Discovery of Novel Allosteric Eg5 Inhibitors Through Structure-Based Virtual Screening
Abstract
Mitotic kinesin Eg5 is an attractive anticancer drug target. Discovery of Eg5 inhibitors has been focused on targeting the ‘monastrol-binding site’. However, acquired drug resistance has been reported for such inhibitors. Therefore, identifying new Eg5 inhibitors which function through a different mechanism(s) could complement current drug candidates and improve drug efficacy. In this study, we explored a novel allosteric site of Eg5 and identified new Eg5 inhibitors through structure-based virtual screening. Experiments with the saturation-transfer difference NMR demonstrated that the identified Eg5 inhibitor SRI35566 binds directly to Eg5 without involving microtubules. Moreover, SRI35566 and its two analogs significantly induced monopolar spindle formation in colorectal cancer HCT116 cells and suppressed cancer cell viability and colony formation. Together, our findings reveal a new allosteric regulation mechanism of Eg5 and a novel drug targeting site for cancer therapy.
Kinesins represent a family of cytoskeletal motor proteins that utilize the energy from ATP hydrolysis to perform mechanical work along microtubules (MT) and mediate cellular processes such as cargo transport, spindle assembly, and chromosome movement 1. Mitotic kinesins are required for various aspects of mitosis, including bipolar spindle assembly, chromosome alignment, chromosome segregation, and cytokinesis 2. Mitotic kinesin Eg5, a plus-end-directed member of the kinesin-5 subfamily, is an attractive anticancer drug target. Inhibition of Eg5 function blocks centrosome migration and leads to cell cycle arrest and eventually apoptotic cell death 3. As Eg5 is exclusively involved in mitotic spindle of proliferating cells, Eg5-specific inhibitors exhibit improved safety profiles compared to other traditional antimitotic drugs which target the multifunction relevant MT. Discovery of small molecule inhibitors of Eg5 has attracted much attention in the past decade, and several Eg5 inhibitors have advanced into clinical trials 4-18.
Monastrol is the first Eg5 inhibitor that was identified over a decade ago 19. Since then, drug discovery studies targeting Eg5 have been mainly focused on an allosteric site where monastrol binds. However, mutagenesis studies have demonstrated that subtle changes at this ‘monastrol-binding site’ conferred resistance to ligand binding without affecting the enzymatic function of Eg5, indicating acquired drug resistance of current Eg5 inhibitors 20, 21. In addition, mutations at the monastrol-binding site were found in Eg5 inhibitor (ispinesib)-resistant cancer cells 22. Therefore, identification of new inhibitors that interact at a different binding site(s) of Eg5 could be a unique and important strategy to complement current Eg5 drug candidates.
Experimental studies have demonstrated the existence of multiple allosteric sites of Eg5 23-25. However, the exact locations of these allosteric sites are not clear. We have previously conducted molecular dynamics (MD) simulation studies of Eg5 and identified several novel allosteric sites (Figure 1) using correlation analysis and binding site mapping based on the simulation-generated structural and dynamics results 26. The identified S1 and S2 sites have been predicted as suitable for tight binding of small molecules and therefore are potential targeting sites for the discovery of novel Eg5 inhibitors. A recently published crystal structure of an Eg5-inhibitor complex27 confirmed the existence of the predicted S2 site and validated our computational strategy for identifying novel allosteric sites.
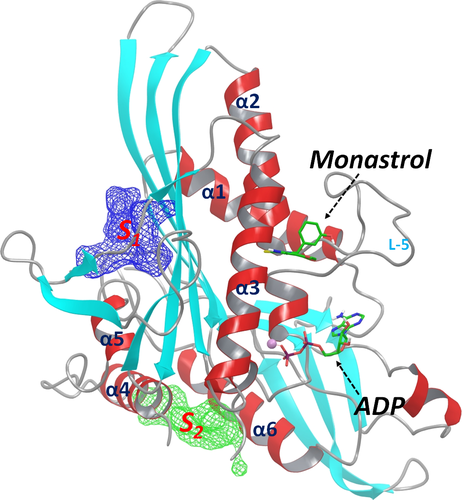
The other identified allosteric S1 site locates nearby the functionally important kinesin neck linker region 28. It has long been recognized that conformational changes of kinesin neck linker are correlated with structural relaxation of kinesin α4–helix, and such a correlation is crucial for the force-generating behavior of kinesin proteins 29-31. A ligand that binds at the S1 site would block the interactions between the neck linker and the kinesin core structure, interrupt the force-generating conformational changes, and consequently inhibit the kinesin functions. So far, no published studies have targeted this allosteric S1 site. In this study, we explored the S1 site of Eg5 and identified several novel inhibitors through structure-based virtual screening (SBVS). These Eg5 inhibitors significantly induced monopolar spindle formation and suppressed cell viability of colorectal cancer cells.
Methods and Materials
Preparation of Eg5 motor protein
Protein expression and purification were based on published protocols 32-34. Eg5 motor domain (amino acids 15-368) was cloned into pEt24a and expressed in Escherichia coli BL21 (DE3) cells for 12 h at 30°C. Protein purification was carried out in two steps with His-tag affinity and gel-filtration chromatography. Specifically, cell pellet was resuspended in the lysis buffer (75 mm Tris–HCI pH 8.0, 300 mm NaCl, 5% glycerol, 20 mm imidazole, 0.1% Triton-X-100, 0.5 mm TCEP) supplemented with a protease-inhibitor cocktail and 1 μg/mL benzonase nuclease and was lysed by two passages through a French press. Cell debris was removed by centrifugation, and the clear supernatant was passed through a nickel-chelating Sepharose column (Amersham Bioscience, Piscataway, NJ, USA) equilibrated with the lysis buffer. The column was then washed extensively with washing buffer (20 mm Tris–HCI pH 8.0, 300 mm NaCI, 5% glycerol, 20 mm imidazole and 0.5 mm TCEP) to remove non-specific proteins. The bound proteins were eluted using a 300-mL linear gradient of 50–400 mm imidazole in an elution buffer (20 mm Tris–HCI pH 8.0, 300 mm NaCI, 5% glycerol, 400 mm imidazole, and 0.5 mm TCEP). Eluted fractions were analyzed by SDS-PAGE, and fractions containing Eg5 were pooled. Further purification was performed with a gel-filtration column (Superdex 200 26/60 Amersham Biosciences) (buffer: 50 mm phosphate pH 7.4, 100 mm NaCI and 1 mm DTT). Purified protein fractions were pooled together and concentrated to 10 mg/mL, frozen quickly in liquid nitrogen, and stored at −20°C.
Virtual compound library
For the purpose of applying structure-based virtual screening, we assembled a compound library consisting of approximately 500 000 structurally diverse compounds selected from different commercial sources. Specifically, structures of a total amount of approximately eight million compounds were downloaded directly from the websites of ten vendors (Asinex, Chembridge, ChemDiv, Enamine, FCH group, InterBioScreen, Life chemicals, TIMTEC, SPECS and Vitas-M). From the eight million commercially available compounds, we first identified the most diverse set of 100 000 structurally representative compounds using the clustering and diversity analysis protocols of Pipeline Pilot 35; then for each of the 100 000 compounds, four structurally most similar analogs were selected based on the Tanimoto coefficients calculated from the 2D structural fingerprints. Such an assembled library covers a large portion of chemical space, contains diverse structures and could provide initial structure–activity relationship (SAR) information.
Molecular modeling
Structural model generation and molecular docking studies were conducted using the programs of the Schrödinger Suite 2014 (Schrödinger, LLC, New York, NY, 2014). The Eg5 model was generated based on the crystal structure of ADP-bound Eg5 complex (PDB ID: 1II6) using the protein preparation wizard of the maestro program1. The sitemap program 25 was used to identify potential binding pocket(s) on Eg5 by mapping the surface of the structural models. The 3D structures of ligands were prepared using the ligprep program 36. The glide program2 was used for docking studies with the default parameters. Specifically, the Induced-Fit-Docking (IFD) protocol of glide 37, which is capable of sampling dramatic side-chain conformational changes as well as minor changes in protein backbone structure, was applied to explore the binding mode of the identified active compounds. Residues within 5 Å of the docked ligands were allowed to be flexible, and the docked results were scored using the extra-precision (XP) mode of glide.
Virtual screening
Structure-based virtual screening was performed using the virtual screening workflow of maestro, which uses a three-step docking/scoring protocol implemented in glide. 3D conformations of the 500 000 assembled compounds were prepared using ligprep, and a total of 923 408 conformers were generated. All of the 923 408 conformers were first docked into the S1 site of Eg5 using the high-throughput virtual screening (HTVS) mode; the top 5% best-scored conformers were then redocked and scored using the standard-precision (SP) mode; and the top 10% of the SP docking resulted conformers were further docked and scored using the extra-precision (XP) mode. Finally, the top 25% of the best XP-scored conformers were outputted for visual examination.
Enzyme inhibition assay
The ADP Hunter™ Plus assay from DiscoveRx, which detects the MT-stimulated enzymatic ATPase activity of Eg5, was used to evaluate compound activity. Compounds were first tested in triplicates at 100 μm; hit compounds were further tested with a 10-point twofold serial dilution to confirm their activity and determine their IC50 values. S-Trityl-L-cysteine (STLC), a selective Eg5 inhibitor 38, 39, was used as the control compound. Specifically, 20 μL of 15 mm PIPES (pH 7.0) containing 1 mm MgCl2, 50 nm MT, 20 μm paclitaxel, 200 μm ATP, 5% DMSO, 60 nm Eg5 proteins, and 1:2 serial dilutions of each individual compound starting from 1000 μm were added to each well of a 96-well plate. The plate was incubated at room temperature for 0.5 h, and then, the ADP Hunter™ Plus reagents were added. The plate was further incubated for 0.5 h and then read for fluorescence (ex.530/em.590) on Synergy 4 (BioTek, Winooski, VT, USA).
NMR spectroscopy
STD-NMR data were collected following established protocols 40, 41 using a Bruker DRX500. Samples containing SRI35566 and Eg5 protein at a concentration ratio of 20:1 were prepared in D2O. STD-NMR spectra were recorded with a total of 32 K points, 80 scans, and selective saturation of protein resonances at 0, 0.65, 1.67, and 7.61 ppm (−8.18 ppm for the reference spectra), using a series of SEDUCE pulses (1000 points, 50 mseconds), for a total saturation time of 10 seconds (SEDUCE-1 pulse is similar to a Gaussian pulse and has been used by other laboratories 42. Reference experiments using the free ligands themselves (i.e. without Eg5) were performed under the same experimental conditions to verify true ligand binding.
Cell culture
Colorectal cancer cell line HCT116 was obtained from ATCC and cultured in RPMI1640 medium containing 10% fetal bovine serum, 2 mm of l-glutamine, 100 units/mL of penicillin, and 100 μg/mL of streptomycin. Cells were grown under standard cell culture conditions at 37°C in a humidified atmosphere with 5% CO2.
Confocal microscopy
HCT116 cells were grown on glass coverslips. After overnight incubation, the cells were treated with each individual compound for 16 h. The cells were fixed with 4% paraformaldehyde in phosphate-buffered saline for 20 min and permeabilized with 0.2% Triton-X-100 for 10 min at room temperature. The cells were then incubated with anti-α-tubulin-FITC antibody (Sigma) for 45 min, and nuclei were stained with NucRed647 for 10 min. The cells were then examined by a laser scan confocal microscope (Leica DMI 4000 B, Buffalo Grove, IL, USA). All images were captured with an HCX PL Apo 63x oil immersion objective. Images were processed and analyzed using Leica's LAS Image Analysis software. The percentage of mitotic cells with monopolar spindles was calculated out of a total number of a minimum of 20 mitotic cells per coverslip that were detected in different and randomly chosen microscopic fields.
Cell viability assay
Cells were seeded into 96-well tissue culture-treated microtitre plates at a density of 4000 cells/well. After overnight incubation, the cells were treated with compounds for 96 h. Cell viability was measured by the CellTiter-Glo Assay (Promega, Madison, WI, USA).
Colony formation assay
HCT116 cells were seeded at a density of 500 cells/well into 6-well plates. After overnight culture, the cells were treated with compounds at 80 μm, and media were replenished every 3 days. After being incubated for 14 days, colonies were fixed with 4% formaldehyde, stained with 0.5 mg/mL crystal violet, and imaged on a FluorChem HD2 Imager System (Alpha Innotech, San Leandro, CA, USA).
Results
Structural characterization of the allosteric S1 site
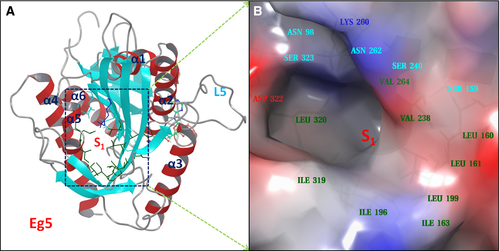
Our previous study based on MD simulations demonstrated that the residues at the S1 site of Eg5 correlate dynamically with the active site (nucleotide-binding site) residues, suggesting compounds that bind to the S1 site could allosterically affect the function of Eg5 26. We analyzed the Eg5 crystal structure (PDB ID: 1II6) using the sitemap program, which was designed for mapping and scoring potential binding site(s) based on properties such as binding pocket size, exposure/enclosure, contact, hydrophobic/hydrophilic balance, donor/acceptor character. A SiteScore value of 0.80 has been shown to accurately distinguish a drug-binding site from non-drug-binding sites 25. The predicted SiteScore value of the Eg5 S1 site is 0.98, suggesting an excellent site for tight binding of small molecule drugs. As shown in Figure 2, the S1 site locates at the opposite side of the active site and consists of residues from the short α5-helix and the surrounding beta-sheets. It is an open pocket formed mainly by hydrophobic residues, including Leu160, Leu161, Ile163, Ile196, Leu199, Val238, Val264, Ile319, and Leu320, with several polar (Ser159, Ser240, Asn262, and Ser323) and charged (Asp322, Lys260) residues at the entrance area.
Identification of an Eg5 inhibitor through structure-based virtual screening
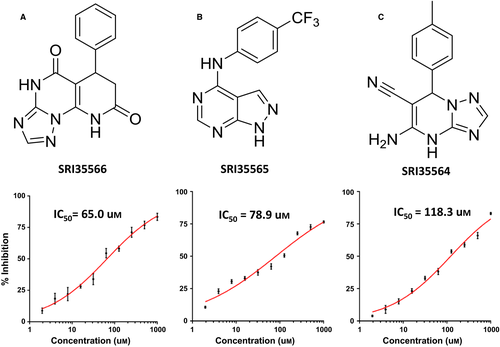
To explore whether the S1 pocket is a real binding site that can be targeted to modulate Eg5 function, we conducted SBVS to identify compounds that can potentially bind to the S1 site. We assembled 500 000 structurally diverse compounds from approximately eight million commercially available compounds. Using the Eg5 crystal structure as the receptor template, we screened these 500 000 compounds through a three-stage docking process. From the top-scored SBVS results, we selected 50 compounds that showed sufficient structural complementarity to the S1 site based on visual examination of the docked Eg5-compound complex models. Among the 50 selected compounds, 37 commercially available compounds were finally purchased and tested in the Eg5 ATPase assays. One compound, SRI35566 (Figure 3A), was confirmed by the serial dilution assay as an Eg5 inhibitor with an IC50 value of 65 μm.
SRI35566 binds directly to Eg5 without involving microtubule
As the ADP Hunter™ plus assay detects the MT-stimulated Eg5 ATPase activities, there is a chance that the identified active compounds may inhibit Eg5 function through a MT-related mechanism, which is unwanted due to the side-effects of interfering MT 43. For instance, compounds that bind to MT (such as taxanes44) or bind to kinesin-MT complex (such as AZ8245) will be detected as actives in this assay. To examine whether the inhibitory effect of SRI35566 is related to MT, we applied STD-NMR to test the Eg5-SRI35566 binding in the absence of MT. STD-NMR is a technique that not only detects transient binding, but can also provide information regarding which part(s) of a ligand interacts directly with a receptor 40, 42, 46. In the case that the ligand does not bind to the protein in a ligand–protein mixture sample, no STD-NMR signal will be detected. The observed STD-NMR spectrum of the SRI35566/Eg5 mixture sample proved that SRI35566 bound directly to Eg5 (Figure 4). As a control, no STD signals were present in the free ligand sample (data not shown), which confirmed that the observed STD signals in the presence of Eg5 were due to a true saturation transfer from the protein.
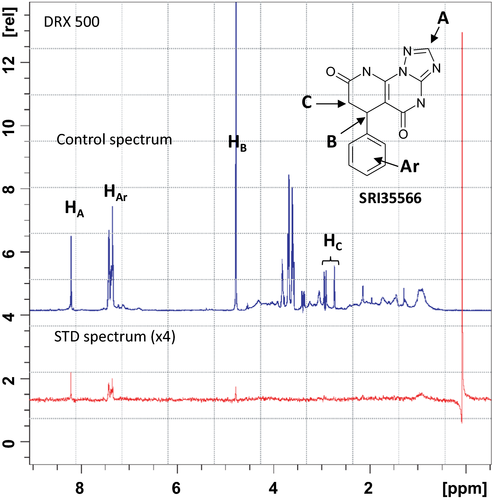
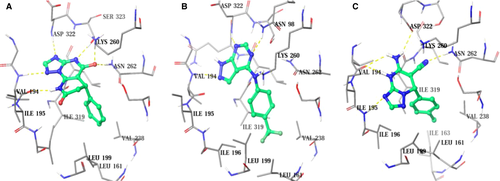
To further explore the structural insight of Eg5-SRI35566 interactions, we redocked the compound using an Induced-Fit Docking (IFD) protocol to take into consideration structural flexibility of the binding site residues. The predicted Eg5-SRI35566 complex model (Figure 5A) shows that SRI35566 fits well into the S1 pocket: The phenyl ring of SRI35566 is buried deeply into the hydrophobic core, and the heterocyclic fragment of SRI35566 further stabilizes its binding by forming multiple hydrogen bonds with the side chain and/or main chain atoms of the residues near the entrance. Interestingly, these hydrogen atoms that showed clear STD-NMR signals (Figure 4) are in close proximity to the Eg5 residues in the docked model (Figure 5A). The distances of the closest protein–ligand atom pairs for HAr, HA, and HB are 2.24, 2.68, and 2.22 Å, respectively. As a comparison, the closest distance for the HC pairs, which did not show observable STD-NMR signal, is 4.21 Å. The consistency between the docked model and the STD-NMR results supports the predicted binding mode at the S1 site. Taken together, these results suggest that SRI35566 inhibits Eg5 through a direct binding at the allosteric S1 site.
Analog exploration
A hit expansion effort based on structural similarity and substructure searches was further devoted to identifying commercial analogs of SRI35566. About 5000 compounds were selected using different search strategies. Those compounds were then docked into the S1 site of Eg5 and visually examined for their structural complementary to the binding site. Thirty-five SRI35566 ‘analogs’ that docked well into the S1 site were purchased and tested in the Eg5 ATPase assay. Two compounds, SRI35565 and SRI35564, were confirmed as Eg5 inhibitors with IC50 values of 78.9 and 118.3 μm, respectively (Figure 3B,C). For both compounds, docking studies using the IFD protocol resulted in similar binding modes as SRI35566, with strong hydrophobic interactions at the main pocket and additional polar interactions at the gate region (Figure 5B,C).
Identified Eg5 inhibitors trigger monopolar spindle formation in cancer cells
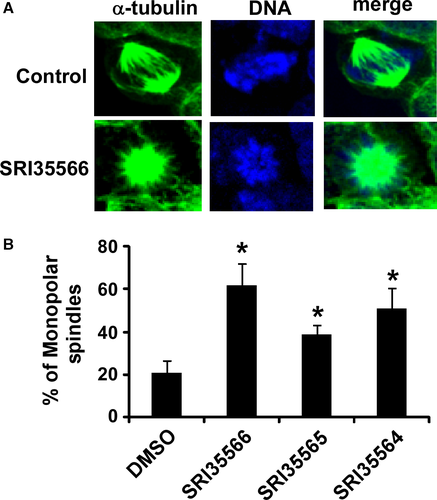
Eg5 is a highly conserved plus-end-directed MT motor that plays a critical role in bipolar spindle assembly, and loss of Eg5 induces monopolar spindle formation 38, 47-49. Therefore, we performed spindle formation assay in colorectal cancer HCT116 cells. As shown in Figure 6, SRI35566, SRI35565, and SRI35564 at 80 μm significantly enhanced the percentage of HCT116 cells containing monopolar spindles.
Identified Eg5 inhibitors suppress cancer cell viability
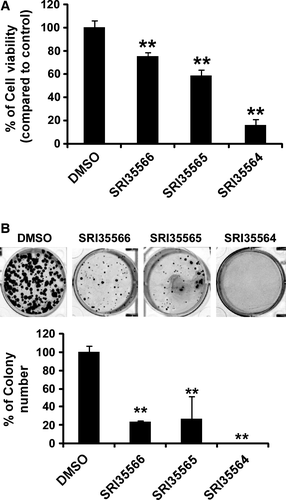
Given that our identified Eg5 inhibitors can induce monopolar spindle formation in colorectal HCT116 cells, we then examined the effect of the Eg5 inhibitors on HCT116 cell viability. As shown in Figure 7, SRI35566, SRI35565, and SRI35564 inhibited HCT116 cancer cell viability at a concentration shown to suppress Eg5 activity and induce monopolar spindle formation in colorectal cancer cells. Moreover, we performed colony formation assays and found that SRI35566, SRI35565, and SRI35564 at 80 μm significantly suppressed colony formation in HCT116 cells.
Discussion
Allostery is one of the most common and powerful means to regulate protein function 50, 51. Residues at the allosteric site(s) of a protein are evolutionarily less conserved compared to its active site residues. Allosteric ligands thus have better chances to bind specifically to the target protein and modulate its functions. While targeting allosteric site has been proven a promising strategy for the discovery and development of target-selective ligands, identifying allosteric site remains a challenging task. Most known allosteric sites, including the monastrol site of Eg5, were found after the discovery of allosteric ligands which were identified either by serendipity or through a high-throughput screening campaign 52. Currently, there are no established experimental strategies for the discovery of novel allosteric sites.
Protein flexibility and conformational dynamics are the key elements of allosteric regulations. To this end, MD simulation is a very powerful tool for studying protein dynamics as well as the related intramolecular interactions 53-61, and simulation-based computational methods have already been utilized to understand how a known allosteric ligand(s) regulates protein functions 58, 59, 62-64. In our previous study, we utilized such computational approaches to predict unknown allosteric sites and identified several Eg5 allosteric sites as potential drug targets 26. However, such theoretically predicted allosteric sites need to be experimentally validated. Notably, the crystal structure of Eg5-BI8 complex (PDB ID:3ZCW) 27, which was solved about two years after our predictions, confirmed the existence of the allosteric S2 site of Eg5 and demonstrated the usefulness of such computational predictions.
The validation of the predicted S2 site of Eg5 is a rare case in that a ligand (BI8) that binds to the S2 site is available (although its binding site was unknown). In most cases, to confirm a novel allosteric site, a ligand that binds to this site needs to be identified first. In the current study, we applied SBVS to identify inhibitors that bind to the S1 site of Eg5. While a compound may bind to a certain site(s) of a protein without affecting its function, our results demonstrated that the identified compounds bound directly to the Eg5 protein, induced monopolar spindle formation, a phenotype of Eg5 inhibition, in colorectal cancer cells, and suppressed cancer cell viability and colony formation. These results confirmed the existence of the S1 site and its feasibility as a potential drug targeting site for modulating Eg5 functions. The identified Eg5 inhibitors provide a useful starting point for the development of lead compounds.
To the best of our knowledge, this is the first experimental demonstration of the allosteric S1 site. So far, three allosteric sites of Eg5 have been identified, including the monastrol site, the S2 site and this S1 site. Unlike the monastrol site, which is Eg5-specific due to a unique long loop-5 that forms part of the site, both S1 and S2 sites locate at the structurally conserved regions of kinesins, thus may be ubiquitous for the kinesin family. Several kinesin proteins have been demonstrated as promising drug targets for cancer treatment in recent years 65-67. The identified allosteric S1 and S2 sites thus provide an opportunity to develop selective kinesin inhibitors for drug development. We conducted multiple sequence alignment using ClustalX program68 and compared the S1 site residues of several representative human kinesins, including KHC, the conventional N-terminal kinesin 1, KIFC1, a C-terminal kinesin 69, CENP-E, which possesses a long, kinesin-7 family-specific neck region 1, 70, and Eg5. As shown in Table S1, while residues at the core and the gate regions of the S1 site keep their hydrophobic or polar properties, only a few of them are conserved, other binding site residues are also not conserved among these kinesins, indicating that compounds could be designed to selectively inhibit a specific kinesin by targeting the S1 site. By saying so, the existence of S1 and S2 on other kinesins needs to be further validated.
While the present study demonstrated the existence of the allosteric S1 site, whether the identified compounds selectively inhibit Eg5 needs to be further evaluated. Among the three Eg5 inhibitors identified in this study, SRI35564 displayed the highest cytotoxic effect on colorectal cancer HCT116, but has a relatively weaker potency on Eg5 ATPase inhibition, indicating a potential off-target effect of these compounds. Further experimental studies, such as mutagenesis and crystallographic studies, could provide additional structural and biological details of the interactions between Eg5 and its inhibitors and help to develop selective inhibitors of Eg5 for drug discovery purposes.
Acknowledgments
We thank Dr. N. Rama Krishna and Dr. Ronald Shin of the UAB High Field NMR Facility for their supports on the collection of NMR spectrum. This work was partially supported by grants from Southern Research Institute, the National Institutes of Health R21CA182056 and Alabama Innovation Fund.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.




