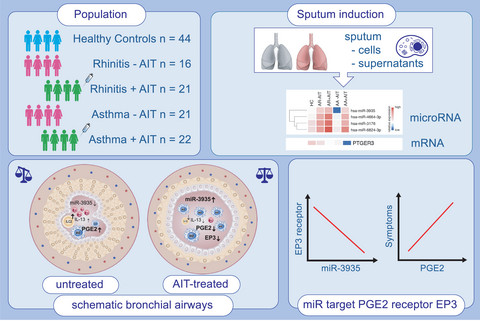
Sputum microRNA-screening reveals Prostaglandin EP3 receptor as selective target in allergen-specific immunotherapy
Funding information
German Center for Lung Research (DZL); Comprehensive Pneumology Center (CPC) Munich; German Research Foundation (DFG): 398577603.
Abstract
Background
Several microRNAs (miRs) have been described as potential biomarkers in liquid biopsies and in the context of allergic asthma, while therapeutic effects on the airway expression of miRs remain elusive. In this study, we investigated epigenetic miR-associated mechanisms in the sputum of grass pollen-allergic patients with and without allergen-specific immunotherapy (AIT).
Methods
Induced sputum samples of healthy controls (HC), AIT-treated and -untreated grass pollen-allergic rhinitis patients with (AA) and without asthma (AR) were profiled using miR microarray and whole-transcriptome microarray analysis of the same samples. miR targets were predicted in silico and used to identify inverse regulation. Local PGE2 levels were measured using ELISA.
Results
Two hundred and fifty nine miRs were upregulated in the sputum of AA patients compared with HC, while only one was downregulated. The inverse picture was observed in induced sputum of AIT-treated patients: while 21 miRs were downregulated, only 4 miRs were upregulated in asthmatics upon AIT. Of these 4 miRs, miR-3935 stood out, as its predicted target PTGER3, the prostaglandin EP3 receptor, was downregulated in treated AA patients compared with untreated. The levels of its ligand PGE2 in the sputum supernatants of these samples were increased in allergic patients, especially asthmatics, and downregulated after AIT. Finally, local PGE2 levels correlated with ILC2 frequencies, secreted sputum IL-13 levels, inflammatory cell load, sputum eosinophils and symptom burden.
Conclusions
While profiling the sputum of allergic patients for novel miR expression patterns, we uncovered an association between miR-3935 and its predicted target gene, the prostaglandin E3 receptor, which might mediate AIT effects through suppression of the PGE2-PTGER3 axis.
Graphical Abstract
miR-3935 is upregulated in sputum cells of allergic asthma patients, who received allergen-specific immunotherapy treatement, while mRNA levels of its predicted target, the prostaglandin E2 receptor, is downregulated as its ligand PGE2 as well. PGE2 is strongly upregulated in sputum supernatants of untreated allergic patients and is reduced in patients, who received allergen-specific immunotherapy. PGE2 levels correlate with clinical parameters, like eosinophils and symptom score.
Key Messages
- AIT induces miR-3935 in induced sputum cells of allergic asthmatics, while mRNA of its target, prostaglandin E3 receptor, is downregulated.
- EP3 ligand PGE2 is reduced in sputum supernatants of AIT-treated patients.
- PGE2 correlates with local clinical parameters, like eosinophils and inflammatory cell load.
1 INTRODUCTION
Allergic asthma (AA) comprises different disease forms that are classified either according to their clinical manifestations into phenotypes1 or to their biological mechanisms into endotypes.2, 3 One of the best-described endotypes is the eosinophilic or type-2 endotype.4 A number of sputum markers associated with type-2 endotypes were found to be elevated at both the protein and transcript levels, including secreted mediators of allergic inflammation such as IL-4, IL-5, IL-13, CCL17, CCL26,5, 6 IL-247 and periostin.8 The concept of the “united airway” postulates that the pro-inflammatory mediator signature is also present and regulated in the airways of patients with allergic rhinitis without concomitant asthma (AR), suggesting that upper and lower airways are interdependently regulated.6, 9 This concept postulates that the upper and lower airways form a morphological and functional unit, sharing common features related to physical barrier, mucociliary clearance and immune processes. These mechanisms can be studied in the lower airways using induced sputum samples, which consists of components derived from the respiratory epithelium of the central airways.10-12
MicroRNAs (miRs) are small nucleic acid molecules that suppress gene expression post-transcriptionally and therefore via an epigenetic mechanism.13 Individual miRs can have wide-ranging regulatory functions, as they can each target multiple mRNA transcripts along a particular signalling pathway.14 As microRNAs are quite resistant to degradation, they can be easily be detected in body fluids including samples from the lower airways such as induced sputum. Along this line, deregulated microRNAs have been described in asthma as well as in allergic rhinitis (reviewed in15). For example, levels of miR-122-5p were increased in sputum and serum of severe, uncontrolled asthmatics in comparison to healthy controls,16 and miR-223-3p was associated with increased neutrophil counts and T-helper cells of type 17 signalling in the sputum of asthmatics.17 In allergic rhinitis, miR-206, −338-3p, −329 and −26a were found to be elevated in patients with AR, but not in healthy individuals or those with asthma.18 Recent studies underscored the importance of miRNAs in the regulation of airway epithelial responses in asthma. MiR-23a was postulated to target IL-4 in epithelial cells.19 Finally, also airway epithelium-derived miRs (miR-34a, miR-92b and miR-210) were implicated to play a role in the early development of the local immune processes in asthma.20
Interestingly, miR-mediated expression inhibition also plays a role in eicosanoid biosynthesis.21 Eicosanoids are key mediators of type-2 inflammation,22-25 with miRs intervening at different steps on the eicosanoid pathway. In fact, it has been reported for the cyclooxygenase-2 (COX-2) that 28 miRs intervene in its gene regulation and thereby affect subsequent PGE2 production,26, 27 in particular, miR-155, the expression of which is increased in smooth muscle cells of the asthmatic airways.28 In addition, miR-155 is systemically increased in severe asthmatics with cockroach allergy compared with non-asthmatics and has been shown to enhance COX-2 and the concomitant Th2/Th17-associated lung inflammation in asthmatic mice as well as in patients allergic to cockroaches.29
IL-13, produced by Th2 cells, but also by group 2 innate lymphoid cells (ILC2),30-33 was found to increase neutrophil-derived PGE2 levels through upregulation of COX-2 gene expression.34 The suppressive effects of allergen-specific immunotherapy (AIT) on type-2 immune responses not only affect Th2 cells but also induce a shift from ILC2 to ILC1.35
The effect of allergen-specific immunotherapy on local miR expression and the corresponding gene transcript level, however, remains to be studied in patients, particularly in the mucosal context. In order to resolve local miR-related mechanisms including regulatory and anti-inflammatory mechanisms of allergic airway diseases, in this study, we profiled the genome-wide miR and simultaneously predicted miR-target mRNA signatures in induced sputum of patients with allergic rhinitis with (AA) and without (AR) concomitant asthma.
2 METHODS
2.1 Patients and IRB statement
All interventions were performed in the Allergy Section, Department of Otolaryngology, TUM School of Medicine, Munich, Germany. We used pooled data from two observational, non-interventional studies within three subsequent years, which were approved by the Institutional Review Board (IRB), namely the local Ethics Commission of the TUM School of Medicine, Munich, Germany (5534/12 and 5156/11). Each participant gave informed, written declaration of consent. Samples were taken from 44 healthy individuals and from 37 patients with grass pollen-related allergic rhinitis without concomitant asthma (AR), from whom, 21 had received grass pollen-specific AIT (patient characteristics; Table 1). In addition, 43 patients with grass pollen-allergic rhinitis who suffered from concomitant asthma (AA) were included, for whom the GINA scores were evaluated. In the group of 43 AA patients, 22 patients had received grass pollen-specific AIT. Further patient details can be found in the Supplemental Information. Asthma patients were classified as asthmatic based on a previous doctor's diagnosis and a reported history of shortness of breath, tightness in the chest and coughing during natural allergen exposure, that is during pollen flight, and / or a previously documented positive bronchodilation test. A GINA score was assessed from asthma patients. Patients with a grass pollen allergy with a history of moderate and chronic persistent allergic rhinitis according to the definition of allergic rhinitis and its effect on asthma (ARIA) criteria36 for more than two years during the grass pollen season, a wheal with a diameter of more than three millimetres and grass pollen-specific IgE levels above 0.70kU/l, were included. To avoid effects of, for example, inhaled corticosteroids, patients were asked to stop medication intake seven days before sampling to have an adequate washout period. AIT treatment was not interrupted for the study. All patients had good lung function (FEV1% > 70%). Patients in the AIT-treated groups received AIT for at least one year. All subjects completed a questionnaire on the quality of life in rhinoconjunctivitis (mRQLQ), received a pulmonary function test and underwent a sputum induction using the method described below.
| Controls (n = 44) | Allergic rhinitis w/o AIT (n = 16) | Allergic rhinitis with AIT (n = 21) | Allergic asthma w/o AIT (n = 21) | Allergic asthma with AIT (n = 22) | |
|---|---|---|---|---|---|
| Age [years] | 24.77 (±0.56) | 24.00 (±0.77) | 26.75 (±0.92) | 25.90 (±1.26) | 31.82 (±1.83) |
| Sex (m/f) | 27/17 | 8/8 | 10/11 | 10/11 | 16/6 |
| GINA score | n.d. | n.d. | n.d. | 1.17 ± 0.27 | 0.38 ± 0.16 |
| mRQLQ Score | 0.13 (±0.05) | 2.64 (±0.19) | 1.49 (±0.19) | 2.73 (±0.22) | 1.20 (±0.18) |
| Total IgE [IU/L] | 28.44 (±5.10) | 208.1 (±51.16) | 70.46 (±26.38) | 287.0 (±53.94) | 107.50 (±27.37) |
| sIge (kUA/l) | n.d. | 20.83 (±5.98) | 15.03 (±5.37) | 27.34 (±7.62) | 19.68 (±4.64) |
| (PT/CAP) to | |||||
| Grass | 0 | 16/16 | 21/21 | 21/21 | 22/22 |
| Birch | 0 | 14/13 | 15/12 | 17/15 | 15/14 |
| HDM | 0 | 10/9 | 8/9 | 9/9 | 6/6 |
| Cat | 0 | 5/3 | 8/5 | 13/11 | 10/8 |
| FVC [L] | 4.49 (±0.15) | 4.92 (±0.30) | 4.64 (±0.17) | 4.56 (±0.17) | 4.82 (±0.19) |
| FVC [%] | 98.60 (±1.77) | 103.40 (±2.69) | 104.40 (±3.16) | 101.40 (±3.01) | 102.60 (±2.69) |
| FEV [L] | 3.88 (±0.14) | 4.29 (±0.27) | 4.00 (±0.11) | 3.78 (±0.11) | 3.90 (±0.18) |
| FEV1 [%] | 98.89 (±1.87) | 105.00 (±2.59) | 105.00 (±2.32) | 98.86 (±2.82) | 98.00 (±2.99) |
| FEV1/FVC ratio | 85.93 (±1.08) | 87.06 (±1.23) | 87.00 (±1.46) | 82.99 (±1.74) | 77.66 (±3.48) |
| Sputum cells / ml (x104) | 89.34 (±12.62) | 250.5 (±47.42) | 71.29 (±11.72) | 282.3 (±61.99) | 80.46 (±10.52) |
| Macrophages (%) | 84.15 (±2.07) | 83.36 (±2.54) | 86.68 (±2.34) | 77.00 (±4.71) | 79.29 (±4.87) |
| Lymphocytes (%) | 1.09 (±0.21) | 1.43 (±0.23) | 0.95 (±0.18) | 2.81 (±0.97) | 0.93 (±0.31) |
| Neutrophils (%) | 14.79 (±2.05) | 12.21 (±1.98) | 11.68 (±2.21) | 15.38 (±4.48) | 18.57 (±4.45) |
| Eosinophils (%) | 0.23 (±0.11) | 3.36 (±0.78) | 0.79 (±0.33) | 5.13 (±1.23) | 1.50 (±0.69) |
Note
- Data are presented as mean ± SEM.
- Abbreviation: n.d., not determined.
2.2 Data collection and statistical analysis
All experiments were performed by researchers blinded to the diagnosis. Significant miR/mRNA expression changes of sputum cells were identified in the microarray data analysis by an initial principal component analysis (PCA) with a homogeneous component distribution, and by subsequent filtration by means of Volcano plot analysis using the moderated t test with the cut-off criteria of FC ≥ 1.5 (fold change) and p ≤ .05 using the GeneSpring Software GX 14.9 (Agilent Technologies). In the Genespring software, the miRNA microarrays were normalized using the percentile shift method. According to the manufacturer, it normalizes the microRNA array data using the 90th percentile. False-positive intensity values will most likely to fall into the lower percentile range of the array and are rated as noise, according to the manufacturer. Therefore, in this case, the 90th percentile is used as a robust intensity value to normalize the data. The details of the microarray analysis and all remaining techniques are described in detail in the supplemental information. The relative expression levels (log2 fold change) in the range from low in blue to high in red were shown as colour codes for the heat maps of the microarray analyses. Data in Figure 1A-G, 5A and B were tested statistically using Mann-Whitney U tests calculated with the GraphPad Prism software (GraphPad Software Inc.). In Figures 4G, 5C–F and in Figure S1A–C, Spearman correlation was used to correlated sputum PGE2 levels with clinical parameters and sputum cell counts. All statistically significant differences are shown as *p ≤ .05, **p ≤ .01, ***p ≤ .001, ****p ≤ .0001.
3 RESULTS
Induced sputum was obtained from allergic rhinitis patients with (AA) and without (AR) concomitant asthma, some of whom had received AIT for more than a year (AA+AIT; AR+AIT), and some of whom did not (AA-AIT; AR-AIT) as well as from healthy controls (HC) in order to gain access to local mediators of the lower airways.
3.1 Treatment-associated patient cohort characteristics and symptoms
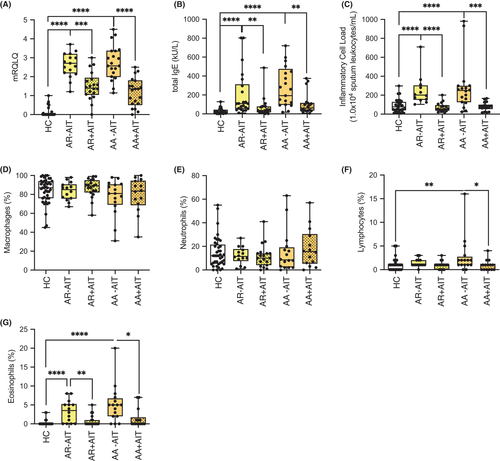
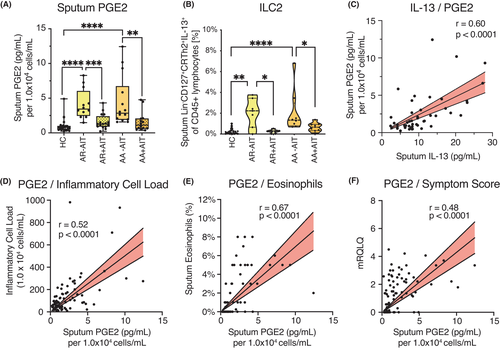
The patients who were recruited for this study were identified based on clinical parameters, and the symptom score was assessed by the validated disease-related mini quality of life questionnaire (mRQLQ) (Figure 1A). Successful AIT was confirmed both in AR and AA patients by reduced total serum IgE levels (Figure 1B). The local inflammatory cell load, identified by sputum leucocytes per millilitre, showed a pattern similar to total serum IgE and mRQLQ, with untreated AR and AA patients having an increased IgE titre, but reduced quality of life compared to controls and to AIT-treated patients (Figure 1C). The composition of sputum macrophages and neutrophils did not differ among the groups (Figure 1D,E), except for the frequency of lymphocytes, which was reduced in AIT-treated AR patients compared with untreated and was slightly decreased in treated AA patients (AR+AIT and AA+AIT; Figure 1F). Sputum eosinophils were practically absent in healthy subjects, while their number was strongly increased in AR-AIT patients and even more so in AA-AIT patients (Figure 1G). As expected, these numbers were reduced in AR+AIT and AA+AIT compared with untreated patients. These data confirm that the patient cohort in this study was clearly defined not only on the basis of the symptom burden and the clinical picture, but also on the basis of local inflammation parameters.
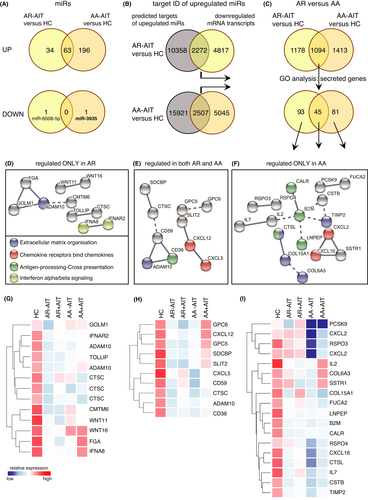
3.2 miR-target networks involve extracellular matrix organization, chemotaxis and antigen-presentation in allergic rhinitis with concomitant asthma
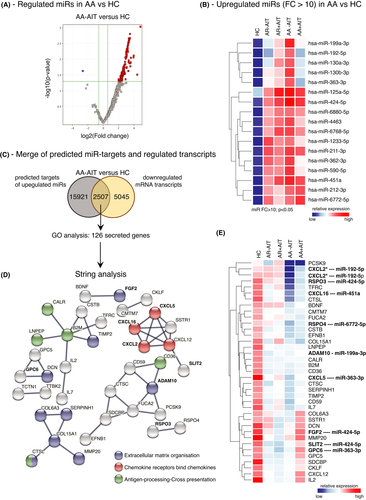
Since miRs are important regulators of airway epithelial responses, we focussed on the differential analysis of the expression of miRs in sputum cells from patients with allergic rhinitis with and without concomitant asthma. The cellular miR expression profile of induced sputum cells was measured using human microRNA microarray technology. Interestingly, 259 miRs out of 2549 miRs analysed were upregulated in sputum cells of AA-AIT patients, but only one miR was downregulated in comparison with healthy controls (Figure 2A; fold change (FC) ≥ 1.5; p ≤ .05). Figure 2B shows the top upregulated miRs with the cut-off at FC ≥ 10 in AA-AIT samples compared with HC samples (Table S1; Figure S2; Table S27). Next, in order to put the detected miRs into functional context in this disease background, we used an open-source miRNA database (mirdb.org) to predict transcript targets of the identified 259 cellular miRs. A whole-transcriptome microarray analysis of the same samples was performed, and the results from the miRdb database were inversely merged with significantly altered mRNA transcripts. The resulting cut set was mapped in order to define a potential miR-mRNA-network. 7552 mRNA transcripts were downregulated in AA-AIT patients compared with HC (FC≥1.5; p ≤ .05), of which 2507 were predicted targets of the 259 upregulated miRs (Figure 2C, Table S2). Using a GO-term filter, 126 out of this cut set were considered as secreted factors, which were then subjected to string network analysis to identify enriched cellular mechanisms (Figure 2D, Table S3). In AA-AIT patients, target genes formed enriched networks that are involved in extracellular matrix organization, in antigen-processing/cross-presentation and in chemotaxis. All of those genes were significantly downregulated in sputum cells from AA-AIT patients compared with HC (Figure 2E; Table S4; Figure S3; Table S28; FC ≥ 1.5; p ≤ .05) and 10 of them were predicted targets of miRs that were upregulated more than 10-fold.
3.3 Overlapping miR-associated gene regulation underpins the united airway concept in patients with allergic rhinitis
Similar to AA-AIT patients, in the sputum cells of AR-AIT patients, 97 miRs were increased and one miR was decreased compared with HC. Almost two-thirds of the elevated cellular miRs were also increased in asthmatic patients (Figure 3A, Tables S5–S6), although the main site of clinical manifestation of allergic rhinitis is the upper respiratory tract. The downregulated miR-6508 in the sputum of AR patients differed from the one downregulated miR in AA patients, miR-3935. We focussed on the miR targets, which showed a patient group-specific expression pattern, as well as on the genes that were synergistically downregulated in both patient groups. We identified 12630 predicted targets of locally upregulated miRs using the miRdb and 7089 downregulated mRNA transcripts in AR-AIT patients compared with HC (Figure 3B, Tables S7–S8). In this analysis, 2272 transcripts met both conditions. In a similar analysis in AA-AIT patients, 18428 predicted targets of upregulated were identified and 7552 mRNA transcripts were downregulated in the same samples, of which 2507 mRNA transcripts met both criteria. Next, a Venn analysis of the gene lists resulting from the analyses of AA-AIT versus HC and AR-AIT versus HC was performed to identify mRNA transcripts that are supposedly downregulated by miR (Figure 3C, Tables S9–S10), filtered for secreted genes and submitted to string network. The focus of this analysis was set on the above-mentioned regulated pathways in AA-AIT patients within the three subdivisions: targets only changed in rhinitis patients, targets changed in both patient groups and targets only changed in asthmatic patients (Figure 3D-I, Tables S11–S16; Figure S4–S6; Tables S29–S31).
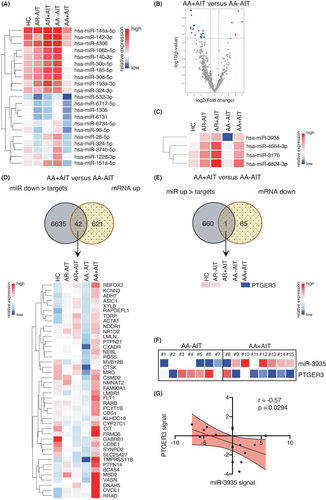
3.4 AIT-related induction of miR-3935 corresponds to suppression of transcripts of its target PTGER3
In comparison to miR regulation comparing allergic patients and healthy subjects, the analysis of the effect of AIT on cellular miR regulation revealed the opposite. Only four cellular miRs were locally upregulated, while 21 cellular miRs were downregulated in AA+AIT patients compared with AA-AIT patients (Figure 4A-C; Tables S17–S18; Figure S7–S8; Tables S32–33). In total, a number of 6677 genes were predicted as targets of the downregulated miRs (Figure 4D, Table S19; Figure S9; Table S34), of which 42 were significantly upregulated in AA+AIT patients compared with AA-AIT patients. On the other hand, 661 genes were predicted as targets of the upregulated miRs (Figure 4E, Table S20; Figure S10; Table S35), of which only one gene (PTGER3) was also significantly downregulated on the mRNA level in the same samples. PTGER3, the prostaglandin EP3 receptor (EP3), is one of four receptors for prostaglandin E2 (PGE2) and is involved in bronchoconstriction.37, 38 PTGER3 showed a counter regulation to miR-3935 as expected (Figure 4F, Figure S1E-F), which was underlined by the inverse correlation between miR-3935 and its predicted target PTGER3 (Figure 4G; r = −0.57; p = .0294). Of the four known PGE2 receptors EP1, EP2, EP3 and EP4, only EP3 was significantly changed on the mRNA level upon AIT. Although EP2 mRNA in sputum cells of asthmatics was slightly, but not significantly increased and EP4 was significantly reduced in comparison to healthy controls, no significant regulation was found comparing AA+AIT with AA-AIT patient samples (data not shown). In addition, immunofluorescence microscopy of the EP3 protein was performed on cytospin samples of the induced sputa of AA-AIT and AA+AIT patients, which revealed on the one hand that EP3 was mainly detected in macrophages and on the other hand, however, mainly in specimens from AA-AIT patients, while the staining of samples of the treated asthma group (AA+AIT) was weak (Figure S11). This reflects the above-described results on the mRNA expression levels of PTGER3 (EP3) (Figure 4F, Figure S1E&F).
3.5 Reduction of sputum protein levels of EP3 ligand PGE2 correlates with clinical parameters and symptoms in AIT-treated patients
Finally, we measured the levels of the EP3 ligand PGE2 in the sputum supernatants of the same samples using ELISA. PGE2 was increased in the supernatants of AA-AIT patients compared with HC and was decreased upon AIT in AA and AR patients (Figure 5A). Since PGE2 has been reported to be induced via IL-13 and COX-2, we examined the frequencies of IL-13-expressing ILC2s using flow cytometry (ILC2: CD45+Lineage−CD127+CRTh2+IL-13+; Gating strategy: Figure S1D) and found that these cells were indeed increased in AR-AIT and AA-AIT patients compared with HC and decreased in AIT-treated compared with untreated AR and AA patients (Figure 5B). IL-13 and IL-22 protein levels in the sputum supernatants of the same samples also correlated positively with sputum PGE2 levels (Figure 5C; Figure S1). Notably, the PGE2 levels in sputum supernatants further correlated positively with local inflammatory cell load (Figure 5D; r = 0.52, p < .0001) and sputum eosinophil frequencies in the same sputum samples (Figure 5E; r = 0.67, p < .0001; for neutrophils and macrophages, please see: Figure S1) as well as with symptom burden assessed with the mRQLQ (Figure 5F; r = 0.48, p < .0001).
4 DISCUSSION
The global profiling of miRs in local inflammation of the lower airways of allergic rhinitis and asthma reveals novel local miR expression patterns and confirms previously described disease-related miRs. These data were collected in the context of therapeutic interventions using allergen-specific immunotherapy.
4.1 Known allergy-associated miRs detected in the lower airways
The miRs detected comprise previously reported miRs in the context of asthma, such as miR-15b-5p, whose systemic levels are associated with lung function,39 miR-16-5p, whose circulating levels correlated with hyper-responsiveness of the airways,18, 40, 41 miR-19a-3p, which has been reported to be expressed in local T cells,42 and miR-21-3p, which was found in exhaled breath condensate from asthmatics, however in reduced levels compared to healthy controls.43 In addition, we detected elevated levels of miR-191-5p, the serum levels of which also correlated with lung function, measured with FEV1/FVC, in a recent paediatric study,39 and miR-210-3p, which is involved in mast cell activation,44 and, finally, miR-223-3p, which was increased in asthmatics compared to healthy controls and correlated with lung function parameters.45 It has further been published that miR-223-3p is involved in the maturation and function of neutrophils.46
In order to understand the role of so far unknown miRs in the regulation of airway diseases, we also performed a transcriptome analysis on the same samples. This transcriptome analysis was then limited to predicted target transcripts of upregulated miRs, which were reversely regulated in the same sample. This analysis revealed a large number of genes whose mRNA levels were downregulated. Since the function of the luminal sputum cells is critically determined by their secreted factors, we focussed on the bioinformatic analysis of secreted and extracellular entities. The unsupervised network analysis revealed three principal mechanisms, namely those of extracellular matrix organization (including genes like DCN, ADAM10, MMP20, TIMP2, LOXL3, FGF2, CTSK, ADAMTS3, COL6A3, CTSL, COL15A1, LAMA1, ADAM9, SERPINH1), antigen-processing/cross-presentation (LNPEP, CALR, CTSL, CD36, B2M) and chemotaxis (CXCL16, CXCL5, CXCL12, CXCL2).
4.2 miRs in upper and lower airways
MiR target genes involved in extracellular matrix reorganization were mainly repressed in asthmatic patients only, which was expected as asthma mainly manifests in the lower respiratory tract and has frequently been associated with extracellular matrix remodelling.47-50 Similarly, the factors CALR, B2M, CTSL and LNPEP associated with antigen-processing/cross-presentation were only affected in sputum cells from AA-AIT patients, but not from AR-AIT patients, while CD36 (predicted target of miR-629-3p, miR-3180-5p, miR-485-3p and miR-3605-5p) was downregulated in both groups. However, this could be also due to inflammatory processes that are reflected by a shift from a macrophage-dominated towards leucocyte- and eosinophil-dominated in cell composition of the sputum. The scavenger receptor CD36 is known to be an important factor for macrophage recognition and phagocytosis of apoptotic fibroblasts51 and to promote house dust mite allergy development.52 Notably, CD36 was also increased in exosomes from asthmatics and associated with leukotriene production.53 On the other hand, the reduction in the chemokine expression was common to both diseases. While CXCL2 (predicted target of miR-192-5p, miR-215-5p, miR-7–1-3p and miR-582-5p) and CXCL16 (predicted target of miR-451a and miR-7–1-3p) were only affected in asthmatics, CXCL12 (predicted target of miR-141-3p, miR-200a-3p, miR-5100, miR-222-3p, miR-5587-5p, miR-3928-5p and miR-3605-5p) and CXCL5 (predicted target of miR-25-3p, miR-363-3p and miR-3163) were also affected in AR-AIT patients. CXCL2 is known to regulate airway smooth muscle cell migration54 and is a predicted target of multiple miRs, including miR-192-5p, which is upregulated more than 10-fold in this data set. CXCL16 on the other hand exhibits a direct chemoattractant effect on ILC2,55 while CXCL12 levels were reported to correlate with decreased ILC2 numbers in blood of asthmatic patients.56 Thereby, rhinitis patients share not only the reduction of an ILC2-attractant chemokine, but also of the scavenger receptor CD36 and ADAM10 with asthmatic patients, which underscores the united airway concept mentioned above. In addition, our data show a decrease of IFNA8 (predicted target of miR-642b-3p) and IFNAR2 (predicted target of miR-6765-3p and miR-4286) in allergic rhinitis patients only, which is in line with the observation that allergic rhinitis is related to the dominance of type-2 cytokines, which is reversed by allergen-specific immunotherapy.5, 7 We have previously shown that in the upper airways of AR patients, the interferon expression levels (IFNGR2) were restored after more than three years of AIT.
4.3 miR-3935-target PTGER3 and associated PGE2 regulation
Studying miR expression in AIT-treated patients compared to untreated patients, we found that the AIT-associated upregulated miRs unexpectedly targeted structural genes rather than immunoregulatory factors. However, the AIT-associated downregulated miR-3935 targets prostaglandin EP3 receptor (PTGER3), the receptor for prostaglandin E2 (PGE2). It was originally described to have bronchodilatory effects mediated through an inhibitory effect on smooth muscle cells57-59 and mast cells that mainly depends on EP2 signalling.60 However, PGE2 also influences the behaviour of several immune cells, namely macrophages,61 T cells62 and B cells,63 thus suppressing type-2 inflammation. In this study, the levels of secreted PGE2 in the sputum supernatant of the same samples resembled the mRNA levels of its receptor PTGER3. Depending on tissue and condition, PGE2 has either pro- or anti-inflammatory effects.64-71 In this study, sputum PGE2 was associated with a pro-inflammatory state in patients with allergic asthma, since it correlated with pro-inflammatory clinical parameters like sputum inflammatory cell load and sputum eosinophil counts. Furthermore, sputum PGE2 levels were decreased in patients who had undergone AIT. Further research is needed to elucidate the bilateral roles of PGE2 in inflammatory processes in the airways in different diseases. It might even present itself as a critical factor in controlling inflammation in one versus the other direction. Future research needs to focus on the question, how lipid mediators can contribute to allergen tolerance and selective inhibitors can be utilized to promote tolerance re-induction.
As mentioned above, PGE2 exerts its action via its four G-coupled protein receptors EP1, EP2, EP3 and EP4.72 Previous studies have mainly focussed on the effect of PGE2 in inflammatory processes exerted through the EP2 and EP4 receptors; however, its effect via the EP3 receptor is not well understood and remains to be elucidated in further detail. Studies in human and guinea pig systems suggest a broncho-constrictive effect of PGE2-EP3 signalling,37, 38 thus suggesting that miR-mediated downregulation of EP3 expression upon AIT may have beneficial effects on lung function in asthmatic patients.
The finding that the levels of PGE2 were regulated synergistically to EP3 leads us to believe that the pro-inflammatory, bronchoconstrictor effects of the PGE2 via EP3 are suppressed in AIT-treated patients upon upregulation of miR-3935 and downregulation of EP3 and PGE2. An indication of the pro-inflammatory characteristics of PGE2 in this disease context is the finding that the sputum PGE2 levels correlate positively with clinically assessed parameters such as sputum inflammatory cell load, sputum eosinophil infiltration and symptom burden. We therefore assume that this potential miR-3935-related mechanism suppresses the local pro-inflammatory effects of the PGE2 signalling via EP3, which in turn could represent one novel mechanism of AIT in allergic asthma. This hypothesis is supported by a recent publication describing reduced spontaneous release of PGE2 by PBMCs a few months after venom-specific immunotherapy, which negatively correlated with the tolerogenic cytokine IL-10.73 As PGE2/EP3 signalling can trigger mast cell degranulation and pro-inflammatory mediator release,74, 75 miR-3935-mediated downregulation of EP3 may contribute to mast cell desensitization following AIT. Thus, future studies should assess potential correlations between miR-3935, EP3 and markers of mast cell activation (e.g., histamine, CysLTs) in patients undergoing AIT.
In summary, this study shows a one-sided AA-associated upregulation of miR expression in sputum cells, whereas the AIT-associated miR expression revealed the opposite picture. These data provide first evidence for a negative regulatory association of miR-3935 and its predicted target gene, the PGE2 receptor EP3, as a potential AIT-mediated mechanism in the airways of AA patients. In this study, sputum PGE2 correlates with pro-inflammatory cellular and clinical parameters. Sputum PGE2 levels also drop in AIT-treated patients, which provides evidence for an involvement of the prostanoid PGE2 in local inflammatory processes mediated by AIT.
This study therefore provides evidence that associates AIT with epigenetic processes in the lungs of allergic asthmatic patients. Future studies are needed to determine whether miR-3935 is a candidate biomarker for AIT efficacy.
CONFLICT OF INTEREST
Dr. Jakwerth has a patent “A ratio of immune cells as prognostic indicator of therapeutic success in allergen-specific immunotherapy: 17 177 681.8” that is pending. PD Dr. Chaker reports on grants and other from Allergopharma during the conduct of the study; grants and/or other from ALK Abello, Bencard / Allergen Therapeutics, ASIT Biotech, Lofarma, GSK, Novartis, LETI, Roche, Zeller, Sanofi Genzyme/Regeneron, European Institute of Technology, AstraZeneca, Immunotek, all outside of the submitted work. In addition, PD Dr. Chaker has a patent “A ratio of immune cells as prognostic indicator of therapeutic success in allergen-specific immunotherapy: 17 177 681.8” that is pending. Prof. Dr. Schmidt-Weber reports personal fees from Bencard and personal fees from Allergopharma during the conduct of the study. In addition, Prof. Dr. Schmidt-Weber has a patent on nasal secretions that is pending, and a patent “A ratio of immune cells as prognostic indicator of therapeutic success in allergen-specific immunotherapy: 17 177 681.8” that is pending. Dr. Zissler reports grants and personal fees from German Center for Lung research (DZL), grants and personal fees from German Research Foundation (DFG), grants from Helmholtz I&I Initiative, during the conduct of the study. In addition, Dr. Zissler has a patent “A ratio of immune cells as prognostic indicator of therapeutic success in allergen-specific immunotherapy: 17 177 681.8” that is pending and Dr. Zissler received payment for manuscripts from Deutsches Aerzteblatt and funds for travel from the European Academy of Allergy and Clinical Immunology (EAACI) and Collegium Internationale Allergologicum (CIA). The rest of the authors declare that they have no relevant conflicts of interest.
AUTHOR CONTRIBUTIONS
C.A.J., A.M.C., C.S.-W., and U.M.Z. involved in study design; C.A.J., F.G., M.O., L.P., L.G., J.U., J.K., M.P., M.W., A. Ku., A. Kl., and U.M.Z. conducted the experiments; C.A.J., A.M.C. F.G., M.O., L.P., L.G., J.U., J.K., M.P., M.W., A. Ku., A. Kl. and U.M.Z. collected the data; C.A.J., F.G., and U.M.Z. analysed the data; C.A.J., A.M.C., A.E., S.K.-E., J. EvB, C.S.-W., and U.M.Z. involved in data interpretation; C.A.J.,, A.M.C., A.E., S.K.-E., J. EvB, C.S.-W., and U.M.Z. involved in literature search; all authors wrote the article.
Open Research
DATA AVAILABILITY STATEMENT
The data discussed in this publication are accessible via the accession number GSE184382 deposited in NCBI's Gene Expression Omnibus (GEO). All other methods are described in the supplemental information.