNA
-
- ribonucleic acid
-
- RT
-
- reverse transcription
-
- USA
-
- United states
-
- WHO
-
- World health organization
-
- YAP1
-
- Yes1 associated transcriptional regulator
Introduction
Skin adnexal tumours are a heterogeneous group of neoplasms showing differentiation towards cutaneous adnexal structures, including eccrine and/or apocrine-folliculo-sebaceous units, with 24 benign and 25 malignant neoplasms included in the current World Health Organisation (WHO) classification.1 To date, the definition of these neoplasms has been based mainly on morphology, although recurrent genetic alterations have been identified in a number of entities and can serve as diagnostic markers in difficult-to-diagnose cases.2 In particular, several gene fusions have been identified in sweat gland tumours,3 including YAP1::MAML2 and YAP1::NUTM1 in poromas and porocarcinomas3 or CRTC1/3::MAML2 in hidradenomas4 and hidradenocarcinomas.5 In 2022, our group identified recurrent FOXK1::GRHL1/2 and GPS2::GRHL1/2/3 gene fusions in trichogerminoma, a rare neoplasm with hair follicle differentiation, characterised by immature tumour cells and ‘cell ball’ formation.6-8 More recently, we demonstrated recurrent PAK2 gene fusions in a subset of poromas showing follicular and sebaceous differentiation, such alteration being mutually exclusive with YAP1-rearrangement.9
Sebaceoma is a benign sebaceous neoplasm mainly composed of immature basophilic cells associated with sparse mature sebocytes. Rare morphological variants included sebaceoma with organoid pattern,10, 11 apocrine differentiation,12 infundibulocystic structures13 or squamous cell metaplasia.13 Although mismatch repair (MMR) deficiency may be observed in some cases of sebaceoma, the nature of the genetic alterations contributing to the development of the majority of sebaceomas is still unknown.14, 15
In the present study, we describe the clinical, histological and molecular features of eight sebaceomas genetically characterised by somatic GRHL genes rearrangements with partners differing from those previously observed in trichogerminoma, i.e. BCL6 (B cell lymphoma 6, two cases), RCOR1 (REST co-repressor 1, four cases), BCOR (BCL6 co-repressor, one case) and TLE1 (TLE family member 1, one case).
Methods
Patients and samples
Following the identification of one index case, eight cases of sebaceoma associated with BCL6::GRHL1, RCOR1::GRHL2, RCOR1::GRHL1, BCOR::GRHL2 or TLE1::GRHL1 fusion transcripts were identified from the consultation files of the authors (D.V.K., M.L./T.K, E.F./M.B., N.M./M.B., E.C., S.T.). The diagnosis of sebaceoma in these cases was based on the criteria defined by the current WHO classification of skin tumours16: ‘Sebaceoma is a benign adnexal neoplasm showing sebaceous differentiation. It is characterised by rounded, cellular lobules and microcystic spaces and consists mostly of immature basaloid cells admixed with occasional mature sebocytes (less than 50% of the lesion)’.
Other adnexal tumours used as controls were obtained from the archives of the authors (E.C., T.K., M.B., P.S., B.C., N.M.) with some of these lesions (i.e. trichogerminoma, poroma with folliculo-sebaceous differentiation) already published in the previous studies.7, 9 The design of this European multicentre retrospective study was in accordance with the requirements for the use of biological material in research proposed by the ethical guidelines of our institution (Local Ethics Committee, Tours, France; no. ID RCB2009-A01056-51).
The following clinical data were obtained from the patient files: sex, age, tumour location and tumour size.
Morphological analysis
The following morphological criteria were evaluated by two of the authors (M.L., T.K.): epidermal connection, location, delimitation, architecture [macro or micronodular, cribriform, strands or cords, organoid pattern (extremely complex and often tortuous arrangement of closely packed cords or strands of neoplastic cells), columnar, cyst or Pinkus-like], cytology [nucleocytoplasmic ratio (high, moderate or low), nuclear shape (round, oval), chromatin (dusty, nucleoli), atypia (mild, severe), clear cell change, follicular differentiation, small infundibular cyst, cell balls, peripheral palisading, cleft between tumour cells and stroma and keratin calcification], sebaceous differentiation (mature sebocyte, holocrine section or eosinophilic cuticle), poroid differentiation (duct formation, decapitation secretion), stroma (hyalinised, myxoid and/or fibroblastic), intracystic cholesterol cleft, necrosis ‘en masse’ and mitotic count.
Immunohistochemistry
Immunohistochemical staining for EpCAM (clone BerEP4, dilution 1/200; Dako, Wiesentheid, Germany), PHLDA1 (clone RN-6E2, dilution 1/80; Santa Cruz, CA, USA), cytokeratin 20 (clone M7019, dilution 1/100; Dako), cytokeratin 7 (clone OV-TL 12/30, dilution 1/200; Dako), epithelial membrane antigen (EMA) (clone E29, dilution 1/100; Dako), carcino embryonic antigen (CEA) (clone CEA 31, prediluted; Cell Marque, Rocklin, CA, USA), p63 (clone 4A4, dilution 1/200; Biocare Medical, Pacheco, CA, USA), transcription factor (SOX10) (clone SP267, prediluted; Cell Marque), androgen receptor (clone SP107, prediluted; Ventana, Tucson, AZ, USA), somatostatin receptor 2a (SSTR2A) (clone SSTR2A, dilution 1/20; CliniSciences, Nanterre, France), mismatch repair protein Mlh1 (MLH1) (clone M1, prediluted; Ventana), postmeiotic segregation increased 2 (PMS2) (clone A16-4, prediluted; Roche Diagnostics GmbH, Penzberg, Germany), homo sapiens mutS homologue 6 (MSH6) (clone AP59, dilution 1/50; BioSB, Goleta, CA, USA), MSH2 (clone G219-1129, dilution 1/100; Cell Marque), RB (clone 13A10, dilution 1/50; Leica, Wetzlar, Germany), p16 (clone MX007, dilution 1/100; Zeta Corporation, Monrovia, CA, USA), p53 (clone D07, dilution 1/50; Dako) were performed using a BenchMark XT Platform (Roche) as instructed. SSTR2A immunohistochemical expression was evaluated by two pathologists using a semiquantitative score, as previously described.17 For p16 and p53 immunohistochemical expression, an estimation of the percentage of cells showing staining was used.18
DNA/RNA extraction
RNA and DNA extraction was performed on our samples and sebaceous neoplasms (sebaceous adenoma, five; sebaceoma, 9; sebaceous carcinoma, five) from formalin-fixed paraffin embedded (FFPE) tissue samples by using a Maxwell 16 Instrument (Promega, Madison, WI, USA) with the Maxwell 16 LEV RNA FFPE kit and the Maxwell 16 FFPE Plus LEV DNA purification kit, according to the manufacturer's protocol. Concentration (ng/μl) and purity (260/280 nm ratio) of DNA and RNA were determined using the NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
HPV genotyping
HPV genotyping was performed using the INNO LiPA version extra II (Fujirebio, Tokyo, Japan) according to the manufacturer's instructions. This test allows the detection of 32 human papilloma virus (HPV) genotypes, including high-grade (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68), potentially high-grade (26, 53, 66, 70, 73 and 82) and low-grade (HPV6, 11, 40, 42, 43, 44, 54, 61, 62, 67, 81, 83 and 89) genotypes. Ct values and polymerase chain reaction (PCR) amplification curves are available for the different HPV targets. Results were reported as ‘positive’ or ‘negative’, according to the manufacturer's instructions.
DNA sequencing
Molecular analysis was performed retrospectively using a custom-designed next-generation sequencing (NGS) panel (ATLAS panel; NCI, Bethedsa, MD, USA) including 92 genes involved in cutaneous carcinomas pathogenesis. Fragmentation of FFPE-derived DNA samples and library production were conducted with SureSelect XT HS technology (Agilent, Santa Clara, CA, USA). Qualification of purified libraries was assessed with TapeStation (high sensitivity D1000 ScreenTape; Agilent) and Bioanalyzer (DNA 1000 kit for 2100 Bioanalyzer Systems; Agilent). NGS was performed on a Nextseq 2000 platform (Illumina, San Diego, CA, USA). Coding regions, 5'UTR regions and exon-intron junctions (−20 to +20) of the targeted genes were sequenced with coverage > ×100. Sequencing results were obtained after fastq alignment to the GRCh37 human reference sequence (hg19), variant calling and copy number variations (CNV) estimation using our custom bioinformatics pipeline. Pathogenic or probably pathogenic variants were identified using at least 3/5 in-silico prediction software (CADD, SIFT, PolyPhen2, AlignGVGD, Mutation Taster) and had a frequency in general population databases fewer than 1%.
RNA sequencing
Next-generation sequencing libraries were prepared with anchored multiplex PCR-based methodology using the Archer® FusionPlex® Comprehensive Sarcoma Panel (Archer DX, Boulder, CO, USA) customised to include the GRHL1, GRHL2 and GRHL3 genes. Libraries were generated according to the manufacturer's protocol using 250 ng of total RNA. Amplifiable cDNA quality was assessed using the PreSeq RNA QC assay. Final libraries were quantified with a Qubit fluorometer (dsDNA HS assay kit; Life Technologies, Carlsbad, CA, USA). Illumina paired-end indexed libraries were sequenced 8-plex on a MiSeq with the MiSeq Reagent Kit V2 (300 cycles) from Illumina and sequencing data were analysed via the Archer® automated analysis pipeline (version 6.2.7).
Confirmation of RNA sequencing findings by reverse transcription PCR and Sanger sequencing
Reverse transcription (RT) was performed using the GoTaq® Probe 2-step RT-qPCR system kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. It was followed by PCR using custom primers and 5 μl of cDNA. PCR products were purified by a mix of FastAP thermosensitive alkaline phosphatase and exonuclease I (Thermo Fisher Scientific, Waltham, MA, USA) under the following conditions: 15 min at 37°C and 15 min at 85°C. The sequencing reaction was carried out using Big Dye terminator version 3.1 cycle sequencing kit (Thermo Fisher Scientific), according to the manufacturer's protocol. Products were purified as recommended by the BigDye XTerminator™ Purification Kit (Thermo Fisher Scientific) and were sequenced on a Genetic Analyser 3500 Dx (Applied Biosystems, Waltham, MS, USA). The sequencing data were analysed using the Sequencing Analysis software (Thermo Fisher Scientific).
Analysis of DNA methylation
Genomic DNA was extracted from FFPE tissues using Maxwell apparatus (Promega, Madison, WI, USA) with Maxwell 16 FFPE Plus LEV DNA purification kits. Methylation profiling employed the methylation EPIC BeadChip (Illumina, San Diego, SA, USA); 300 ng of DNA were processed according to the manufacturer's protocol, as previously described.19 Data were generated in the Department of Neuropathology of the University Hospital, Heidelberg. Computational analyses were based on R version 4.6.1 (https://www.R-project.org). tSNE plot was generated on 10.000 most variable cytosine–phosphate–guanine (CpG) sites upon standard deviation, 3.000 iterations and using a perplexity parameter of 10.
Results
Eight cases of sebaceomas with GRHL fusion were included in the present study. The clinical features are summarised in Table 1. Briefly, four patients were female and four were male, median age 63 years (age range = 29–89 years). In concordance with previous descriptions of sebaceoma,20 tumours were located in the head and neck area [either the scalp (one) or the ear (n = one, case 4)] with a median tumour size of 12 mm (range = 6–20 mm). Follow-up data were available for seven cases (median follow-up duration = 19 months, range = 2–120). No local or distant recurrence was detected.
| Clinical features | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 |
|---|---|---|---|---|---|---|---|---|
| Age (years) | 89 | 60 | 70 | 65 | 74 | 29 | 49 | 38 |
| Sex | F | M | F | M | F | M | F | M |
| Location | Scalp | Scalp | Scalp | External auditory canal | Scalp | Scalp | Scalp | Scalp |
| Tumour size (mm) | 20 | 7 | 6 | 12 | 15 | 13 | 12 | 10 |
| Follow-up (months) | NA | 20 | 2 | 19 | 8 | 9 | 120 | 53 |
| Excision | Excision without margins | Wide excision | Excision without margins | Excision with margins | Wide excision | Excision without margins | Excision without margins | Partial excision |
| Recurrence | NA | No | No | No | No | No | No | No |
| Morphological features | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 |
|---|---|---|---|---|---|---|---|---|
| Epidermal connection | Yes | No | No | No | Yes | No | Yes | No |
| Location | ||||||||
| Dermis | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Subcutaneous | No | No | Focal | No | Yes | Yes | No | No |
| Delimitation architecture | Well | Well | Well | Well | Well | Well | Well | Well |
| Massive | Yes (P) | Yes (P) | Yes (P) | Yes (P) | Yes (P) | Yes (P) | Yes (P) | Yes (P) |
| Macronodular | Yes (P) | Yes (P) | Yes (P) | Yes (P) | Yes (P) | Yes (P) | Yes (P) | Yes |
| Micronodular | Yes | No | No | No | Yes | Yes | Yes | Yes |
| Cribriform | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
| Stand or cord | Focal | Yes | No | No | Focal | Yes | Yes | Yes |
| Organoid pattern | Yes | Focal | No | No | Yes | Yes | Yes | Yes (P) |
| Columnar | No | No | No | No | No | No | No | No |
| Cyst | No | No | No | No | No | No | No | No |
| Pinkus-like | No | No | No | No | No | No | No | No |
| Cytology | ||||||||
| Nucleo-cytoplasmic ratio | High | High | Mild | High | Mild | Mild | High | Mild |
| Nuclear shape | Oval | Oval | Oval | Round | Round-oval | Round–oval | Round–oval | Round–oval |
| Chromatin compaction | Small nucleoli | Small nucleoli | Dusty/small nucleoli | Small nucleoli | Small nucleoli | Small nucleoli | Prominent nucleoli | Small nucleoli |
| Atypia | Mild | Mild | Mild | Mild/high | Mild | Mild | Mild | Mild |
| Clear cell changes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Focal |
| Follicular differentiation | ||||||||
| Small infundibular cyst | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
| Cell balls | No | No | No | No | No | No | No | No |
| Peripheral palisading | No | No | No | No | No | No | No | No |
| Cleft (tumour/stroma) | No | No | Focal | Focal | No | No | No | No |
| Keratin calcification | No | No | No | No | No | No | No | No |
| Sebaceous differentiation | ||||||||
| Mature sebocytes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Focal |
| Holocrine secretion/eosinophilic cuticle | Focal | Focal | Yes | Yes | Yes | Focal | Yes | Focal |
| Duct formation | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Focal |
| Decapitation secretion | No | No | No | No | No | No | No | No |
| Stroma | ||||||||
| Hyalinised | Focal | Yes | No | Focal | Yes | Yes | Yes | Yes |
| Myxoid | No | No | No | No | No | No | No | No |
| Fibroblastic | Yes | No | Yes | Yes | No | No | No | No |
| Others | ||||||||
| Intra-cystic cholesterol cleft | Yes | No | No | No | No | No | No | No |
| Necrosis ‘en masse’ | No | No | No | No | No | No | No | No |
| Mitotic count/mm2 | <1 | < 1 | < 1 | 2 | 2 | <1 | 4 | <1 |
| Perineurial or vascular invasion | No | No | No | No | No | No | No | No |
| IHC featuresa | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case #6 | Case 7 | Case 8 |
|---|---|---|---|---|---|---|---|---|
| Follicular markers | ||||||||
| EpCam (BerEP4) | NA | Ducts | Focal | Focal | Ducts | NA | NA | NA |
| PHLDA1 | NA | – | Focal | – | – | NA | NA | NA |
| Merkel cell markers | ||||||||
| Cytokeratin 20 | NA | – | + Merkel cells | + Merkel cells | – | NA | NA | NA |
| Merkel cell density | – | 14/mm2 | 16/mm2 | – | ||||
| Other | ||||||||
| CK7 | NA | Ducts | Ducts | Focal | Ducts | NA | NA | NA |
| EMA | NA | Ducts and sebocytes | Ducts and sebocytes | Ducts and sebocytes | Ducts and sebocytes | Ducts and sebocytes | NA | NA |
| CEA | NA | – | Ducts | – | Ducts | – | NA | NA |
| P63 | NA | Diffuse | Diffuse | Diffuse | Diffuse | NA | Diffuse | Diffuse |
| SOX10 | NA | – | – | – | – | NA | NA | NA |
| SSTR2A | NA | – | 5% | – | 5% | NA | NA | NA |
| Androgen Receptor | 80% | 100% | 50% | 100% | 100% | NA | 97% | 97% |
| MSH2 | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse |
| MSH6 | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse |
| PMS2 | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse |
| MLH1 | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse | Diffuse |
| P53 | NA | 45% | 75% | 55% | 75% | NA | NA | NA |
| P16 | NA | 30% | 3% | 25% | 5% | NA | NA | NA |
| RB | NA | Heterogenous | Heterogenous | Heterogenous | Heterogenous | NA | NA | NA |
| Genetic features | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 |
|---|---|---|---|---|---|---|---|---|
| RNA-seq | ||||||||
| GRHL fusion transcripts | BCL6::GRHL1 | RCOR1::GRHL2 | BCOR::GRHL2 | RCOR1::GRHL2 | TLE1::GRHL1 | RCOR1::GRHL2 | BCL6::GRHL1 | RCOR1::GRHL1 |
| Accession numbers | NM_001706.4 (BCL6):e.5:: NM_198182.2(GRHL1):e.6 | NM_015156.3 (RCOR1):e.11:: NM_024915.3(GRHL2):e.5 | NM_017745.5 (BCOR):e.10:: NM_024915.3 (GRHL2):e.3 | NM_015156.3 (RCOR1):e.11:: NM_024915.3 (GRHL2):e.5 | NM_005077.4 (TLE1):e.14:: NM_198182.2 (GRHL1):e.6 | NM_015156.3 (RCOR1):e.11:: NM_024915.3:e.5 | NM_001706.4 (BCL6):e.5:: NM_198182.2 (GRHL1):e.2 | NM_015156.3 (RCOR1):e.11: NM_198182.2 (GRHL1):e.3 |
- P, predominant; e, exon; F, female; IHC, immunohistochemical; M, male; NA, non-available data.
- a Results are expressed as percentage of positive cells.
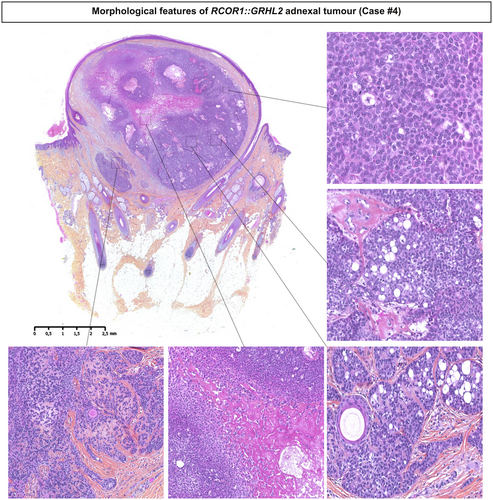
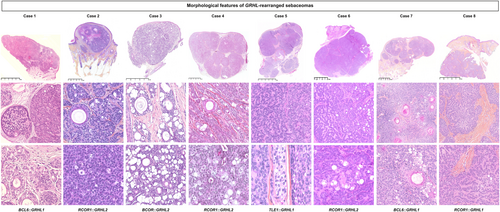
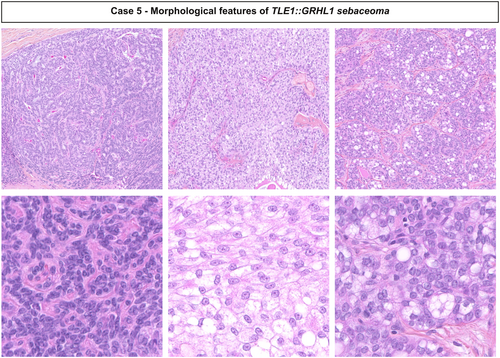
Histologically, all tumours were well-demarcated and located in the dermis with focal extension into the subcutaneous tissues in three cases (cases 3, 5 and 6) (Figures 1 and 2). Connection to the epidermis was present in three cases (cases 1, 5 and 7). The neoplasms showed a combination of macronodular (eight), cribriform (seven), and organoid (six) growth patterns and were surrounded by a fibrous or hyalinised stroma. Cystic spaces containing acellular eosinophilic deposits were present in one case (case 2). Lesions were mainly composed of immature medium-sized basophilic monotonous cells with large oval nuclei, a single small nucleolus and dusty chromatin. Scattered mature sebocytes, ductal structures associated with an eosinophilic cuticle sometimes forming clusters and focal clear cell change were observed in all cases. Numerous small infundibular cysts with lamellar keratin were identified in all except one case. Mitotic figures were rare (none/one to four mitoses/mm2). No tumour showed necrosis, perineurial or vascular invasion.
Using immunohistochemistry, all cases showed positivity for androgen receptor and p63 (seven of seven and six of six, respectively) (Table 1 and Figure 3). As expected for sebaceoma,21 CEA (two of five), EpCam (two of four) and cytokeratin 7 (CK7) (three of four) only highlighted the presence of ducts, while EMA expression was observed in ducts and in mature sebocytes (five of five) (Figure 3). In contrast to GRHL-fused trichogerminoma,7, 8 tumour cells only focally expressed SSTR2A in two cases (cases 3 and 5) and weak PHLDA1 positivity was detected in only one case (case 3). Moreover, CK20 revealed the presence of only few Merkel cells in two cases (density = 14–16/mm2).

RNA-sequencing revealed rearrangements of GRHL1/2 in all cases, with a RCOR1::GRHL2 in three cases (cases 2, 4 and 6), a RCOR1::GRHL1 in one case (case 8), a BCL6::GRHL1 fusion in two cases (cases 1 and 7), a BCOR::GRHL2 fusion transcript in case 3 and TLE1::GRHL1 fusion transcript in case 5 (Table 1, Supporting information, Figure S1). These fusions involve as 5' partner, RCOR1 located in 14q32.32, BCOR located in Xp11.4, BCL6 located in 3q27.3, TLE1 located in 9q21.32, fused to either GRHL1 located in 2p25.1 or its paralogue GRHL2 located in 8q22.3, the dimerisation and DNA-binding domains of GRHL1/2 being constantly preserved in the fusion protein.22, 23 Presence of the fusion was investigated and confirmed by Sanger sequencing in two cases (cases 5 and 6) with sufficient available material (Supporting information, Figure S2) and BCOR expression was demonstrated by immunohistochemistry in case 3 harbouring BCOR::GRHL2 fusion transcript (Supporting information, Figure S3).
Our group had previously demonstrated that, among follicular adnexal tumours, GRHL fusions were specific of trichogerminomas.6 To further evaluate to which extent the GRHL fusion might be found in sebaceous neoplasms, two additional cases of sebaceoma harbouring infundibulocystic structures, including one case with an organoid growth pattern (Supporting information, Figure S4), seven cases of sebaceoma lacking infundibulocystic structures and organoid patterns (including three cases with MMR instability), six cases of sebaceous adenomas (including two cases with MMR instability) and five sebaceous carcinomas (including one case with MMR instability) were submitted to RNA sequencing. This analysis revealed no fusion transcripts, suggesting that GRHL fusions are restricted to a distinctive subset of sebaceomas harbouring frequent organoid growth pattern and infundibulocystic differentiation.
To test whether GRHL fusion might arise in association with other oncogenic drivers previously identified in sebaceous neoplasms, presence of these latter alterations was investigated in our cohort of GRHL-fused sebaceomas. Immunohistochemistry analysis revealed preserved expression of MLH1, PMS2, MSH2 and MSH6 in all cases (eight of eight). RB was conserved in all tested cases (four of four) and patchy expression of P53 and P16 restricted to a minority of cells was evidenced (median percentage of positive cells = 65%, range = 45–75% and 15%, range = 3–30%, respectively, four of four). Accordingly, no high-grade HPV genome was detected in such samples. Next-generation sequencing analysis of five sebaceous neoplasms (cases 2–6) using a 92 genes DNAseq panel, did not evidence any oncogenic alterations previously described in sebaceous tumours; notably, no mutation was detected in CDKN2A, CTNNB1, HRAS, KRAS KMT2D, NOTCH1-3, RB1 and TP53 genes. Case 5 had a probably oncogenic alteration in the EGFR (NM_005228, DNA sequence variation: c.664C>T; protein sequence variation: p.Arg222Cys). Moderate amplifications (copy number of 3–4) of MDM2 and KRAS were detected in cases 2 and 4.
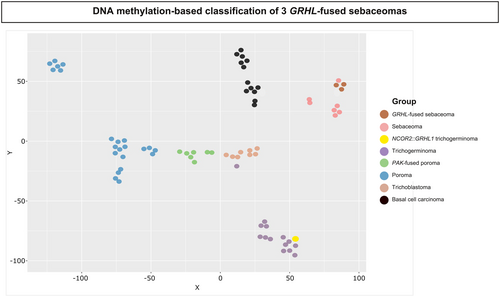
To further clarify the distinction between GRHL1/2 rearranged sebaceomas and other adnexal tumours sharing morphological or molecular similarities with this tumour entity, we analysed the DNA methylation profile of GRHL-fused sebaceomas (three cases with sufficient material, cases 3–5) in comparison to 30 poromas, including eight PAK2-fused poromas showing follicular or sebaceous differentiation,9 15 GRHL-fused trichogerminomas (including 14 cases FOXK1::GRHL1/2 and GPS2::GRHL1/2/3 fusion transcripts7 and one case with a NCOR2::GRHL1 fusion (Supporting information, Figure S5), eight trichoblastomas and 13 basal cell carcinomas. Methylation at CpG dinucleotides is regarded as a conserved marker of cell identity,24 allowing identification of the cell of origin of a neoplasm,25, 26 and therefore might contribute to an accurate classification of closely related tumour entities. This analysis revealed that GRHL-fused sebaceomas represent an independent cluster distinct from GRHL-fused trichogerminoma (including the case with NCOR2::GRHL1 fusion) (Figure 4).
Discussion
Sebaceous neoplasms constitute a group of adnexal tumours assumed to derive from the sebaceous glands of the skin or the eyelid and are subclassified into three categories: sebaceous adenoma, sebaceoma and their malignant counterpart, i.e. sebaceous carcinoma. While sebaceous adenomas are mainly composed of fully differentiated sebocytes associated with a minority of immature cells, the reverse is seen in sebaceomas. In addition to sebaceous differentiation, sebaceous carcinomas harbour evidence of malignancy and are divided into ocular and extra-ocular neoplasms.27-29
From a genetic perspective, it is well established that MMR deficiency arising either sporadically or in a context of Muir–Torre syndrome is observed in a subset of sebaceous neoplasms.27 Morphological clues such as cystic change, intratumoural mucin deposits, squamous metaplasia, ulceration, lymphocytic infiltrates and intertumoural heterogeneity have been described in sebaceous neoplasms within the context of MMR deficiency,20, 30-32 suggesting a correlation between morphology and genetic background. In addition to MMR deficiency, further recurrent mutations in TP53, RB1, CDKN2A, EGFR, CTNNB1, NOTCH1/2/3, KMT2D, HRAS and KRAS have also been reported in sebaceous neoplasms without correlation with specific histological or morphological features.18, 33 In sebaceous carcinomas, co-mutations of TP53 and RB1 related to UV exposure as well as high-grade HPV have been identified as potential oncogenic drivers mutually exclusive of MMR instability.29, 34 By contrast, little is known regarding the genetics background of benign sebaceous neoplasms lacking MMR instability. Herein, we report recurrent GRHL fusions in a subset of sebaceomas.
Interestingly, although GRHL fusions were initially described by our group in trichogerminoma,7 the fusion partners observed in the present study, i.e. BCOR, RCOR1, BCL6 and TLE1, are distinct to the ones observed in the latter (i.e. FOXK1 and GPS2) and belong to the same family of transcriptional co-repressors suggesting a potential impact of these fusion partners on tumour biology. In line with these findings, BCOR and RCOR1 proteins have recently been shown to interact with the androgen receptor and modulate the expression of androgen receptor target genes.35 The BCL6 protein promotes recruitment of the BCOR co-repressor36 and plays a role in DNA demethylation, while the androgen receptor contributes to the establishment or maintenance of DNA methylation.36 Therefore, it is possible that expression of BCOR::GRHL2, RCOR1::GRHL2 or BCL6::GRHL1 modulates the transcriptional profile of androgen receptor target genes, resulting in sebaceous differentiation.
Until the present, GRHL fusions have been only identified in trichogerminomas and sebaceomas and therefore seem to be specific to these tumour entities. Indeed, none of these rearrangements were identified among the 77 850 cases registered in the Mitelman database (https://mitelmandatabase.isb-cgc.org/, last accessed July 2024). Moreover, among the 115 adnexal tumours analysed by our group by whole transcriptome analysis,37 including 37 follicular tumours (20 trichogerminomas, six trichoblastomas, two trichoblastic carcinosarcomas, seven proliferating trichilemmal tumours, two follicular carcinomas) and 77 sweat gland tumours (including 15 poromas, eight porocarcinomas, one cystadenoma, six hidradenomas, two hidradenocarcinomas, 31 mixed tumours, 14 cribriform carcinomas), GRHL fusions were restricted to trichogerminomas (personal data), suggesting that these alterations are specific of few tumour entities.
Although our results need confirmation on a larger cohort, our findings suggest that GRHL fusions are only present in a subset of sebaceomas microscopically characterised by an organoid growth pattern and infundibulocystic structures, morphological features only observed in a minority of cases,10, 11, 13 and having been regarded together with the presence of Merkel cells as a sign of sebaceous mantle differentiation.38, 39 Expression of vimentin has been described as a marker of these subsets of tumours and was observed in one of our cases (data not shown). Importantly, GRHL fusion in our study seems to be mutually exclusive with oncogenic drivers previously described in sebaceous neoplasms, including MMR deficiency or further recurrent mutations in TP53, RB1, CDKN2A, EGFR, CTNNB1, NOTCH1/2/3, KMT2D, HRAS and KRAS.
In addition to GRHL-fused sebaceoma, association of follicular, i.e. infundibulocystic and sebaceous differentiation with a background of monotonous basophilic tumour cells, may be observed in other adnexal tumours, including trichogerminoma7 and poroma with folliculo-sebaceous differentiation with PAK2 fusion.9, 40 In line with these findings, methylation studies revealed close proximity between our cases and the later tumour entities (Figure 4).
Follicular and/or sebaceous differentiation in poroma was first described in 1981 in the so-called ‘infundibular adenoma’.41 The term ‘apocrine/holocrine poroma’ was next used to describe similar lesions42, 43 and it is included in the current WHO classification of skin tumours as a variant of poroma.16 In 2023, our group identified a recurrent rearrangement of the PAK2 gene in a series of 13 cases of poroma with folliculo-sebaceous differentiation, an alteration that, together with YAP1 fusion, is usually observed in eccrine poroma/porocarcinoma.9, 44, 45 In addition to ductal structures with cuticles, a hallmark of poroma, such cases also show few scattered mature sebocytes and follicular cysts, features with close similarity to the entity described in the present manuscript. Although close proximity was highlighted by methylation analysis between GRHL-rearranged sebaceoma and PAK-rearranged poromas, these two entities may be distinguished based on distinct morphological and, particularly, cytological features. Indeed, although small poroid cells sometimes associated with a squamoid component are observed in PAK2-rearranged tumours, GRHL-rearranged sebaceomas are composed of medium-sized cells with a small nucleolus and dusty chromatin lacking ‘poroid cytology’.
Immunohistochemically, both PAK2-rearranged poromas with folliculo-sebaceous differentiation and our cases may display similar profiles with focally positive for BerEP49 and ducts were highlighted by EMA and CEA. However, it is interesting to note that our cases showed strong diffuse expression of the androgen receptor, whereas this is generally focally and/or weakly expressed in poroid neoplasms.46
In addition to GRHL fusions, association of follicular and sebaceous differentiation is also a common morphological feature shared by our cases and trichogerminoma, although sebaceous differentiation is only focal in the latter. Indeed, trichogerminoma is a follicular tumour characterised by basaloid ‘immature’ cytology, follicular squamous epithelium and cell ball formation.6 Recently, recurrent FOXK1::GRHL1/2 and GPS2::GRHL1/2/3 gene fusions have been identified and have been shown to differentiate trichogerminoma from other trichoblastomas,7, 8 as confirmed by the methylation analysis performed herein. Interestingly, in the present study we identified a case of adnexal tumour harbouring trichogerminoma morphology and a previously undescribed GRHL-rearrangement, involving the partner NCOR2, another co-repressor gene (Supporting information, Figure S5). In methylation analysis, this case clustered together with other trichogerminomas. This observation might either suggest that expression of the same fusion protein in different cells could lead to the development of distinct adnexal tumours or, alternatively, may indicate that NCOR2::GRHL fusion harbours biological properties similar to GPS2::GRHL and FOXK1::GRHL and distinct from those of the GRHL fusions proteins detected in sebaceoma.
In conclusion, we report recurrent fusions of the GRHL gene fusions in a distinctive subset of sebaceomas displaying infundibulocystic differentiation, an organoid growth pattern and lack of MMR instability.
Acknowledgements
This study was supported by an Illumina grant. We did not receive specific funding for this study.
Conflicts of interest
The authors declare that no financial benefit from any publication of the material, and they do not have a claim on any possible future uses of this content.
Institutional review board
The local Ethics Committee in Human Research of Tours (France) approved the study (no. ID RCB2009-A01056-51).
Open Research
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.