Androgen receptor mRNA analysis from whole blood: a low-cost strategy for detection of androgen receptor gene splicing defects
Graphical Abstract
Androgen insensitivity syndrome (AIS) is caused by defects in the androgen receptor (AR) gene and is the most common aetiology of 46,XY disorders of sex development. Allelic variants in the AR gene are found in 90% of complete AIS (CAIS), but in only 28% to 50% of cases of partial AIS. Even a single nucleic acid change can disrupt splicing sites or splicing regulatory sequences, resulting in inadequate exon and intron recognition, ultimately leading to an aberrant transcript. Therefore, we tested the feasibility of conducting AR cDNA analysis from whole blood and from gonadal tissue in a patient with CAIS due to AR synonymous mutation (c.1530C > T, p.Ser510Ser; NM_000044.3), which led to an aberrant splicing site causing deletion of 92 nucleotides resulting in a very short transcript. AR cDNA sequencing was similar in the whole blood and in the gonadal tissue, with similar evidence of a consequent altered AR transcript. We propose that analysis of AR RNA extracted from whole blood with AR DNA sequencing can help to improve the frequency of molecular diagnosis, particularly for partial AIS.
Androgen insensitivity syndrome (AIS) is caused by defects in the androgen receptor (AR) gene and is the most common aetiology of 46,XY disorders of sex development.1 Allelic variants in the exonic region of the AR gene are found in almost all cases of complete AIS (CAIS), but in only 28% to 50% of cases of partial AIS, suggesting the presence of other AIS-associated genetic alterations. Even a single nucleic acid change can disrupt splicing sites or splicing regulatory sequences, resulting in inadequate exon and intron recognition, ultimately leading to an aberrant transcript.2 According to the Human Gene Mutation Database (HGMD), 9% of all reported mutations are splicing mutations (HGMD accessed on May 18, 2018); however, this frequency might be underestimated, because the impact of synonymous variants has not been adequately investigated.
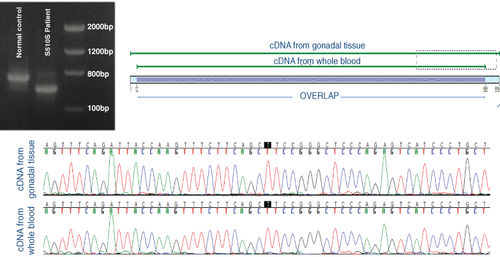
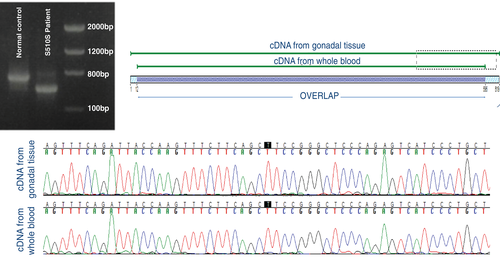
There are various strategies available to test the functional impact of allelic variants in splicing sites, including investigations of DNA, RNA, and RNA-protein interactions as well as protein analyses. One of the most effective options is to conduct RNA analyses from patient tissues following cDNA (complementary DNA) sequencing to confirm the presence of a splicing defect. A minigene assay and the truncated protein test can also be used for this purpose, although these methods are relatively more expensive and complicated than cDNA analysis.3 However, cDNA analysis is limited for AIS, mainly due to the requirement of procuring the patient's tissue sample. AR is also expressed in the whole blood, despite of lower AR expression level than in gonadal tissue, raising the possibility of a less invasive test. We recently reported detection of a synonymous mutation (c.1530C > T, p.Ser510Ser; NM_000044.3) in the AR gene in a patient with CAIS, which led to an aberrant splicing site causing deletion of 92 nucleotides resulting in a very short transcript.4
Therefore, in the present study, we tested the feasibility of conducting AR cDNA analysis from whole blood. To prove the functional effect of this synonymous variant, we analysed the cDNA using the patient's gonadal tissue, and we compared the results of AR cDNA sequencing between RNA extracted from whole blood and from the gonadal tissue of the patient. The gonadal tissue was obtained after gonadectomy, which was performed due to the risk of development of germ cell tumours as is common in AIS. We used a sample from a normal male (46,XY) as a control for AR cDNA sequencing. This study was conducted according to the Declaration of Helsinki, and written consent was obtained from all participants.
As expected, AR cDNA sequencing in the affected patient showed the same allelic variant in the whole blood and in the gonadal tissue, with similar evidence of a consequent altered AR transcript (Figure 1). There was a 5.8% mismatch between cDNA from whole blood and gonadal tissue due to lower quality sequencing, which do not influence on sequencing analysis (Figure 1).
The strategy of extracting RNA and DNA from whole blood to analyse disease-related genes has been previously employed for patients with cardiac diseases such as long QT syndrome, Marfan syndrome, and hypertrophic and dilated cardiomyopathy. These conditions are genetic cardiac diseases, known to be caused by multiexon genes that are principally expressed in the cardiovascular tissues.5 Moreover, the use of cDNA analysis may be helpful for several other types of genetic diseases because it is estimated that 15% to 50% of all monogenic diseases are caused by mutations that affect pre-mRNA splicing.
In conclusion, the parallel analysis of AR RNA extracted from whole blood with AR DNA sequencing can help to improve the frequency of molecular diagnosis, particularly for partial AIS. This strategy can also be helpful to analyse the functional impact of synonymous mutations, rather than generally discarding them as benign or probably non-pathogenic, in AIS and other genetic disorders.
ACKNOWLEDGEMENTS
We thank Monica M. França for reviewing the molecular biology aspects of this paper. This work was supported by the Conselho Nacional de Desenvolvimento Cientíco e Tecnológico (CNPQ) Grant No. 303002/2016-6 (to B.B.M.)
Conflict of interest
The authors declare no conflict of interest