LINC01305 recruits basonuclin 1 to act on G-protein pathway suppressor 1 to promote esophageal squamous cell carcinoma
Abstract
EsophageaL squamous cell carcinoma (ESCC) is one of the most common and lethal tumors, however, its underlying molecular mechanisms are not completely understood and new therapeutic targets are needed. Here, we found that the transcription factor basonuclin 1 (BNC1) was significantly upregulated and closely related to the differentiation and metastasis of ESCC. Furthermore, BNC1, LINC01305, and G-protein pathway suppressor 1 (GPS1) had significant oncogenic roles in ESCC. In addition, in vivo experiments showed that knockdown of BNC1 indeed significantly inhibited the proliferation and metastasis of ESCC. We also revealed the molecular mechanism by which LINC01305 recruits BNC1 to the promoter of GPS1, and then GPS1 could mediate the JNK signaling pathway to promote the proliferation and metastases of ESCC. Taken together, we discovered the novel molecular mechanism by which LINC01305/BNC1 upregulates GPS1 expression to promote the development of ESCC, providing a new therapeutic target for ESCC.
Abbreviations
-
- BNC1
-
- basonuclin 1
-
- ChIP-seq
-
- ChIP sequencing
-
- EC
-
- esophageal cancer
-
- EMT
-
- epithelial–mesenchymal transition
-
- ESCC
-
- esophageal squamous cell carcinoma
-
- GPS1
-
- G-protein pathway suppressor 1
-
- GRHL2
-
- grainyhead like transcription factor 2
-
- lncRNA
-
- long noncoding RNA
-
- NAT
-
- normal tissues
-
- qPCR
-
- quantitative PCR
-
- RIP
-
- RNA immunoprecipitation
1 INTRODUCTION
Esophageal cancer is a common and aggressive malignancy that causes over 400,000 deaths worldwide annually.1, 2 The two main subtypes of EC, ESCC and esophageal adenocarcinoma, have different epidemiological distributions and pathogenesis mechanisms.3, 4 The ESCC accounts for up to 80% of esophageal cancers and is usually diagnosed at advanced stages.5, 6 Unfortunately, despite the development of diagnostic and therapeutic techniques, the treatment and prognosis of ESCC are poor and the 5-year survival rate remains low because of the highly malignant proliferation,7 invasion, and metastasis of ESCC.8, 9 Increasing evidence suggests that transcription factors play an important role in tumor proliferation and metastasis.10, 11 Thus, it is of great significance to identify effective therapeutic targets of transcription factors for better treatment of ESCC.
Based on our sequencing information,12 we found that transcription factor BNC1 plays an important role in the development of ESCC. Basonuclin 1, located on chromosome 15,13 is a transcription factor specific for zinc finger proteins and is more conserved during evolution.14, 15 Basonuclin 1 was first identified in human epidermal keratinocytes cultured in vitro and could play an important role in germ cells.16-18 More studies have shown that BNC1 is closely related to the occurrence of tumors, and BNC1 is upregulated by the transcription factor tumor protein P63 (TP63) in head and neck squamous cell carcinoma19 and highly expressed by Gli protein in basal cell carcinoma.20 Research has shown that transcription factors could not function in the process as protein forms alone, and they usually bind with lncRNAs to control the transcription of target genes.21-24 But whether BNC1 plays its role in binding to lncRNAs in ESCC is unknown.
Through RIP and RNA pull-down experiments, we showed that LINC01305 binds to BNC1 experimentally, and further studies revealed that both LINC01305 and BNC1 are expressed in the nucleus. Cell biological function results showed that LINC01305 exerts oncogenic effects in ESCC. LINC01305 is a newly discovered lncRNA located on human chromosome 2q31.125 and its length was 3122 bp.26 Consistent with our experimental results, Huang et al. also found that LINC01305 played an important role in the proliferation and metastatic ability of ESCC.27 Therefore, we explored the specific mechanism of the complex. Thus, we then investigated the specific mechanism of action of the complex of BNC1 and LINC01305.
We then sought the target gene of BNC1 and LINC01305. GPS1 was identified as a BNC1 regulatory target gene by transcriptome sequencing combined with ChIP-seq combined analysis. Aso known as COP9 signalosome complex 1 (GSN1), GPS1 is a conserved protein complex composed of eight subunits, GNS1–GNS8, of which GPS1 is the largest subunit constituting the COP9 signalosome.28-30 Based on the finding that BNC1 regulates oncogenic target gene GPS1 and inhibition of LINC01305 reduced GPS1 mRNA expression levels, further studies revealed that BNC1 enrichment at the GPS1 promoter was reduced and BNC1 protein expression levels did not change after knockdown of LINC01305, indicating that LINC01305 regulates GPS1 expression by affecting BNC1 binding to the GPS1 promoter. Thus, we identify the molecular mechanism by which LINC01305 recruits BNC1 to act on GPS1.
In this study, we found that BNC1, LINC01305, and GPS1 play important oncogenic roles in ESCC. In vivo experiments further confirmed that knockdown of BNC1 could significantly inhibit the proliferation and migration of ESCC. Mechanistically, studies have shown that LIN01305 binds to recruit BNC1 to act on the promoter of GPS1, and promotes the GPS1-mediated JNK signaling pathway to promote the development of ESCC.
2 MATERIALS AND METHODS
2.1 Clinical samples
The collection and use of patient samples were approved by the ethics committee of Nanchong Central Hospital. Six pairs of ESCC cancer tissues and adjacent normal esophageal tissues were collected from Nanchong Central Hospital for RNA sequencing, and 67 pairs of ESCC cancer tissues and adjacent normal tissues were collected for immunohistochemical staining. Fresh tissue samples for RNA sequencing were immediately frozen in liquid nitrogen 30 min after surgery; none of the six patients had received any prior treatment and all gave signed informed consent. The pathological features of 67 cases of ESCC used for immunohistochemical staining are shown in Table S1.
2.2 Cell lines and cell culture
KYSE-30 and KYSE-410 human ESCC cell lines were purchased from Procell Life Science and Technology and KYSE-70 was purchased from Keygen Biotech. TE-1 and KYSE-150 cell lines were purchased from Genechem and TE-11, KYSE-510, and HET-1A were purchased from Shanghai Xuan Yi Biotechnology Service Center. The ECA109 ESCC cell line was derived from cells maintained at the Institute of Tissue Engineering and Stem Cells, Nanchong Central Hospital.
KYSE-150, KYSE-410, KYSE-510, TE-11, TE-1, and ECA109 ESCC cells were cultured in RPMI-1640 (Gibco), and HET-1A, KYSE-30, and KYSE-70 cells were cultured in DMEM (Gibco), supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. They were routinely cultured at 37°C in a 5% CO2 incubator.
2.3 siRNAs and transfection
The siRNAs si-NC, si-GRHL2#1, si-GRHL2#2, and si-GPS1 were purchased from Keygen Bio and si-LIN01305 was purchased from Gene Pharma. The transfection of siRNAs was undertaken with Lipofectamine 2000 (Invitrogen) and the sequences for siRNAs are listed in Table S2.
2.4 Quantitative RT-PCR
Esophageal squamous cell carcinoma cells were collected 48 h after transfection. The RNA isolation kit (FastPure Cell/Tissue Total RNA Isolation Kit; Vazyme Biotech Co., Ltd) and reverse transcription kit (HiScript III 1st Strand cDNA Synthesis Kit [+gDNA Wiper]; Vazyme Biotech Co., Ltd) were used to extract RNA and reverse transcribe it into cDNA. The cDNA was amplified by qPCR according to SYBR Green Master Mix instructions (ChamQ Universal SYBR qPCR Master Mix; Vazyme Biotech Co., Ltd). The qPCR primer sequences are shown in Table S3. The relative expression levels of target genes were calculated by the 2−ΔΔCt method with GAPDH as internal reference.
2.5 Stable cell line construction
Lentiviruses knocking down BNC1 expression and with puromycin resistance (Shanghai Genechem, Ltd) were constructed using the shBNC1#2 sequence and replaced with 10% FBS medium 16 h after transfection of lentiviruses. After 72 h, 10% FBS medium containing 2.5 μg/mL puromycin was added for culture, and stable ESCC cells (KYSE-30) with knockdown of BNC1 expression were obtained 1 week later.
2.6 Immunohistochemistry
Immunohistochemical analysis was undertaken on 67 pairs of clinically collected paraffin-embedded ESCC tissues and three pairs of paraffin-embedded animal experimental subcutaneous ESCC tissue sections. The primary Abs used were as follows: BNC1 polyclonal Ab (PA5-85984, 1:200; Invitrogen, Thermo Fisher Scientific), anti-CSN1 (ab194359, 1:200; Abcam), Ki-67(ready-to-use, Fuzhou Maxin Biotechnology Development Co., Ltd), and vimentin (ready-to-use, Fuzhou Maxin Biotechnology Development Co., Ltd). Immunohistochemical staining evaluation was carried out separately by two pathologists, and the scoring criteria were divided into the proportion of positive cells and the staining intensity multiplied, with a total score of 20 points. Specific scoring criteria were: cell proportions were scored as follows: 0, no positive cells; 1, 0–10% positive cells; 2, 10–35% positive cells; 3, 35–75% positive cells; and 4, over 75% positive cells. Staining intensity was graded according to the following standard: 1, no staining; 2, weak staining (light yellow); 3, moderate staining (yellow); 4, normal staining (yellow brown); and 5, strong staining (brown).
2.7 Colony formation assays
Cells were plated evenly in 6-well plates (1000 cells/well) for 10 days, fixed in 20% methanol for 30 min, and stained with 0.2% crystal violet for 3 min.
2.8 RNA immunoprecipitation
Cells (KYSE-30) were collected and lysed with lysis solution; target protein Ab anti-BNC1 (PA5-85984; Invitrogen, Thermo Fisher Scientific) and magnetic beads were incubated; protein lysate was added to form Ab-target protein-RNA complexes. Multiple washes were carried out to remove nonspecifically bound chromatin and purify the complexes. Cross-links were dissociated, RNA fragments were purified, and libraries were built for sequencing analysis.
2.9 Chromatin immunoprecipitation-qPCR
The ChIP-DNA enrichment was undertaken first: DNA-protein complexes formed by cross-linking were sonicated to interrupt. BNC1-IP Ab (IgG Ab as control) was added to form Ab-target protein-DNA complexes. Protein A/G beads were used to precipitate the complex, multiple washes were carried out to remove nonspecifically bound chromatin, and the complex was purified. Decross-linking and purification of DNA fragments was carried out, then RT-qPCR quantitative enrichment fold analysis. Quantitative PCR of the GPS1promoterpeak gene locus was undertaken in the input, IP, and IgG groups, and enrichment fold analysis was performed.
2.10 Dual luciferase reporter assays
Basonuclin 1 overexpression vector (pCDNA3.1-BNC1) and GPS1pro vector (pGL3-GPS1pro) were first constructed. They were divided into two groups: pCDNA3.1 + pGL3-GPS1pro and pCDNA3.1-BNC1 + pGL3-GPS1pro. 293T cells were cotransferred into pCDNA3.1 + pGL3-GPS1pro and pCDNA3.1 + pGL3-GPS1pro. Cells were harvested for dual luciferase activity assay after 48 h of transfection.
2.11 RNA pull-down
The first and second exon sequences of linc01305 were synthesized by the whole gene, and the linearized plasmids after digestion of the recombinant plasmids with KpnI and SacI, respectively, were used as templates for sense and antisense probes, respectively. Templates were transcribed in vitro and biotin-labeled with the kit. One milligram of whole cell total protein was added to the refolded RNA probe and mixed and incubated overnight at 4°C. Prewashed streptavidin beads were added to capture RNA-protein complexes. The beads were collected and washed five times with 1 mL RIP buffer. Finally, beads were collected, 30 μL SDS loading buffer was added to resuspend beads, and proteins captured by RNA pull-down were eluted at 95°C for 5 min. Western blot analysis was undertaken to detect enrichment of BNC1 (PA5-85984; Invitrogen, Thermo Fisher Scientific) in the input, sense, and antisense groups.
2.12 Western blot analysis
Proteins were extracted with RIPA lysates containing protease inhibitors, PMSF, and phosphatase inhibitors. The target protein was separated by 10% SDS-PAGE and transferred to a fluorinated membrane on PVDF. Second, after blocking for 1 h at room temperature with 5% nonfat dry milk, the following primary Abs were used overnight at 4°C: BNC1 (PA5-66883, 1:5000 and PA5-85984, 1:7500; Invitrogen), GPS1 (ab194359, 1:5000; Abcam), N-cadherin (ER0503, 1:1000; HUABIO), E-cadherin (ET1607-75, 1:2500; HUABIO), vimentin (AF1975, 1:1000; Beyotime), Snail (AF8013, 1:1000; Beyotime), JNK (AJ518-1, 1:1500; Beyotime), p-JNK (4668 T, 1:1000; CST), and GAPDH (R1108-1, 1:5000; HUABIO). Following four subsequent washes with PBS-T, HRP goat anti-rabbit IgG (1:1000) was added. Gray scale analysis was undertaken with ImageJ (National Institutes of Health, USA) after unannounced exposure using substrate chromogenic reagents (WBKLS0100; Millipore).
2.13 Flow cytometry
Transfected cells from each group were digested with EDTA-free trypsin, collected, washed twice with precooled PBS. and 100 μL of 1× binding buffer was added and gently blown until single cell suspension. Five microliters of annexin V-FITC and 5 μL propidium iodide solution were added, mixed well, and treated at room temperature in the dark for 15 min before upper flow cytometry detection.
2.14 Xenograft model
Six-week-old athymic nude mice were injected s.c. with 2 × 106 KYSE-30/shNC or KYSE-30/shBNC1#2 cells. Mouse weight and tumor volumes were monitored every 3 days, and tumor size was calculated using the following formula: tumor volume (mm3) = width × length × (width + length) × 0.5. Mice were killed on day 21 after injection to isolate tumors for photographing, immunohistochemical analysis, and H&E staining. Animal studies were carried out according to the protocols approved by the ethics committee of the Institutional Animal Care and Use Committee of the North Sichuan Medical College.
KYSE-30 cells (1.5 × 106 cells/nude mouse) with or without altered BNC1 expression were i.v. injected into athymic mice to assess whether BNC1 affects tumor metastasis in vivo. The nude mice were photographed at 35 days to observe fluorescence expression.
2.15 Migration and invasion assays
Cells from each group at 48 h posttransfection were digested, centrifuged, and washed twice with serum-free medium to remove residual serum. For the cell migration assay, the cell density of each group was 2.5 × 105 cells/mL resuspended with serum-free medium. One hundred microliters of cell suspension was inoculated into the upper chamber of each Transwell, and medium containing 10% serum was added into the lower chamber. The upper chamber was removed at 48 h of culture, fixed in 20% methanol, and stained with 0.1% crystal violet for 10 min. Uncrossed cells were carefully wiped off with a moistened cotton swab, washed twice with PBS, and observed, photographed, and counted under a light microscope (100×). Except for the use of the Transwell upper chamber precoated with Matrigel gel and the addition of 100 μL cell density of 7.5 × 105 cells/mL, the subsequent cell invasion assay experimental procedures were consistent with the migration assay.
2.16 Wound healing test
Forty-eight hours after transfection, cells were seeded into 6-well plates, and after the cells were completely attached, scratches were made in each well with a pipette tip. At 0 and 48 h, photographs were taken with an inverted microscope to observe scratch healing, and the scratch healing rate of the cells was calculated using the following formula: scratch healing rate = (scratch width at 1–48 h/scratch width at 0 h) × 100%.
2.17 Cell counting kit-8 method
Cells from each group at 48 h after transfection were digested, resuspended, counted, and seeded into 96-well plates at 3.5 × 103 cells/well for culture. The CCK-8 reagent (10 μL/well) was added at 24, 48, 72, and 96 h, and the culture was continued for 1 h. The optical density value of each well was detected at a wavelength of 450 nm on a microplate reader to reflect the proliferative activity of the cells, and the mean number of three replicate wells was taken from each group of cells for statistical analysis of the proliferative ability of the cells.
2.18 Chromatin immunoprecipitation sequencing
Cells (KYSE-30) were cross-linked with formaldehyde at a final concentration of 1%, and the cross-linking reaction was terminated using glycine. The DNA-protein complex formed by cross-linking was sonicated and the target BNC1 Ab was added to form an Ab-target protein-DNA complex. This complex was precipitated using Protein G beads, and the DNA fragments bound to the target protein were specifically enriched. Multiple washes removed the nonspecifically bound chromatin, and the complex was purified. Cross-linking was unlinked, the DNA fragments were purified, and sequencing analysis was carried out.
2.19 Statistical processing
The above main experiments were independently repeated three times. Statistical analysis and graphs were undertaken using GraphPad 7.0 (GraphPad Software, USA). Measurement data conforming to normal distribution were expressed mean ± SD. The t-test was used to compare data between the two groups. One-way ANOVA was used to compare data between multiple groups. p < 0.05, p < 0.01, p < 0.001, or p < 0.0001 indicated statistically significant differences.
3 RESULTS
3.1 Basonuclin 1 promotes development of ESCC
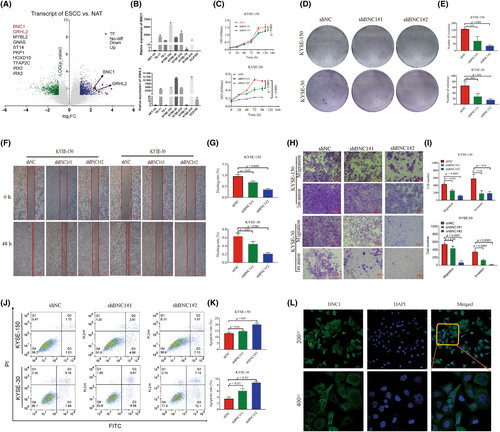
In order to screen new oncogenic transcription factors of ESCC, transcriptome sequencing was carried out in six pairs of fresh ESCC tissues and adjacent normal tissues collected clinically. Ten transcription factors highly expressed in ESCC were selected (Figure 1A), and eight ESCC cell lines and normal esophageal epithelial cells were subjected to qPCR validation (Figure S1A–H). Quantitative PCR results showed that GRHL2 and BNC1 were the most significantly different and could be oncogenic transcription factors in ESCC (Figure 1B). Unfortunately, there was no change in the biological function of ESCC after GRHL2 knockdown using siRNA (Figure S2A–D). It is gratifying that knockdown of BNC1 inhibited the proliferation of esophageal squamous carcinoma cells (Figures 1C–E and S2E,F) and inhibited migration and invasion (Figure 1F–I), promoting apoptosis (Figure 1J,K). We also undertook immunofluorescence localization and found that BNC1 was expressed in both the cytosol and nucleus (Figure 1L). These results indicate that the transcription factor BNC1 plays an important role in the biological function of ESCC cells.
3.2 Basonuclin 1 binds to LINC01305 to promote development of ESCC
At present, a large number of studies have reported that transcription factors do not act as protein forms alone for their transcriptional function, generally by binding lncRNAs to form transcriptional complexes acting on target genes and regulating the expression of downstream target genes.21-24 We hypothesized that BNC1 might bind to lncRNAs to form a transcriptional complex to promote the expression of downstream target genes, based on the above reports and scenarios. We undertook RIP experiments to select 19 lncRNAs that bind to BNC1 (Figure 2A,B, Table S4) conditional on log fold change greater than 4, and then passed the online algorithm RPISeq,31 which predicted the binding likelihood of each lncRNA (Table S4). Three lncRNAs, FLJ30679, LINC01305, and MIS18A-AS1, that may bind to BNC1, were predicted using the criteria that both Prediction using RF and SVM classifiers were greater than 0.8. The RIP results showed that LINC01305 bound to BNC1 protein (Figure 2C), and these results suggest that LINC01305 might bind to BNC1 to promote the development of ESCC. Further qPCR showed that only LINC01305 was highly expressed in ESCC (Figure 2D). To clarify the binding effect of LINC01305 to BNC1, we undertook the RNA pull-down assay to verify LINC01305 binding to BNC1 (Figure 2E). Furthermore, LINC01305 expression was mainly expressed in the nucleus (Figure 2F). To investigate the role of LINC01305 in ESCC, knockdown of LINC01305 expression using siRNA (Figure S3A) significantly inhibited the proliferation (Figure 2G–I), migration and invasion (Figure 2J–M), and slightly promoted the apoptosis of ESCC cells (Figure 2N,O). The above results showed that LINC01305, which binds to BNC1, promoted the development of ESCC.

3.3 Positive regulation of GPS1 by BNC1 promotes progression of ESCC
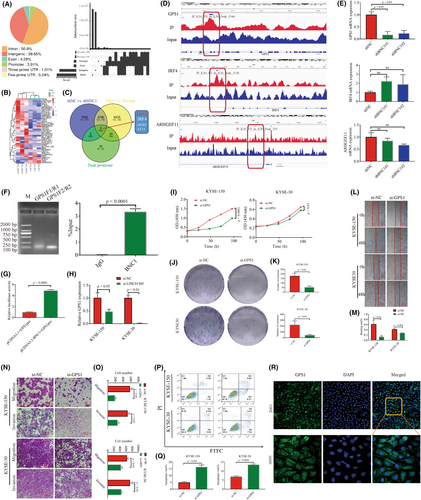
Long noncoding RNAs recruit transcription factors to bind target gene promoters of transcription factors and then mediate oncogenic target genes to promote tumor development. Therefore, it is particularly important to find oncogenic target genes regulated by BNC1 and can also be regulated by LINC01305. The ChIP-seq results revealed that an additional 3.91% of reads were enriched to promoters, and 41 possible downstream target genes were significantly changed by transcriptome sequencing analysis combined with knockdown of BNC1 (Figure 3A,B). Transcriptome sequencing and ChIP-seq results showed that three genes, ARHGEF11, GPS1, and IRF4, could be target genes positively regulated by BNC1 (Figure 3C,D). Further qPCR was undertaken to verify that only the mRNA expression of GPS1 was significantly decreased after knockdown of BNC1 (Figure 3E), so GPS1 was speculated to be a target gene positively regulated by BNC1. Dual luciferase experiments and ChIP-qPCR results also verified that BNC1 positively regulates GPS1 (Figure 3F,G). Excitingly, mRNA levels of GPS1 were also reduced following knockdown of LINC01305 (Figure 3H). Inhibition of GPS1 expression also significantly inhibited the proliferation, migration, and invasion of ESCC cells and promoted apoptosis (Figures 3I–Q and S3B). We found that GPS1 was expressed in both the nucleus and cytoplasm by fluorescence localization (Figure 3R). In conclusion, GPS1 is an oncogenic target gene of BNC1, while mRNA expression of GPS1 is also regulated by LINC01305.
3.4 Basonuclin 1 promotes proliferation and metastasis of ESCC in vivo
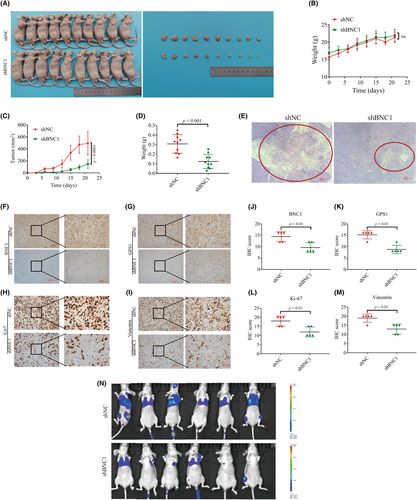
To further investigate the function of BNC1 in nude mice, we successfully constructed a KYSE-30 cell stable strain that inhibited BNC1 expression using lentivirus (Figure S3C). Subcutaneous tumor model experiments were carried out, and the results showed that after inhibiting the expression of BNC1, the tumor volume and tumor weight were significantly reduced (Figure 4A,D), and the necrotic area of the tumor was significantly reduced (Figure 4E). Further immunohistochemical analysis revealed that the expression of BNC1 and GPS1 was decreased in the shBNC1 group, and the expression of proliferation and metastasis markers Ki-67 and vimentin was also lower than that in the shNC group (Figure 4F–M). Metastatic models injected with tumor cells in the tail vein showed a significant decrease in metastatic capacity of ESCC following inhibition of BNC1 (Figure 4N). These data suggest that knockdown of BNC1 expression in vivo inhibits proliferation and metastasis of ESCC.
3.5 LINC01305 binding recruits BNC1 to act on GPS1 promoter to mediate JNK signaling pathway to promote EMT in ESCC
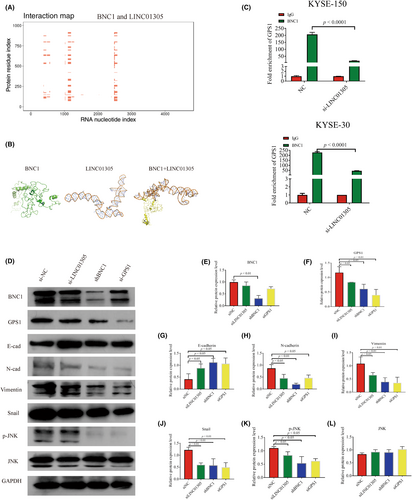
We predicted the binding sites of BNC1 protein and LINC01305 by catRAPID (http://service.tartaglialab.com/page/catrapid_group), and then predicted the sequences with the greatest docking potential by HEX (http://hex.loria.fr/), a macromolecular docking software. The results showed that BNC1 protein could have a molecular docking relationship with LINC01305 (Figure 5A,B). Previously, we have found that BNC1 can specifically bind to LINC01305 and is expressed in the nucleus and BNC1, GPS1, and LINC01305 play an important role in the development of ESCC respectively. Although BNC1 and LINC01305 can regulate the expression of GPS1, the molecular mechanism through which GPS1 is regulated by LINC01305 is unknown. Surprisingly, ChIP-qPCR experiments showed reduced enrichment of BNC1 at the promoter of GPS1 after knockdown of LIN01305 (Figure 5C). Protein levels showed that protein levels of GPS1 gradually decreased after knockdown of LINC01305, BNC1, and GPS1, but BNC1 was unchanged after knockdown of LINC01305 (Figure 5D–F). These results all validate the molecular mechanism conjecture that LINC01305 recruits BNC1 to act on GPS1. Li et al. discovered that GPS1 can mediate JNK to promote the development of EMT.32 Knockdown of LINC01305, BNC1, and GPS1 inhibited JNK phosphorylation and decreased expression of EMT-related proteins N-cadherin, vimentin, and Snail, and increased E-cadherin protein expression (Figure 5D,G–L). This result suggests that LINC01305 recruits BNC1 to act on the GPS1 promoter to mediate the molecular mechanism by which JNK signaling promotes EMT.
3.6 Clinical relevance analysis for BNC1, GPS1, and LINC01305
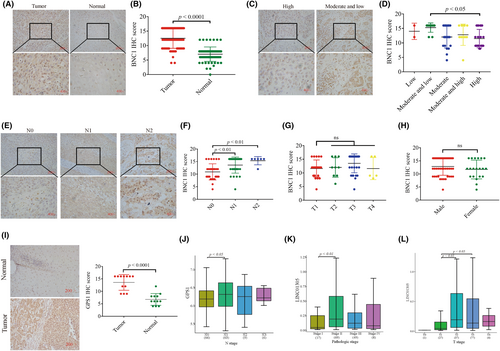
We randomly collected ESCC tissues and adjacent noncancerous tissues from 67 patients with ESCC, and immunohistochemical staining results revealed that BNC1 was highly expressed in cancer tissues (Figure 6A,B). Combined with clinical information analysis, these results showed that although BNC1 expression was not correlated with the stage or gender of patients with ESCC (Figure 6G,H), BNC1 expression was correlated with the differentiation and metastasis of ESCC (Figure 6C–F). These results suggest that BNC1 has potential as a molecular marker for clinical diagnosis. Furthermore, GPS1 was found to be highly expressed in ESCC tumor tissues by clinical samples (Figure 6I, Table S5). In addition, we further undertook clinical data analysis based on The Cancer Genome Atlas ESCA dataset data for LINC01305 and GPS1. The GPS1 gene was significantly more expressed in N1 stage than in N0 stage (Figure 6J), and stage II was significantly more expressed than stage I (Figure 6K). The LINC01305 gene was significantly more expressed at T2 and T3 than at T1 (Figure 6L). These clinical results suggest that BNC1, GPS1 and LINC01305 are closely related to the development of clinical ESCC.
4 DISCUSSION
A number of studies have already shown that BNC1 could be closely related to the development of tumors. For example, the expression level of BNC1 gradually increases with the development of cutaneous squamous cell carcinoma, suggesting that it may be an oncogene.33 Iden et al. also reported that BNC1 might be an oncogene in cervical cancer.34 In the present study, we identified the transcription factors GRHL2 and BNC1 to be highly expressed in ESCC tissues by sequencing six pairs of ESCC tissues and adjacent normal tissues. Only inhibition of BNC1 altered the biological function of ESCC cells. Therefore, we further investigated the role BNC1 plays in ESCC by cell function experiments. The in vitro experiments showed that inhibiting the expression of BNC1 in ESCC cells significantly decreased the proliferation, migration, and invasion of ESCC cells, and promoted the apoptosis of ESCC cells. In vivo experiments also showed that silencing the expression of BNC1 inhibited the malignant proliferation and metastatic ability of ESCC. A number of studies have shown that transcription factors bind to lncRNAs to promote tumor development.21-23, 35 Thus, we found that LINC01305 and BNC1 were specifically bound using RIP assay and RNA pull-down assay.
Long noncoding RNAs can show different regulatory mechanisms because of their different cellular localization, which in turn leads to their different functional roles in tumors36; lncRNAs mainly act as sponges in the cytoplasm to block the effects between miRNAs and their target genes, whereas lncRNAs localized in the nucleus can participate in epigenetic regulation or mRNA transcriptional regulation.37, 38 We undertook immunofluorescence localization and showed that both BNC1 and LINC01305 were expressed in the nucleus, so we speculated that BNC1/LINC01305 plays a transcriptional regulatory role.
LINC01305 is a lncRNA and located on human chromosome 2q31.1.25 LINC01305 has been reported to be closely related to the occurrence and development of tumors; LINC01305 can promote the occurrence and development of cervical cancer through the exosome pathway or regulate the TNXB-PI3K/AKT signaling axis to affect the EMT of cervical cancer and lung cancer.26, 39, 40 Our preliminary experiments found that LINC01035 was highly expressed in ESCC, so it is speculated that LINC01305 might play a carcinogenic role in ESCC. As expected, knockdown of LINC01305 expression in ESCC cells significantly inhibited the proliferation, migration, and invasion of ESCC cells. These results are also consistent with Huang et al.'s finding that LIN01305 can promote metastasis and proliferation of ESCC.27 However, the target genes regulated downstream of BNC1/LINC01305 are not yet clear. In this regard, we undertook transcriptome sequencing analysis of experiments such as knockdown BNC1 and ChIP-seq and found that the promoter of GPS1, IRF4, and ARHGEF11 are target genes directly acted upon by BNC1 and the mRNA expression levels are regulated by BNC1.
Unfortunately, we only found that the mRNA expression level of GPS1 was decreased after knockdown of BNC1 by RT-qPCR. The relationship between GPS1, a target gene positively regulated by BNC1, was verified by dual luciferase, ChIP-qPCR, and immunohistochemical experiments. To further explore whether LINC01305 also affected GPS1 expression, we undertook RT-qPCR experiments after knocking down LINC01305 and found that mRNA expression levels of GPS1 were also decreased in KYSE-150 and KYSE-30 cells.
Although LINC01305 is also known to regulate the mRNA expression of GPS1, the specific molecular mechanism of its regulation is unknown. In order to investigate the specific molecular mechanism of GPS1 regulation by LINC01305, knockdown of LINC01305 by ChIP-qPCR assay and western blot assay revealed that BNC1 enrichment at the GPS1 promoter was significantly decreased, while BNC1 protein levels did not change, indicating that LINC01305 did recruit BNC1 to act on the GPS1 promoter and then regulate GPS1 transcription.
It has been shown that GPS1 is highly expressed in glioblastoma tissues, and it could be an oncogene.41 Consistent with this, we showed that knockdown of GPS1 expression in ESCC cells significantly inhibited the proliferation, migration, and invasion of ESCC cells and promoted the apoptosis of ESCC cells. These experimental results show that GPS1 is a oncogenic target gene regulated by BNC1. Studies have shown that GPS1 can activate the JNK pathway and then mediate the Wnt pathway to promote the development of triple-negative breast cancer,32 and phosphorylation of JNK promotes the progression of EMT.42 In addition, Meng et al. found that after inhibition of JNK phosphorylation, E-cadherin expression increased and vimentin expression levels decreased, inhibiting epithelial cytoplasmic mesenchymal transition in colorectal cancer.43 We thus hypothesize that GPS1 promotes EMT in ESCC through the JNK pathway. Fortunately, we found that knockdown of LINC01305, BNC1, and GPS1 did decrease p-JNK expression, increased EMT-related protein E-cadherin, and decreased N-cadherin, Snail, and vimentin expression.
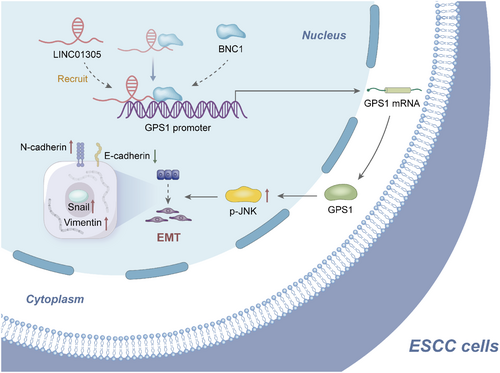
We found that LINC0135 recruits BNC1 to act on the GPS1 promoter to promote GPS1 expression, mediating the JNK signaling pathway to promote EMT in ESCC, revealing a previously unknown mechanism (Figure 7). High BNC1 expression in human ESCC specimens correlates with differentiation and metastasis in ESCC patients, demonstrating the clinical relevance and significance of our findings. The finding that LINC01305 recruits BNC1 to act on GPS1 could provide promising avenues for the treatment of patients with ESCC. However, there are many factors that affect gene transcription levels, such as enhancer regions, and because this study only focuses on promoter regions, whether BNC1 will bind enhancers of GPS1 or other genes and then play a role has not been explored, which could be a focus of our subsequent studies.
AUTHOR CONTRIBUTIONS
Li Xiong, Jinsong Tan, and Ruolan Zhang performed the experimental tests. Li Xiong and Kang Liu drafted the manuscript. Rong Xiong and Yanqun Liu reviewed histological specimens and prepared histological sections for immunohistochemistry. Qiongxian Long assessed immunohistochemistry. Yun Liu and Jiancai Tang analyzed the data. Yun Liu, Gang Feng, Guiqin Song, Yan Li, and Kang Liu worked and revised the manuscript.
ACKNOWLEDGMENTS
The authors thank the members of the laboratories at the Institute of Tissue Engineering and Stem Cells, Nanchong Central Hospital, for technical assistance and discussion; and the laboratories at the Institute of Tissue Engineering and Stem Cells, Nanchong Central Hospital, and the Institute of Basic Medicine and Forensic Medicine, North Sichuan Medical College for technical support.
FUNDING INFORMATION
This work was supported by grants from the National Natural Science Foundation of China (No.82203851), Natural Science Foundation of Sichuan Province (2023NSFSC0731), Sichuan Science and Technology Program (2023YFSY0045), Doctoral Fund of North Sichuan Medical College (CBY22-QDA01), and Nanchong Science and Technology Program (No. 22SXQT0336, 22SXQT0334, 20SXQT0328, 20SXPTJS0003).
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest.
ETHICS STATEMENTS
Approval of the research protocol by an institutional review board: The research protocol was approved by the Ethics Committee of Nanchong Central Hospital (Ethical Approval No.: 2019 Review No. 095).
Informed consent: Informed consent was obtained from the subject(s) and/or guardian(s).
Registry and the registration no. of the study/trial: N/A.
Animal studies: The use of animals for animal experiments was approved by the Ethics Review Committee of North Sichuan Medical College (Ethics No.: NSMC Ethical Animal Review [2021] No. 44).
Open Research
DATA AVAILABILITY STATEMENT
All authors approved all versions including the final version, and are responsible for the accuracy and integrity of all aspects of the manuscript.




