Mouse IgE clone SPE-7 can contain functional mouse IgG
The hybridoma-produced mouse IgE anti-DNP antibody clone SPE-7 is one of the most widely used molecules to study IgE biology and function. At the same time, SPE-7 has exhibited some unique characteristics such as the ability to trigger mast cell activation and survival independent of a cross-linking antigen,1, 2 the potential to bind DNP-unrelated antigens through conformational diversity,3 and an increased capability to bind mouse Fcγ receptors.4 Here, we compared IgE clone SPE-7 to other mouse IgE clones using enzyme-linked immunosorbent assays (ELISA), biolayer interferometry (BLI), liquid chromatography—mass spectrometry (LC–MS), and cell-based in vitro assays with mouse bone-marrow derived mast cells (BMMCs) and spleen-derived mouse B cells.
Surprisingly, we found that in contrast to other mouse IgE clones, SPE-7 was recognized by an anti-IgG detection antibody (Figure 1A–D, Figure S1). These IgG signals were lost by purification of SPE-7 using protein G columns. IgG present in SPE-7 could be captured with an anti-IgG2a/b antibody (but not with anti-IgG1) and detected with an anti-IgE antibody, suggesting the presence of IgG2a/b-IgE complexes in SPE-7 (Figure 1E,F). Similarly, SPE-7 ultracentrifugation reduced the contaminating IgG in SPE-7 (Figure S1).
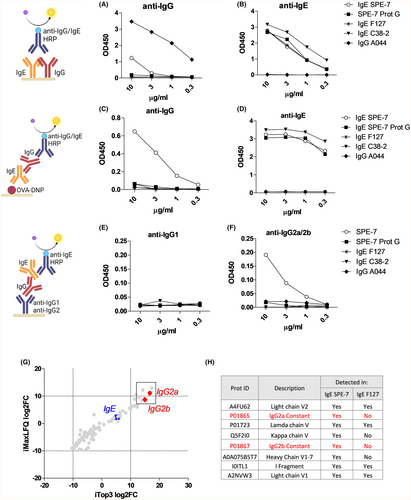
The presence of IgG in SPE-7 was confirmed by LC–MS. We compared the cumulative distribution between samples using MaxLFQ and Top3 normalization protocols (suppl. Methods, Figure S1). Figure 1G shows an increased prevalence of IgG2a/b in IgE SPE-7 compared to IgE F127, and both IgG2a and IgG2b showed up in the top 8 up-regulated hits summarized in Figure 1H.
To test if IgE SPE-7 can functionally bind to IgG receptors, we displayed FcγRI (CD64) on BLI chips (Figure 2A) or on ELISA plates (Figure 2B). In both cases SPE-7, but no other IgE clone, showed binding activity to FcγRI. Using flow cytometry analysis, we next tested whether IgG influences the binding of IgE SPE-7 to the IgE receptor CD23 expressed on spleen-derived B cells. Indeed, Figure 2C shows that binding of IgE SPE-7 to CD23 was increased compared to other IgEs, or compared to IgG-depleted SPE-7.
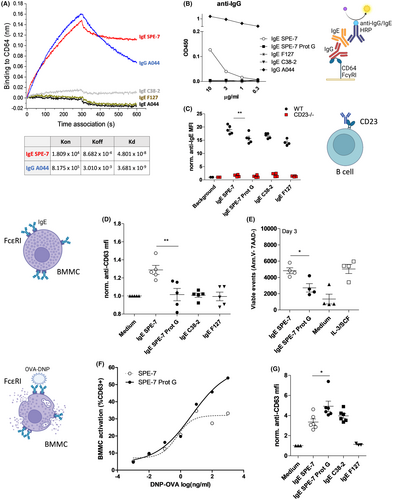
The binding of SPE-7 to FcεRI-expressing BMMCs at 4°C was not significantly altered by IgG removal (Figure S2). When BMMCs were incubated with the different IgEs at 37°C, activation marker CD63 was slightly but consistently up-regulated by SPE-7 but not by IgG-depleted SPE-7 or control IgEs (Figure 2D). In survival tests over 3 days in absence of IL-3/SCF, IgE SPE-7 but not IgG-depleted IgE SPE-7 increased BMMC survival compared to medium controls (Figure 2E). Finally, in classical antigen-dependent degranulation assays, IgG depletion led to a striking increase in the ability of IgE SPE-7 to de-granulate mast cells (Figure 2F,G).
In conclusion, we unveil that purified monoclonal IgE SPE-7 can contain mouse IgG antibodies that form complexes with IgE and alter its functional profile. Our results are in line with previous studies that attributed the unique effects of SPE-7 to the presence of trimeric aggregates.1 Our findings challenge the concept that IgE exerts antigen-independent effects on mast cell degranulation and survival. Moreover, the extent to which mouse IgE can bind to FcγRs needs to be carefully re-evaluated. The physiological relevance for IgG-IgE complexes is still given, as natural IgG anti-IgE autoantibodies and IgG-IgE complexes occur in mice and humans.5 Our findings are important to consider in the production of therapeutic IgE antibodies, which are gaining interest in cancer therapy.6 Contaminating IgGs may alter the functional and pharmacological profile of these therapeutic IgEs. Future studies will investigate how IgG makes its way into hybridoma-produced IgE clones.
AUTHOR CONTRIBUTIONS
P.E. conceptualized and supervised the study. M.S. and M.Z. purified IgE and performed ELISA. M.S. and P.E. performed cell-based assays. M.V. performed biolayer interferometry. A.C.U. and S.B.L. performed mass spectrometry. P.E. wrote the manuscript, A.C.U., M.S. and M.V. edited the manuscript.
ACKNOWLEDGMENTS
We thank Prof. Manfred Heller and the Proteomics and Mass Spectrometry Core Facility of the University of Bern. We thank Aleksandra Nonic, Simon Zinkhan, Alessandro Pardini, for technical assistance and Prof. Martin F. Bachmann for critically reading the manuscript (all University Hospital Bern). Open access funding provided by Inselspital Universitatsspital Bern.
FUNDING INFORMATION
This work was funded by a grant from Holcim Stiftung zur Förderung der wissenschaftlichen Fortbildung, and a grant from Novartis Foundation for Medical-Biological Research number 23B097 awarded to P.E.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest in relation to this work.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




