Allergen-specific immunotherapy induces the suppressive secretoglobin 1A1 in cells of the lower airways
Schmidt-Weber and Chaker these authors equally contributed to the manuscript.
Funding information
This study was supported by the German Center for Lung Research (DZL) to UMZ, HG, and CSW; Helmholtz Inflammation&Immunology (I&I) to CSW; a Grant of the German Research Foundation (DFG) No. 398577603 to CSW and UMZ.
Abstract
Background
While several systemic immunomodulatory effects of allergen-specific immunotherapy (AIT) have been discovered, local anti-inflammatory mechanisms in the respiratory tract are largely unknown. We sought to elucidate local and epithelial mechanisms underlying allergen-specific immunotherapy in a genome-wide approach.
Methods
We induced sputum in hay fever patients and healthy controls during the pollen peak season and stratified patients by effective allergen immunotherapy or as untreated. Sputum was directly processed after induction and subjected to whole transcriptome RNA microarray analysis. Nasal secretions were analyzed for Secretoglobin1A1 (SCGB1A1) and IL-24 protein levels in an additional validation cohort at three defined time points during the 3-year course of AIT. Subsequently, RNA was extracted and subjected to an array-based whole transcriptome analysis.
Results
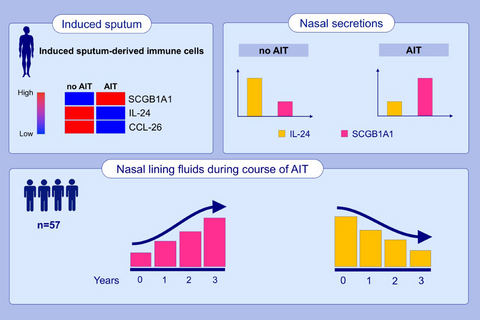
Allergen-specific immunotherapy inhibited pro-inflammatory CXCL8, IL24, and CCL26mRNA expression, while SCGB1A1, IL7, CCL5, CCL23, and WNT5BmRNAs were induced independently of the asthma status and allergen season. In our validation cohort, local increase of SCGB1A1 occurred concomitantly with the reduction of local IL-24 in upper airways during the course of AIT. Additionally, SCGB1A1 was identified as a suppressor of epithelial gene expression.
Conclusions
Allergen-specific immunotherapy induces a yet unknown local gene expression footprint in the lower airways that on one hand appears to be a result of multiple regulatory pathways and on the other hand reveals SCGB1A1 as novel anti-inflammatory mediator of long-term allergen-specific therapeutic intervention in the local environment.
Graphical Abstract
Allergen-specific immunotherapy inhibited pro-inflammatory CXCL8, IL24 and CCL26 mRNA expression in alveolar sputum cells. SCGB1A1 was induced in lower airway cells independently of the asthma status and allergen season. The epithelial type-2 (E2) cytokine IL-24 was reduced following 3 years of AIT-treatment, while SCGB1A1 was significantly increased and identified as a suppressor of epithelial gene expression.
Abbreviations
-
- AIT
-
- allergen-specific immunotherapy
-
- CCL
-
- C-C motif chemokine ligand
-
- CXCL
-
- C-X-C motif chemokine ligand
-
- SCGB1A1
-
- secretoglobin 1A1
1 INTRODUCTION
Allergen-specific immunotherapy (AIT) for allergic airway diseases such as hay fever is the only causative treatment of underlying immune-pathology and has proven to be safe and effective on long-term. However, biomarkers with predictive value for therapy efficacy are not yet available. It can be considered as a model for tolerogenic vaccination and understanding its mechanisms may provide insight for rational vaccine design not only in the field of allergy but also in autoimmunity, or transplantation. Albeit largely driven by Th-cells, type 2 responses are mainly involved in the late phase reaction to allergens. These Th2 responses have been hypothesized to be reduced during AIT, probably via inhibition by de novo recruited Tregs, as was shown in biopsies of affected nasal mucosa.1 Recent studies reported a shift in Th1/Th2 profiles under AIT, suggesting potentially competitive mechanisms being responsible for AIT-mediated reduction of Th2 cells and increased IFN-γ-expression of Th1 cells2-4 and was discussed as a potential antagonistic mechanism of suppression.2
Increasing evidence indicates that tissue-resident cells are key regulators of critical elements in the development of immune tolerance, such as tissue-resident regulatory T cells5 and myeloid regulatory cells.6, 7 We have recently suggested a 3-phase model (initiation, conversion, and tolerance-mounting phase) to describe changes in the lymphocyte compartment during allergen immunotherapy, characterized mainly by spatially separated populations.4 While these phenotypic changes of undulating Th17 responses, increasing levels of B-/Tregs and Th1-cells with simultaneous decrease of Th2 cells were mainly observed in peripheral blood we were able to detect these patterns also in upper airway brushings pointing to an interplay of systemic and local epithelial mechanisms in the development of immunological tolerance during AIT. This is in line with earlier published studies that were able to show reduced IL-5 mRNA levels in the airways8 and reduced number of sputum eosinophils.9 In vitro transcriptomic data and samples from disease cohorts provide evidence that airway epithelium mirrors type1 and type2 responses, we thereby accordingly categorized E1 and E2: IL-4 primes epithelial cells toward an E2 phenotype characterized by the expression of CCL26 and IL24, as well as a number of transcription factors, mucins, and anti-microbial peptides.10 Therefore, CCL-26 and IL-24 can be considered as tissue-derived biomarkers for type-2 immune responses.11 Th1 cells impede the E2-phenotype,10 whereas IFN-γ has been reported to potentiate TNF-α release of alveolar macrophages.12 Similar to Th1/Th2-epithelial cell interaction Tregs were shown to regulate epithelial repair functions by production of amphiregulin and restore tissue homeostasis.13 In addition, Tregs support tissue regeneration by promoting basal stem cell growth.14
Respiratory tract macrophages represent the major population within sputum cells and do not only represent phagocytic scavengers and microbial sensors, but also exert tissue homeostatic functions. A recent murine study provided experimental evidence that depletion of alveolar macrophages favors development of type-2 dominated immune responses.15 Alveolar macrophages and epithelial cells interact via CD200R and CD200 respectively, as CD200 expressed by epithelial cells is associated with tolerogenic and inhibitory functions,16 thereby restoring epithelial homeostasis.17 As a consequence of this interaction, we hypothesize that macrophage and epithelial cytokines may serve as markers for treatment efficacy and local tolerance induction.
2 METHODS
2.1 Clinical study
We investigated in this non-interventional observational study two (independent) cohorts of study participants. For the cross-sectional seasonal cohort, we recruited and induced sputa in 12 healthy controls and 40 grass pollen allergic participants with allergic rhinitis (AR) in (May-July) and off the pollen season (October-December). Within the group of grass pollen allergic patients, 21 received effective AIT since at least 1 year or longer (see Table 1). In the group of untreated AR patients, nine of them suffered from asthma comorbidity, while in the group of AIT-treated AR patients 12 showed asthma comorbidity. For details of all methods see Appendix S1.
|
Controls (n = 12) |
Allergic rhinitis w/o AIT (n = 19) |
Allergic rhinitis with AIT (n = 21) |
|
|---|---|---|---|
| Age [years] | 26.6 (±0.93) | 28.8 (±2.51) | 31.05 (±2.20) |
| Sex (m/f) | 7/5 | 10/9 | 12/9 |
| Asthma [%]comorbidity | 0 | 9/19 [47.3%] | 12/21 [57.1%] |
| mRQLQ score | 0 (±0.15) | 2.37 (±0.34)### | 1.5 (±0.26)##, a |
| Total IgE [IU/L] | 87.46 (±57.14) | 372.2 (±88.26)### | 123.4 (±24.33)###, a |
| (PT/CAP)to | |||
| Grass | 0 | 19/19 | 21/21 |
| Birch | 0 | 16/12 | 16/15 |
| HDM | 0 | 12/9 | 13/8 |
| Cat | 0 | 11/7 | 5/7 |
| Clinical symptoms to | |||
| Grass | 0 | 19 | 18 |
| Birch | 0 | 9 | 8 |
| HDM | 0 | 3 | 5 |
| Cat | 0 | 4 | 11 |
| Initially unclear anamnesis | 0 | 2 | 1 |
| FEV1 [L] | 4.56 (±0.44) | 4.13 (±0.27) | 4.32 (±0.25) |
| FVC [L] | 5.31 (±0.68) | 4.51 (±0.27) | 4.84 (±0.26) |
| MEF25 [L] | 3.52 (±0.36) | 2.80 (±0.27)# | 2.99 (±0.29)# |
Note
- Data are presented as mean ± SEM.
- # means significance in relation to healthy controls, # = p < .05; ## = p < .01; ### = p < .001; #### =p < .0001.
- a significance in relation to untreated patients.
2.1.1 Data acquisition and statistical analysis
All experimental procedures and analyses of this exploratory study were conducted by blinded research staff. Data are included in parenthesis throughout the results section as mean ± s.e.m. Non-parametric statistical test were chosen, as the data points were not normally distributed. Kruskal-Wallis tests were performed initially to avoid multiple testing, and, only when medians across patient groups varied significantly, multiple single comparisons were performed using two-tailed Mann-Whitney U tests. Statistically significant differences are depicted as *p < .05, **p < .01, ***p < .001, ****p < .0001. For further details, see Appendix S1.
3 RESULTS
To understand local mechanisms of airway inflammation and the impact of AIT, we analyzed sputum cells from patients with allergic rhinitis (Table 1) and from healthy controls and discovered a previously unknown footprint in sputum cells. Patients during AIT for at least 1 year were included into the AIT group.
3.1 Sputum cell composition and patient characteristics
Differences were assessable for total IgE, RQLQ, and cell load (number of sputum cells per ml, Figure 1A) between treated and untreated patients and healthy controls as shown in Table 1. Of note, substantial reductions in total IgE are usually only visible in larger sample sizes and later phases of up to 3 years following initiation of AIT.18 No differences were observed for lung function parameters (in season), assessed during the study. Some differences in composition of sputum cells became visible between untreated rhinitis patients and healthy controls, which were, however, in the range of previously reported sputum studies (Table 1).19
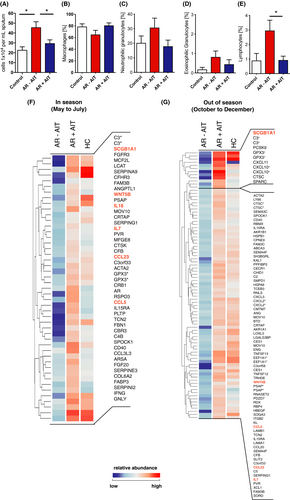
3.2 Differential expression of genes encoding for secreted proteins
Sputum cells of AIT-treated and untreated patients revealed significant gene expression differences which were found at a 1.5-fold threshold and a p-value of ≤ .05 with 1,680 downregulated genes and 485 upregulated genes in the pollen season and 7,916 observed downregulated and 1,288 upregulated off-season (Table S1,2,3,and 4). Sputum leukocytes infiltrate and interact with the lung via soluble mediators, we focused further analysis of AIT-induced expression changes specifically on secreted gene products using Gene Ontology (GO) terms. For in season comparison, this group contained genes such as type-1 cytokine genes IFN-γ (2.90-fold increase) and IL18 (1.62-fold increase), tissue remodeling factors (FGF20,COL6A2,ACTA2, FGFR3, ANGPTL1, RSPO3), enzymes (MOV10,GPX3), complement factors (C3,C4B,Serpins,CFB) and receptors that can be shedded or secreted (CD40,IL15RA; Figure 1F). In contrast, CXCL2 (1.75-fold increase), CXCL10 (3.75-fold increase), CXCL11 (5.59-fold increase), and CCL20 (1.99-fold increase) were exclusively found to be differentially regulated off-season (Figure 1G).
However, of particular interest are such genes that are differentially expressed in response to AIT independent of seasonal influences and pollen exposure as they may represent more stable and robust (bio)markers for a successful AIT treatment. Twenty-two genes encoding for secreted mediators were upregulated after AIT independently of the pollen season. Among these, we identified cytokines SCGB1A1, IL7, and WNT5B as well as the chemokines CCL5 and CCL23 (Figure 2A,C). Interestingly, the CCL-5 scavenger receptor ACKR2 was downregulated by AIT in season (Table S1). Among them, SCGB1A1showed the most robust change over the whole observation period, with a 3.34-fold increase even off-season compared with untreated allergic rhinitis patients.
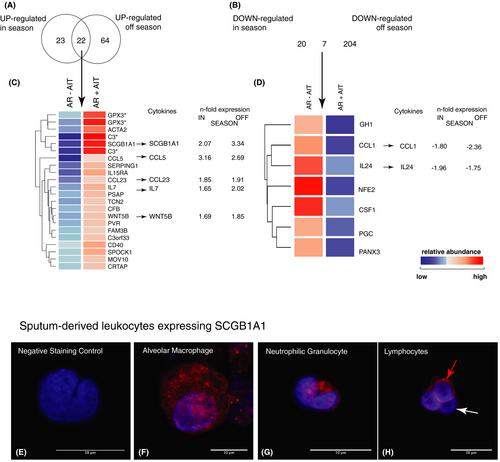
We identified SCGB1A1 positive cells in sputum leukocytes by confocal microscopy, including mononuclear cells such as macrophages (Figure 2E–H). Interestingly, SCGB1A1-positive lymphocytes were observed as well, which we currently investigate using a flow cytometric protocol. We verified the staining of SCGB1A1 in healthy turbinate tissues, which showed - as expected - SCGB1A1 expression in alpha-tubulin negative goblet cells, but also cells close to the basal membrane (Figure S1E–H). Furthermore, the expression of SCGB1A1 in airway epithelial cells was further confirmed in NHBE cells (Figure S1A,B) and ex vivo nasal polyp tissue (Figure S1C,D).
In a subgroup analysis, we investigated treated rhinitis patients who also suffered from asthma comorbidity and detected consistently higher expression of SCGB1A1, IFNγ (only in season), CCL5 and CCL23 compared with untreated patients in induced sputum transcriptomes (Figure S2).
In addition, we also found some important factors in the group of downregulated genes such as CCL1, IL24, CSF1, and Pannexin-3 (PANX3; Figure 2D). In particular, the anti-microbial gene CCL1 and the known type-2-related epithelial cytokine IL24 are of great interest, as IL-24 represents an E2 cytokine that marks the type-2 polarization of epithelial cells independent of the infiltration of immune cells.
3.3 Key cytokines confirmed on transcriptome and protein level in induced sputum
Selected genes, such as SCGB1A1 andIL24, which were significantly regulated in the array analysis (in comparison with known type-2 cytokines IL4 and IL13), were validated by real-time quantitative reverse transcription PCR (Figure 3A–H) and at protein level (Figure 3I–P). AIT-treated allergic patients showed significant differences in SCGB1A1expression levels compared with untreated patients suffering from rhinitis with asthma comorbidity. Up to sevenfold mRNA-expression and significant differences were found when comparing untreated (rhinitis: 0.24 ± 0.05; rhinitis+asthma: 0.42 ± 0.13) vs. AIT-treated rhinitis patients (7.64 ± 4.49; p = .041) or rhinitis patients with asthma comorbidity (1.36 ± 0.44; p = .048) in the pollen season (Figure 3A). For IL24, differences were shown in season for treated (1.16 ± 0.37) and untreated (7.27 ± 3.42; p = .03) rhinitis as well as for treated (0.41 ± 0.14) and untreated (11.31 ± 8.84; p = .026) rhinitis patients with asthma comorbidity (Figure 3B,F).
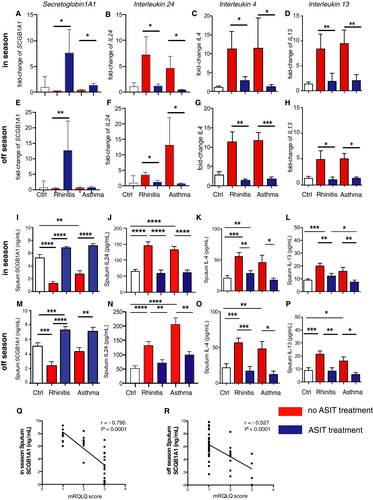
Protein levels for SCGB1A1 and IL-24 in- and off-season (Figure 3I,J,M,N) were validated using the MSD Mesoscale platform. Interestingly, SCGB1A1-levels in sputum supernatants were significantly lower in rhinitis (in and out of season) and rhinitis patients with asthma comorbidity (in season) as compared to healthy controls while IL-24 (as well as IL-4 and IL-13) levels were rather increased. Consistent with array and qPCR data, rhinitis patients receiving AIT treatment showed elevated protein levels of SCGB1A1 in sputum supernatant both in (7.24 ng/ml ± 0.46) and off-season (6.86 ng/ml ± 0.23) compared with untreated patients (in season: 2.38 ng/ml ± 0.60, p < .0001; off-season: 1.23 ng/ml ± 0.38, p < .0001; Figure 3I,M). The difference in asthma patients off-season is not visible on mRNA level of sputum cells (Figure 3E), and therefore, the protein level may represent SCGB1A1 (Figure 3M) originating from airway epithelial cells, which is excreted into airway lumen and collected during sputum induction. Similar changes were seen in treated (in season: 7.09 ng/ml ± 0.52; off-season: 7.22 ng/ml ± 0.31) and untreated rhinitis patients with asthma comorbidity (in season: 4.38 ng/ml ± 0.55, p = .002; off-season: 2.74 ng/ml ± 0.49, p < .0001). AIT-treated patients showed decreased levels of secreted IL-24 in sputum supernatants in (59.89 pg/ml ± 9.14) as well as out of season (71.07 pg/ml ± 11.43) compared with untreated patients (in season: 146.5 pg/ml ± 10.33, p < .0001; off-season: 133.1 pg/ml ± 12.98, p < .0001; Figure 3J,N). Similar changes became visible for AIT-treated (in season: 62.03 pg/ml ± 7.42; off-season: 98.95 pg/ml ± 12.49) and untreated rhinitis patients with asthma comorbidity (in season: 133.1 pg/ml ± 9.73, p < .0001; off-season: 206.6 pg/ml ± 22.03, p = .0011). SCGB1A1 protein levels were negatively strongly correlated with mRQLQ in season (r = −.795, p < .0001; Figure 3Q) and showed moderate negative correlation out of season (r = −.527, p < .0001; Figure 3R). Taken together, these data show that SCGB1A1 levels are inversely regulated by AIT compared with decreased levels of pro-inflammatory type-2 cytokines IL-24, IL-4, and IL-13.
Unexpectedly, increased SCGB1A1 levels were not only observed in sputum, but also in serum in and out of grass pollen season (Figure S3A,B), however with lower differences in magnitudes between groups. In rhinitis patients receiving AIT treatment, elevated serum SCGB1A1 levels were observed in (2.25 ng/ml ± 0.09) as well as off-season (2.79 ng/ml ± 0.02) compared with untreated patients (in season: 1.91 ng/ml ± 0.01, p = .035; off-season: 2.00 ng/ml ± 0.01, p = .006). In rhinitis patients with asthma comorbidity, similar levels for SCGB1A1 in serum were detected for treated (in season: 2.55 ng/ml ± 0.01; off-season: 2.82 ng/ml ± 0.01) and untreated patients (in season: 1.67 ng/ml ± 0.01, p = .002; off-season: 2.22 ng/ml ± 0.01, p < .0001; Figure S3).
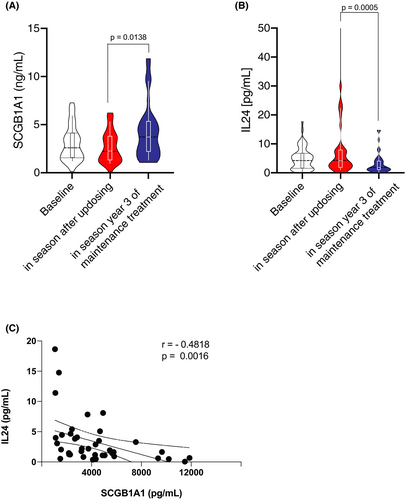
3.4 Secretion of SCGB1A1 and IL-24 during the course of AIT detected in nasal lining fluids
In order to cross-validate SCGB1A1 and its regulation during AIT and to assess another, yet easy accessible airway sample, we measured SCGB1A1 (and IL-24) using non-invasive nasal sampling at baseline of grass specific immunotherapy in a cohort of 57 patients, after pre-seasonal up-dosing period20 and after 3 years of AIT.4 SCGB1A1 remained unchanged immediately after up dosing (2.68 ng/ml ± 0.26) compared with baseline (3.02 ng/ml ± 0.24; Figure 4A). However, after completion of 3 years AIT SCGB1A1 levels were significantly increased compared with the time point after up dosing (4.27 ng/ml ± 0.45, p = .0138), even though changes were not significant to off-season baseline. The epithelial type-2 (E2-) cytokine IL-24 was significantly reduced following 3 years of AIT (3.23 pg/ml ± 0.56) compared with the time point after up dosing (9.32 pg/ml ± 2.78, p = .0005; Figure 4B). IL-24 and SCGB1A1 showed a significant and reverse correlation (r = −.4818, p = .0016; Figure 4C).
3.5 Function of SCGB1A1 as luminal immunotherapy target in vitro
To investigate the possible regulatory role of SCGB1A1 in AIT on respiratory tract epithelial function, recombinant human protein SCGB1A1 was added to primary epithelial cells in culture. Since we intended to demonstrate regulatory/suppressive effects of SCGB1A1, we needed to activate airway epithelial cells. While no intrinsic mechanisms of epithelial activation were so far described for grass pollen extracts, we decided to use house dust mite (HDM) extract, where intrinsic mechanisms of epithelial activation were previously described.21, 22 The concentration applied in the in vitro cultures was titrated ahead of the experiments in cell cultures (qPCR of CCL26 and IL8; Figure S4) and confirmed the concentration range that is observed in nasal secretions. Both, resting and HDM-activated primary human bronchial epithelial cells (NHBE) were exposed to SCGB1A1. For this experiment, we used unprimed NHBEs and also applied natural HDM-extracts as they are used in the clinical routine for diagnosis and treatment of patients. Of note, HDM-extracts naturally contain lipopolysaccharide (LPS; controlled amounts); however, at these concentrations NHBEs did not exhibit an LPS-typical footprint (data not shown). No cytotoxic effects were observed and a live metabolic assay showed no effect on altered oxygen consumption rates or extracellular acidification rates (data not shown). In a whole transcriptome analysis, 75 genes in resting NHBEs were upregulated, while 539 genes were downregulated after addition of SCGB1A1 (Figure 5). HDM-primed NHBEs revealed 91 upregulated genes, while 650 genes were downregulated. Subsequent pathway analysis revealed no significant footprints that could uncover the signaling mechanism of SCGB1A1-mediated cellular suppression. Thus, both resting and HDM-activated NHBEs responded to SCGB1A1 with a generalized downregulation of gene expression. We analyzed the top 50 downregulated genes for SCGB1A1-stimulated and HDM-activated, SCGB1A1-stimulated NHBEs, revealing suppression of pro-inflammatory mediators and pathways, such asCARD9 and MEG3 respectively (Table S13 and S14). Interestingly, between only SCGB1A1-stimulation and HDM-activation followed by SCGB1A1-stimulation, 24 genes including mucin-3a were found to be downregulated thus independently of the activation status (Figure 5).
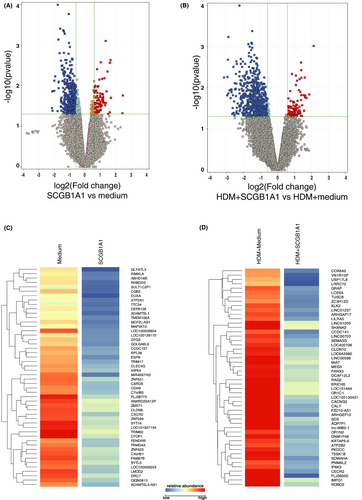
The effect of SCGB1A1 on immunoregulatory epithelial cytokine mRNA expression such as IL8 and IL24 revealed decreased levels, however did not reach statistical significance in the microarray experiment. In BEAS2B cells, SCGB1A1 displayed a significant difference for both genes, IL8 (HDM fold change: 1064.0-fold; HDM+SCGB1A1: 324.8-fold; p = .0142) and IL24 (HDM fold change: 58.8-fold; HDM+SCGB1A1: 22.67-fold; p = .0016; Figure S4).
Future studies need to further characterize epithelial genes and pathways to facilitate the molecular understanding of the SCGB1A1 induced silencing effect.
4 DISCUSSION
This study reveals the effect of AIT on lower airway cells and identifies SCGB1A1 as mediator of AIT-induced anti-inflammatory mechanisms in the epithelium. This finding is novel in a sense that anti-inflammatory mechanisms were so far mainly attributed to regulatory T- and B-cells3, 4 but not to alveolar macrophages.
4.1 Type-2local responses suppressed by AIT
Substantial knowledge exists about AIT mechanisms in systemic lymphocyte compartments. Our current data reveal yet unknown mechanisms namely those represented by sputum cells generated from the airways, where exposure to allergen, inflammation, and symptoms locally occur. Consistent with previous studies, we observe suppressed CXCL8 (IL8) expression, but also show the reduction of type-2-related mediators IL24 and CCL26.10, 23We have previously shown that both cytokines are induced by IL-4 or IL-13.10 Therefore, the seasonal unresponsiveness of CCL26 and IL24 in the course of AIT could be the consequence of diminished IL4 and IL13 production. Interestingly, this effect was also observed not only in sputum samples of rhinitis patients with asthma comorbidity but also in patients limited to upper airway symptoms only. In fact, all analytes were principally indistinguishable between these two patients groups with the exception of SCGB1A1 that is generally higher in rhinitis patients compared with rhinitis patients with asthma comorbidity. This finding underlines the validity of the "united airway" concept, which claims that epithelial activation of the lower airways is mirrored by the upper airways.24, 25 Consequently, the inflammatory mediators in the lower airways of rhinitis patients observed in this study could therefore indicate for a form of "latent" asthma even in rhinitis patients, which requires further investigation in longitudinal cohorts.
4.2 AIT-induced local changes in off-season
Of particular interest in understanding the local mechanisms of AIT in the luminal airways are genes that are induced independently of the allergen season, because they may indicate potential long-acting mechanisms independent of the seasonal influence. Interleukin-7 is an AIT-induced cytokine, as previously described as being important for the survival of tissue-resident B- and memory T-cells26 and thereby also modulate the balance of regulatory T cells.27 Two AIT-induced chemokines, CCL-5 (RANTES), and CCL-23 (MIP3), which are known to be involved in macrophage chemotaxis and were reported to be elevated in serum samples of patients receiving (insect venom) immunotherapy, were also detected in- and off-season.28CCL-5 is a major regulator of suppressive immune programs29 and may contribute to antagonize pro-allergic type-2 immunity by recruiting Th1 cells as well as regulatory T cells.30 The atypical chemokine receptor ACKR2,30 the major CCL-5 scavenger receptor, is decreased through AIT and further increases the biological activity of CCL5. CCL23 was also described to enhance IL10 production in monocytes and T cells.31
4.3 AIT-induced changes in tissue repair mechanisms
Largely unknown are mechanisms of immunotherapy that affect tissue repair and homeostasis. Among those, the epithelial differentiation cytokine WNT5B was induced by AIT in- and out of season and is known to be tightly regulated by the TGF-β signaling pathway.32 Angioarrestin (ANGPTL1), also induced by AIT, can block the angiogenic cascade,33 while AIT-induced FGFR3 (in contrast to other FGFRs) has been shown to negatively regulate epithelial cell proliferation and therefore has oncogenic capabilities.34 These observations suggest that AIT promotes tissue homeostasis in combination with matrix remodeling processes.
4.4 AIT-induced local expression in and off-season
The strongest AIT-mediated induction in and out of season was, however, observed for secretoglobin1A1 (SCGB1A1), a tetrameric glycoprotein of the secretoglobin family, with homologies toFel d1 and Fel d2. It has been previously described to be induced by glucocorticoids35 and IFN-γ.36 In fact, IFN-γ is induced by AIT, but becomes only visible in transcriptomes during the pollen season particularly in the subgroup of rhinitis patients with asthma comorbidity. Further, it has been shown that SCGB1A1 exerts immunosuppressive functions in the cyclooxygenase (COX)-2 pathway37 and gene defects in the SCGB1A1 gene are associated with susceptibility to asthma.38 Also in SCGB1A1 knockout mice, anti-inflammatory and immunomodulatory functions of SCGB1A1 were observed, for example, these mice showed increased type-2 immune responses and exaggerated respiratory tract inflammation upon allergen challenge.39 Further, SCGB1A1 knockout mice show increased airway infiltration by eosinophils upon streptococcus pneumoniae exposure.40 In addition, SCGB1A1 has been shown to attenuate LPS- or IL-13-induced activation of airway epithelial cells.41 Known sources of SCGB1A1 are airway and testicular club cells. Cross-talk between airway cells and macrophages in the conducting airways suggest that ANXA1 was involved in the immunoregulatory functions of SCGB1A1 via the regulation of the activity of pro-inflammatory enzymes such as inducible nitric oxide synthase (iNOS), cPLA2, and potentially cyclooxygenase.42 It is of particular interest that SCGB1A1 was found to be diminished in asthma patients in both BAL43 and serum.44 Serum levels of SCGB1A1 correlate with FEV1 and the Tiffeneau coefficient (FEV1/FVC).44 Detection of SCGB1A1 in serum could originate from the lung or from SCGB1A1-producing lymphocytes, which were shown in this study. Due to well-controlled clinical symptoms, our patients showed no correlation between SCGB1A1 levels and lung function parameters. However, we were able to show a significant correlation to their symptom load assessed by mRQLQ in- and off-season. The mechanisms of AIT-induced SCGB1A1 reconstitution to levels of healthy homeostatic conditions are entirely unclear; however, it is known that IL4 and IL13 are downregulating SCGB1A1.45 Therefore, it can be speculated that either the AIT-reduced expression of IL4 and IL13 or the increase of IFN-γ as natural antagonist of these type-2 cytokines could play an important role in underlying mechanisms of AIT.
4.5 SCGB1A1 as local silencer of epithelial cells
In addition, our data demonstrate for the first time that SCGB1A1 gene expression of resting as well as HDM-activated primary respiratory epithelial cells. In this experiment, primary bronchial epithelial cells were activated with house dust mite (HDM)-extracts in order to use a physiological IgE-independent mechanism. We observed a decrease of IL8 and IL24, which however did not reach statistical significance as using primary NHBEs; however, in BEAS2B cells a robust suppression was observed. SCGB1A1 downregulated expression of CARD9, an adaptor molecule in the NFκB pathway that can have pro- but also anti-inflammatory functions.46, 47 The SCGB1A1-mediated suppression of CARD9 can suppress the NFκB and ERK pathways resulting in decreased levels of pro-inflammatory mediators, such as IL-6, IL-12, GM-CSF, TNF, and IL-1β upon activation, the latter of which also involves the RASGRF1–H-Ras pathway.48 HDM-activation of epithelial cells resulted in downregulation of MEG3, a known target of CARD9 downstream signaling,49 which was described to be positively correlated with basal cell markers TP63, KRT5, KRT17, KRT14, and ITGB4.50Further, MEG3 regulates basal epithelial cell identity50 and inhibits SOX2,51 which is related to SCGB1A1 expression. In this study, transgenic deletion of SOX2 in airway epithelial cells prevented SCGB1A1 expression, consistent with the requirement of SOX2 in differentiation of both club and ciliated cells.51SCGB1A1 is also known to bind and neutralize lipid mediators, which are also triggered by HDM-stimulation of NHBEs. As SCGB1A1 acted independently of activation, it appears unlikely that SCGB1A1 inhibits airway epithelial cells by neutralization of lipid mediators. Future studies need to clarify the role of SCGB1A1 in epithelial lipid turnover and signaling. Our findings on the role of SCGB1A1 in AIT-induced immune suppression are of particular interest, as SCGB1A1 is the first AIT-induced gene involved in epithelial biology which is not a typical immune regulator produced by infiltrating cells.11 Overall, AIT-induced genes that do not underlie seasonal changes (IL7, CCL5, CCL23, WNT5B, and SCGB1A1) are associated with tolerogenic pathways that appear to cumulate in the lumen tissue. Future studies are needed to clarify, whether these changes persist after discontinuation of AIT as it is known for T cell–derived factors.
5 CONCLUSION
In conclusion, the current study demonstrates that AIT is reducing features of local airway inflammation including a reduction in generic inflammatory genes such as IL8, but also a reduction of epithelial type-2 inflammation such as IL-24 and CCL-26. Secretoglobin1A1 is identified as a local mediator induced in the response to AIT. In addition, secretoglobin1A1 is shown to be a factor that can restore intact lung tissue homeostasis and may thus be of relevance for future therapeutic approaches.
CONFLICT OF INTEREST
UMZ received payment for manuscripts from Deutsches Aerzteblatt and funds for travel from the European Academy of Allergy and Clinical Immunology (EAACI) and Collegium InternationaleAllergologicum (CIA). CSW received support for research projects from PLS Design, LETI, Zeller AG, and Allergopharma and accepted honoraria for consultancy and seminars from LETI and Allergopharma. He also received travel support from EAACI. AMC has consultancy arrangements through Technical University Munich with Allergopharma, ALK-Abello, AstraZeneca, GSK, HAL Allergy, Immunetek, Lofarma, Regeneron, Sanofi Genzyme; has conducted clinical studies and received research grants through Technical University Munich from Allergopharma, Novartis, the German Federal Environmental Agency, Bencard/Allergen Therapeutics, ASIT Biotech, GSK, Roche, and Zeller AG; has received payment for lectures from Allergopharma, ALK-Abello, AstraZeneca, GlaxoSmithKline, Leti, Bencard/AllergenTherapeutics, and SanofiGenzyme; has received payment for manuscript preparation from Bayerisches Aerzteblatt; and has received travel support from the European Academy of Allergyand Clinical Immunology (EAACI) and DGAKI. The rest of the authors declare that they have no relevant conflicts of interest.