Functional intronic variant of SLC5A10 affects DRG2 expression and survival outcomes of early-stage non-small-cell lung cancer
Abstract
RegulomeDB is a new tool that can predict the regulatory function of genetic variants. We applied RegulomeDB in selecting putative functional variants and evaluated the relationship between these variants and survival outcomes of surgically resected non-small-cell lung cancer. Among the 244 variants studied, 14 were associated with overall survival (P < 0.05) in the discovery cohort and one variant (rs2257609 C>T) was replicated in the validation cohort. In the combined analysis, rs2257609 C>T was significantly associated with worse overall and disease-free survival under a dominant model (P = 2 × 10−5 and P = 0.001, respectively). rs2257609 is located in the SLC5A10 intron, but RegulomeDB predicted that this variant affected DRG2, not SLC5A10 expression. The expression level of SLC5A10 was not different with the rs2257609 genotype. However, DRG2 expression was different according to the rs2257609 genotype (Ptrend = 0.03) and was significantly higher in tumor than in non-malignant lung tissues (P = 1 × 10−5). Luciferase assay also showed higher promoter activity of DRG2 in samples with the rs2257609 T allele (P < 0.0001). rs2257609 C>T affected DRG2 expression and, thus, influenced the prognosis of early-stage non-small-cell lung cancer. This study was approved by the Institutional Review Broad of Kyungpook National University of Hospital (Approval No. KNUMC 2014-04-210-003).
1 INTRODUCTION
Lung cancer is the leading cause of cancer-related death, and the 5-year survival rate remains poor.1 Curative surgery is the best treatment option for early-stage non-small-cell lung cancer (NSCLC).2 However, a large number of patients experience recurrence after surgery and eventually die. Although the pathological stage is the most important prognostic factor, patients with the same stage sometimes show heterogeneous outcomes.3 Therefore, it is necessary to develop additional prognostic markers to more precisely predict the survival of patients with lung cancer.
A number of studies reported that variants in the protein-coding region could affect the prognosis of lung cancer.4-6 However, genome-wide association studies found that more than 90% of variants that contributed to diverse human diseases were located outside of protein-coding regions.7, 8 This suggests that the regulation of genes is far more complex than previously thought, and non-coding DNA located nearby or distant from the coding genes can influence gene expression.9 This also suggests that variants in non-coding DNA may affect the prognosis of lung cancer.
Recently, Boyle et al10 developed a novel approach and database, RegulomeDB, that guides interpretation of regulatory variants in the human genome. RegulomeDB provides a scoring system, ranging from categories 1 to 6, based on the functional confidence of variants. A lower score indicates a higher possibility that the variant affects transcription factor binding and gene expression.10 Therefore, RegulomeDB can be used at the stage of selecting variants in genetic association studies. Evaluating the relationship between variants with a lower score in RegulomeDB and disease susceptibility or prognosis can be a method for genetic association studies. In the current study, we hypothesized that variants of non-coding DNA may affect gene expression and, thereby, the prognosis of patients with early-stage NSCLC after surgery. To test this hypothesis, we evaluated the association between putative functional variants selected using RegulomeDB and the survival outcome of patients with surgically resected NSCLC.
2 MATERIALS AND METHODS
2.1 Study populations
This study was conducted in two stages. The discovery phase of the study included 376 patients with early-stage NSCLC who underwent surgical resection from September 1998 to July 2007 at Kyungpook National University Hospital (KNUH). An independent validation cohort included 428 patients with surgically resected NSCLC for curative purposes collected by Seoul National University Hospital between September 2005 and October 2010. All patients in the discovery and validation cohorts were ethnic Koreans. Written informed consent was obtained from all participants. Blood samples for genotyping were obtained before surgery. Patients who received chemotherapy or radiotherapy before surgery were excluded to avoid the effects of these agents on DNA. Tissue samples from the tumors and corresponding non-malignant lung tissue specimens were provided by the National Biobank of KNUH, which is supported by the Ministry of Health, Welfare, and Affairs. All materials derived from the National Biobank of Korea-KNUH were obtained (with informed consent) under institutional review board-approved protocols. This study was approved by the Institutional Review Boards of KNUH and Seoul National University Hospital.
2.2 Polymorphism selection and genotyping
As a lower score indicated a higher possibility of variants being functional, category 1 polymorphisms were extracted from RegulomeDB (http://regulome.stanford.edu). A total of 39 433 polymorphisms were category 1 in the RegulomeDB scoring system. Subcategories of these 39 433 polymorphisms were as follows: 352 1a, 2568 1b, 85 1c, 1668 1d, 54 1e, and 34 706 1f. Because category 1a has the strongest evidence of variants affecting gene expression, 352 category 1a polymorphisms were selected in this study. Among these 352 polymorphisms, 97 with low minor allele frequency (<10%) in Asians, based on data from the NCBI single nucleotide polymorphism (SNP) database (http://www.ncbi.nlm.nih.gov/SNP), were excluded. Eleven polymorphisms with strong linkage disequilibrium (r2 >0.8) using HapMap genotyping data were also excluded (Figure S1). Therefore, a total of 244 polymorphisms were genotyped using the MassARRAY iPLEX (Sequenom, San Diego, CA, USA) or RFLP assays. Approximately 5% of samples were selected randomly and again genotyped. The results were in 100% concordance.
2.3 Quantitative reverse transcription polymerase chain reaction
Expression of solute carrier family 5, member 10 (SLC5A10) and human developmentally regulated GTP-binding protein 2 (DRG2) mRNAs was measured by qRT-PCR in a LightCycler 480 (Roche Applied Science, Mannheim, Germany). Total RNA was isolated from paired NSCLC and nonmalignant lung tissues using TRIzol (Invitrogen, Carlsbad, CA, USA) and was reverse transcribed using the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany). qRT-PCR was carried out using the QuantiFast SYBR Green PCR Master Mix (Qiagen) according to the manufacturer's instructions. SLC5A10 and DRG2 primer pairs were as follows: SLC5A10: 5′-CAACATCGCCTACCCCAAG-3′, 5′-CCAGATGTCCATAGTGAAGAGG-3′; DRG2:5′-CTGACCTGCATCTACACCAAG-3′, 5′-CAGGGCGTACTTGAACTGG-3′. Each sample was run in duplicate. Relative expression of SLC5A10 and DRG2 was calculated following normalization with human beta-actin.
2.4 Plasmid constructs
For the in vitro functional study, the pGL3-Basic reporter vector from Promega (Madison, WI, USA) was used to construct luciferase reporter plasmids using the manufacturer's protocols. The sequence of the human DRG2 genomic locus at 17P11.2 (GenBank accession number NC_000017.11) was used to engineer PCR cloning primers. Briefly, promoter regions of DRG2 (−1090 to +203 base pairs; the transcriptional start site is designated as +1) were amplified from human genomic DNA and then cloned into the KpnI and XhoI sites of the pGL3-Basic vector to generate pGL3-DRG2pro. The pGL3-DRG2pro plasmid was used as a template to synthesize the plasmid that included polymorphism fragments. Amplified 190 base pair products containing the C allele of rs2257609 or the T allele of rs2257609 were inserted into the BamHI and SalI sites of pGL3-DRG2pro, respectively. All constructs were verified by direct sequencing before use.
2.5 Transient transfection and the luciferase reporter assay
Transfections were done using Effectene (Qiagen) according to the manufacturer's protocol. Human NSCLC cells (H1299 and H1373) were maintained at 37°C in 5% CO2 in RPMI-1640 medium containing 10% heat-inactivated FBS. Cells were transfected with 300 ng of each plasmid DNA (pGL3-DRG2pro, pGL3- DRG2pro_C and pGL3- DRG2pro_T) and 30 ng pRL-SV40. Luciferase activity was measured on an Orion L Microplate Luminometer (Berthold Detection Systems, Bad Wildbad, Germany) using the Dual-Luciferase Reporter Assay System Kit (Promega). Firefly luciferase activity measurements were normalized to pRL-SV40 Renilla luciferase activity to correct for variations in transfection efficiency. Each experiment was conducted in triplicate at least three times.
2.6 Statistical analyses
Hardy-Weinberg equilibrium was tested using a goodness-of-fit χ2 test. Overall survival (OS) was counted from the day of surgery to the date of death or last follow up. Disease-free survival (DFS) was measured from the day of surgery until recurrence or death from any cause. Estimated survival rate was calculated using the Kaplan-Meier method. Log-rank test was used to compare the difference in OS and DFS across different genotypes. Multivariate Cox proportional hazards models were used to estimate the hazard ratio (HR) and 95% confidence intervals (CI) after adjusting for age (<64 years vs ≥64 years), gender (male vs female), smoking status (never vs ever), tumor histology (squamous cell carcinoma vs adenocarcinoma), adjuvant therapy (yes vs no), and pathological stage (I vs II-IIIA). All analyses were carried out using Statistical Analysis System for Windows, version 9.4 (SAS Institute, Cary, NC, USA).
3 RESULTS
3.1 Patient characteristics
Patient characteristics of the discovery and validation cohorts are described in Table S1. Pathological stage was significantly associated with OS and DFS in both cohorts (P < 0.001, both). In the discovery cohort, younger patients showed better OS (P = 0.01). In the validation cohort, age, gender, and smoking status were associated with OS and DFS upon univariate analysis (Table S1).
3.2 Association between polymorphisms and survival outcomes
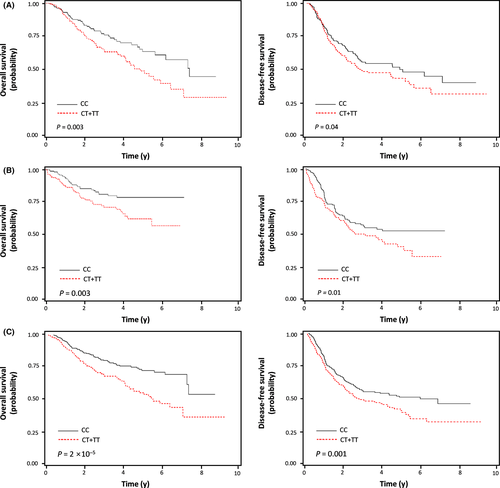
Among the 244 polymorphisms used for survival analysis, 14 were associated with survival outcomes in the discovery cohort (Table S2). Information on 244 polymorphisms and the original results of analyses are shown in Tables S3 and S4. The 14 polymorphisms were further evaluated in the validation cohort. rs2257609 C>T was significantly associated with OS and DFS in the same direction with the discovery cohort (Table 1). In the combined analysis, patients with rs2257609 CT or TT genotypes showed worse OS and DFS than those with the rs2257609 CC genotype (HR = 1.87, 95% CI = 1.41-2.48, P = 2 × 10−5 and HR = 1.44, 95% CI = 1.16-1.79, P = 0.001, respectively; Table 1 and Figure 1).
| Genotypea | No. of cases (%)b | Overall survival | Disease-free survival | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of deaths (%)c | 5-y OSR (%)d | Log-rank P | HR (95% CI)e | P e | No. of events (%)c | 5-y DFSR (%)d | Log-rank P | HR (95% CI) e | P e | |||
| Discovery cohort | CC | 174 (47.0) | 47 (27.0) | 64 | 0.02 | 1.00 | 75 (43.1) | 47 | 0.11 | 1.00 | ||
| CT | 159 (43.0) | 61 (38.4) | 48 | 1.62 (1.09-2.41) | 0.02 | 77 (48.4) | 45 | 1.26 (0.91-1.75) | 0.17 | |||
| TT | 37 (10.0) | 19 (51.4) | 51 | 2.31 (1.33-3.99) | 0.003 | 25 (67.6) | 31 | 1.99 (1.25-3.18) | 0.004 | |||
| Dominant | 0.01 | 1.76 (1.21-2.55) | 0.003 | 0.15 | 1.39 (1.02-1.89) | 0.04 | ||||||
| Recessive | 0.09 | 1.84 (1.10-3.05) | 0.02 | 0.06 | 1.79 (1.16-2.77) | 0.01 | ||||||
| Codominant | 1.54 (1.19-1.99) | 0.001 | 1.37 (1.10-1.71) | 0.01 | ||||||||
| Validation cohort | CC | 208 (50.1) | 31 (14.9) | 79 | 0.02 | 1.00 | 72 (34.6) | 52 | 0.04 | 1.00 | ||
| CT | 165 (39.8) | 43 (26.1) | 62 | 2.07 (1.29-3.32) | 0.003 | 75 (45.5) | 37 | 1.61 (1.16-2.25) | 0.01 | |||
| TT | 42 (10.1) | 9 (21.4) | 65 | 1.80 (0.84-3.85) | 0.13 | 16 (38.1) | 41 | 1.35 (0.77-2.34) | 0.30 | |||
| Dominant | 0.01 | 2.02 (1.28-3.19) | 0.003 | 0.03 | 1.56 (1.13-2.15) | 0.01 | ||||||
| Recessive | 0.92 | 1.23 (0.61-2.50) | 0.56 | 0.74 | 1.06 (0.63-1.80) | 0.82 | ||||||
| Codominant | 1.51 (1.10-2.07) | 0.01 | 1.29 (1.03-1.62) | 0.03 | ||||||||
| Combined analysis | CC | 382 (48.7) | 78 (20.4) | 71 | 4 × 10−4 | 1.00 | 147 (38.5) | 50 | 0.03 | 1.00 | ||
| CT | 324 (41.3) | 104 (32.1) | 53 | 1.82 (1.35-2.44) | 8 × 10−5 | 152 (46.9) | 41 | 1.40 (1.11-1.76) | 0.004 | |||
| TT | 79 (10.1) | 28 (35.4) | 58 | 2.08 (1.34-3.23) | 0.001 | 41 (51.9) | 37 | 1.65 (1.16-2.34) | 0.01 | |||
| Dominant | 8 × 10−5 | 1.87 (1.41-2.48) | 2 × 10−5 | 0.01 | 1.44 (1.16-1.79) | 0.001 | ||||||
| Recessive | 0.17 | 1.54 (1.03-2.31) | 0.04 | 0.24 | 1.40 (1.01-1.95) | 0.05 | ||||||
| Codominant | 1.53 (1.26-1.85) | 2 × 10−5 | 1.32 (1.13-1.54) | 6 × 10−4 | ||||||||
- CI, confidence interval; DFSR, disease-free survival rate; HR, hazard ratio; OSR, overall survival rate.
- a Patients with missing data (6 in discovery cohort, 13 in validation cohort, and 19 in combined analysis) were not included in the analysis.
- b Column percentage.
- c Row percentage.
- d 5-y OSR and 5-y DFSR, proportion of survival derived from Kaplan-Meier analysis.
- e HR, 95% CI and corresponding P-values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status, tumor histology, adjuvant therapy, and pathological stage.
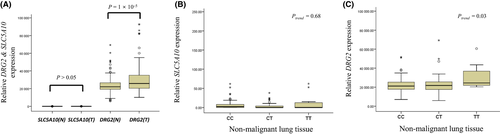
3.3 Expression of SLC5A10 and DRG2 mRNA
Genomic structure regarding SLC5A10, DRG2 and rs2257609 are shown in Figure S2. rs2257609 is located in the intron (ivs5 + 1216) of SLC5A10. However, rs2257609 C>T was predicted to affect human DRG2 expression in RegulomeDB. Therefore, we evaluated SLC5A10 and DRG2 mRNA levels in 144 tumor and paired non-malignant lung tissues. Expression level of SLC5A10 mRNA was not different between tumor and non-malignant lung tissues (Figure 2A). In addition, relative SLC5A10 mRNA expression was not different according to the rs2257609 genotype (Figure 2B). DRG2 mRNA expression was significantly higher in tumor than in non-malignant lung tissues (P = 1 × 10−5; Figure 2A). Relative DRG2 mRNA expression showed an increasing trend with the number of rs2257609 C>T polymorphic alleles in non-malignant lung tissues (Ptrend = 0.03, Figure 2C).
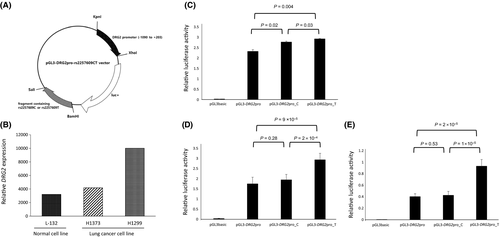
3.4 Effect of rs2257609 C>T on promoter activity
We investigated whether rs2257609 C>T modulated the activity of the DRG2 promoter using a luciferase assay. DRG2 expression levels were different by cell lines (Figure 3B). DRG2 expression was high in H1299 but low in H1373 and L-132. As shown in Figure 3C, luciferase activity was significantly higher in H1299 cells transfected with pGL3-DRG2pro_C or pGL3-DRG2pro_T compared with that in cells transfected with pGL3-DRG2pro. This suggests that the fragment containing rs2257609 C>T enhanced the activity of the DRG2 promoter. The rs2257609T allele was associated with significantly higher activity of the DRG2 promoter compared to the rs2257609C allele, suggesting that rs2257609 C>T may alter DRG2 expression (P = 0.03; Figure 3C). Relative luciferase activities in other cell lines (H1373 and L-132) were analyzed and the results were similar to that in H1299 (Figure 3D,E).
3.5 DRG2 expression and prognosis
When analyzed using Kaplan-Meier Plotter (http://www.kmplot.com/analysis/), the patients with high DRG2 expression showed worse overall survival than those with low DRG2 expression (HR = 1.21, 95% CI: 1.07-1.37, P = 0.003, Figure S3).
4 DISCUSSION
The present study was conducted to investigate whether variants of non-coding DNA affect the survival outcomes of patients with surgically resected NSCLC. Among the 244 variants evaluated, rs2257609 C>T was significantly associated with worse OS and DFS. The rs2257609 C>T variant did not affect the expression of SLC5A10 mRNA, but did alter the mRNA expression and promoter activity of DRG2.
Among the 3.3 billion nucleotides contained in the human genome, the proportion of sequences that code for proteins is very small.11 However, recent data show that non-protein-coding DNAs can be functional, and polymorphisms of these regions can change gene activity or alter the risk of diseases.12-14 In the current study, rs2257609 C>T affected the prognosis of NSCLC. rs2257609 C>T is located in the intron region of SLC5A10. RegulomeDB predicted that this variant affected the expression of DRG2, but not SLC5A10. In concordance with the annotation of RegulomeDB, we found that rs2257609 C>T influenced mRNA expression and promoter activity of DRG2. Expression of SLC5A10 mRNA was not changed by rs2257609 C>T.
DRG2 encodes a GTP-binding protein and acts as a critical regulator of cell growth and differentiation.15 Song et al16 reported that overexpression of DRG2 increased G2/M phase cells and decreased sensitivity to nocodazole-induced apoptosis. Changes in apoptotic capacity can affect carcinogenesis or the prognosis of NSCLC.17-20 In the present study, the expression of DRG2 mRNA was significantly higher in tumor than in non-malignant lung tissues. This result suggests that DRG2 may be associated with lung cancer carcinogenesis. Jang at el21 found that the growth rate of DRG2 knockdown in HeLa cells was substantially lower than that of control cells. They also found that in nude mice xenografts, DRG2-depleted cells were less tumorigenic.21 These results suggest that DRG2 overexpression may be associated with tumorigenesis. These results are consistent with our study. In our study, the rs2257609 variant allele showed higher DRG2 mRNA expression than the rs2257609 wild-type allele. DRG2 overexpression by changes in promoter activity may have affected tumorigenesis and prognosis. Further studies are needed to determine the biological functions and tumorigenesis of DRG2.
Applying RegulomeDB to evaluate lung cancer prognosis is a novel approach. RegulomeDB is useful to predict the regulatory function of variants in non-coding DNA. To date, RegulomeDB has been used to interpret the putative regulatory function of non-coding variants that were identified in genome-wide association or candidate gene studies,22-24 or to assess expression of the quantitative trait locus.25, 26 In the present study, we used RegulomeDB to select more putative functional variants to evaluate lung cancer prognosis. After selecting 244 variants in category 1a, because a lower score in RegulomeDB suggests a higher probability of affecting transcription factor binding and gene expression, we conducted a two-stage study to evaluate the effect of these variants on the survival outcomes of early-stage NSCLC.
The study design involved two independent cohorts, one for discovery and the other for validation. This is one of its major strengths that largely reduces false-positive findings from the genetic association study. In the present study, rs2257609 C>T was significantly associated with worse survival outcomes in both the discovery and validation cohorts with the same direction. In addition, the P value in the combined analysis was compatible with the P value to avoid most of the false-positive associations arising from multiple comparisons.27 Furthermore, the expression level of DRG2, which RegulomeDB predicted would be influenced by rs2257609 C>T, was significantly higher in tumor than in non-malignant lung tissues and was different according to the rs2257609 genotype.
A functional study using the luciferase assay showed that a minor allele of rs2257609 significantly increased the activity of the DRG2 promoter. This result suggests that increased activity of the DRG2 promoter with the minor allele of rs2257609 leads to overexpression of DRG2, resulting in decreased apoptosis and a worse survival outcome. Therefore, the significant effect of rs2257609 C>T on the survival outcome of patients with NSCLC may not be observed by chance. To the best of our knowledge, this study is the first to report that rs2257609 C>T, which can affect DRG2 expression, may affect the survival outcome of NSCLC.
There are several limitations in the present study. The modest sample sizes of the discovery and validation cohorts, lacking optimal statistical power, should be considered. Although the expression level of DRG2 was different according to the rs2257609 genotype, this did not provide direct evidence of the prognostic effect of rs2257609 C>T on NSCLC. Additional studies with a larger sample size, and investigations into the biological mechanism for DRG2, are needed to understand the role of rs2257609 C>T in the survival outcomes of lung cancer.
In conclusion, we applied RegulomeDB to evaluate the survival outcome of NSCLC. rs2257609 C>T, which exists in the intron region of SLC5A10, affected DRG2 expression and the survival outcome of early-stage NSCLC.
ACKNOWLEDGMENTS
This research was supported in part by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C0402), in part by the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea (grant number: 1720040).
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.




