Variants in DOK7 results in fetal akinesia deformation sequence: A case report and review of literature
Abstract
We report the third case of FADS due to biallelic DOK7 variants, which further strengthens the association of DOK7 with this lethal phenotype and lack of genotype phenotype correlation.
Fetal akinesia deformation sequence (FADS) is characterized by reduced or absent fetal movements, intrauterine growth restriction (IUGR), multiple joint contractures, craniofacial anomalies, and pulmonary hypoplasia. It can be due to various intrinsic fetal disorders involving muscle, nervous system, connective tissue, fetal vasculature etc. and extrinsic causes such as IUGR, maternal diseases and drug intake.1 Downstream of tyrosine kinase 7 (DOK7) is a postsynaptic cytoplasmic protein involved in MuSK auto-phosphorylation and activation which further induces clustering of acetylcholine receptors (AChRs) at the NMJ.2 Biallelic variants in DOK7 leading to complete loss of function was identified as a cause of FADS3 (OMIM#618389) in 2009,3 whereas incomplete loss results in congenital myasthenic syndrome 10 (CMS 10, OMIM#254300). Here, we report the third case of DOK7 related FADS3.
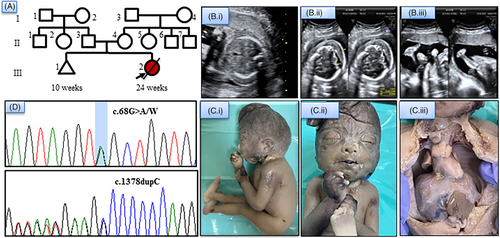
We ascertained a non-consanguineously married couple with features of FADS in antenatal ultrasound. At 22 weeks, ultrasound showed increased nuchal fold thickness (NFT-12.5 mm), subcutaneous edema, absent stomach bubble, flexion contractures at finger and wrist joint, hyperextended knee, rocker bottom feet and polyhydramnios (AFI-18 cm). Postnatal examination showed a female fetus with facial dysmorphism, joint contractures, short neck, subcutaneous edema and hypoplastic lungs (Figure 1). Fetal karyotype was normal. Peripheral blood of parents and fetal amniotic fluid were collected after written, informed consent. The study was approved by the institutional ethics committee.
Trio exome sequencing was performed using Illumina NovaSeq 6000 sequencing platform following enrichment and capture using Twist Comprehensive Library Prep kit according to the manufacturer's instructions. Bioinformatic analysis was done using an in-house pipeline and the sequences reads were aligned to human reference genome hg38. Variant prioritization was based on minor allele frequency <1% in population databases, disease databases, location of variants, in-silico prediction tools, possible modes of inheritance and phenotypic relevance. ACMG/AMP guidelines were used for classification of the candidate variants.
We identified a likely pathogenic nonsense variant in exon 2 of DOK7, NM_173660.5:c.68G > A/p.Trp23Ter (ClinVar accession number: VCV001802676.1 (this study)). This variant is absent in population databases. The second variant in exon 7, NM_173660.5:c.1378dupC/p.Gln460Profs*59, is a likely pathogenic frameshift variant in the COOH terminal region, leading to elongation of protein, with a minor allele frequency of 0.007% in ExAC database and had been reported previously in individuals with CMS.2 The variants were confirmed by Sanger sequencing. DOK7 is a postsynaptic skeletal muscle protein involved in neuromuscular synaptogenesis. It has pleckstrin-homology (PH) and phosphotyrosine binding (PTB) domains in the N-terminal portion and Src homology 2 (SH2) domain target motifs in the C-terminal region.4 Vogt et al. observed three fetuses with FADS from a single family who harbored biallelic splice variant c.331 + 1G > T in DOK7. They hypothesized that complete loss of DOK7 function due to variants in PH or PTB domains result in lethal fetal akinesia phenotype.3 However, this observation was refuted by Radhakrishnan et al., who reported the second case of FADS with biallelic variant, c.1263dupC reported earlier in homozygous state in patients with CMS, suggestive of phenotypic variability.5 This report further strengthens the association of DOK7 with FADS and lack of genotype–phenotype correlation.
ACKNOWLEDGMENTS
We thank the family for their support and Indian Council of Medical Research for funding the study (Grant no. 33/2/2019-TF/Rare/BMS).
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




