Curcumin suppresses lung cancer progression via circRUNX1 mediated miR-760/RAB3D axis
Abstract
Background
Curcumin is a natural chemical component that has an anticancer effect. The aim of this study was to explore the potential molecular mechanism of curcumin regulating lung cancer (LC) progression.
Methods
The expression of circRUNX1, miR-760 and Ras-like GTPase 3D (RAB3D) was detected by qRT-PCR. Cell proliferation were determined by CCK8 assay and colony formation assay. Cell apoptosis, migration and invasion were detected by flow cytometry, wound healing and transwell assays. Protein levels were examined by western blot (WB) analysis. RNA interaction was confirmed by dual-luciferase reporter assay. LC xenograft tumors were constructed using BALB/c nude mice.
Results
CircRUNX1 was upregulated in LC and its expression could be inhibited by curcumin. Curcumin reduced LC cell proliferation, metastasis, and accelerate apoptosis, while circRUNX1 overexpression reversed these effects. MiR-760 was confirmed to be a target of circRUNX1, which could reverse the effects of circRUNX1 on curcumin-treated LC cell functions. RAB3D was a target of miR-760, and its knockdown reversed the promotion effect of miR-760 inhibitor on the progression of curcumin-treated LC cells.
Conclusion
Curcumin suppressed LC progression via circRUNX1/miR-760/RAB3D axis.
INTRODUCTION
Lung cancer (LC) refers to malignant tumors originating from the trachea, bronchus and lungs, and has a high morbidity and mortality rate worldwide.1, 2 The pathogenesis of LC has so far not been fully clarified, but there is evidence that genetic factors and environmental factors are involved in the occurrence of LC.3, 4 Surgery, medication and radiation therapy are the main methods of LC treatment. Although many efforts have been made to improve survival rate, LC patients' prognosis is still unsatisfactory.5, 6 Therefore, it is very important to clarify the molecular mechanism of LC and develop new targets for the treatment of LC.
Curcumin is a natural chemical component extracted from the rhizome of turmeric.7, 8 Curcumin has antioxidant, antibacterial, anti-inflammatory, and anticancer pharmacological effects, and it therefore has the potential to become an effective drug for treating arthritis, cardiovascular disease and cancer.9, 10 Studies have reported that curcumin exerts its anticancer effect by inhibiting cancer cell proliferation, metastasis, and promoting apoptosis, including LC.11, 12 Unfortunately, the anticancer molecular mechanism of curcumin in LC has not yet been fully elucidated.
Circular RNA (circRNA) is noncoding RNA with a circular structure, which is formed by covalent bonds.13, 14 Many studies have confirmed that circRNA functions as a potential biomarker and treatment target for cancer.15, 16 Previous studies have suggested that curcumin could regulate the radiosensitivity of nasopharyngeal carcinoma via regulating the circRNA network.17, 18 Therefore, the curcumin-circRNA axis may be an important way to elucidate the anticancer molecular mechanism of curcumin. Circ_0002360 is derived from RUNX1 gene, and is also known as circRNUX1. Studies have discovered that circRUNX1 is overexpressed in LC and might therefore be a potential target for LC treatment.19 In our study, we determined that curcumin could effectively inhibit circRUNX1 expression, and we speculated that curcumin restrained LC progression by regulating circRUNX1. Through experimental analyses, we revealed the molecular mechanism of curcumin and confirmed the speculation that curcumin participates in the progression of LC by regulating the circRNA network.
METHODS
Sample collection
Thirty patients with LC who received surgery at Central Hospital Affiliated to Shenyang Medical College were enrolled into our study. A total of 39 paired LC tumor tissues and adjacent normal tissues were kept at −80°C until use. All the volunteers signed informed consent, and the study was approved by the Ethics Committee of Central Hospital Affiliated to Shenyang Medical College.
Cell culture and curcumin treatment
Human normal lung epithelial cells (BEAS-2B) and LC cells (H1975 and PC9) were purchased from Biovector NTCC (Beijing, China) and cultivated in RPMI-1640 medium containing 10% FBS (Gibco) and 1% penicillin–streptomycin Solution (Sangon) at 37°C with 5% CO2. For curcumin treatment, H1975 and PC9 cells were treated with different concentrations of curcumin (Amresco) for 24 h.
Cell transfection
H1975 and PC9 cells were seeded in six-well plates and grown to 60% confluences. The circRUNX1 overexpression vector (circRUNX1), miR-760 mimic or inhibitor (anti-miR-760), Ras-like GTPase 3D (RAB3D) small interference RNA (siRNA) (si-RAB3D), or their negative controls were synthesized from Sangon. Cell transfection was carried out using lipofectamine 3000 (Invitrogen).
qRT-PCR
TRIzol reagent (Invitrogen) was used to extract RNA. Using a cDNA synthesis kit (Takara), the RNA was reverse-transcribed into cDNA. Then, qRT-PCR was performed with SYBR Green PCR Master Mix (Takara). Relative expression was calculated using 2−ΔΔCt method with GAPDH or U6 as an internal control. Primer sequences are shown in Table 1.
| Name | Primers (5′-3′) | |
|---|---|---|
| circRUNX1 | Forward | CCACTCCACTGCCTTTAACC |
| Reverse | TGATTTTGATGGCTCTGTGG | |
| RUNX1 | Forward | CTGCCCATCGCTTTCAAGGT |
| Reverse | GCCGAGTAGTTTTCATCATTGCC | |
| miR-760 | Forward | GCCGAGCGGCTCTGGGTCTG |
| Reverse | GTGCAGGGTCCGAGGT | |
| RAB3D | Forward | GATACGCGGACGACTCCTTC |
| Reverse | GGATTCCTGATTGGCGATGTC | |
| GAPDH | Forward | CTCTGCTCCTCCTGTTCGAC |
| Reverse | CGACCAAATCCGTTGACTCC | |
| U6 | Forward | CGCTTCGGCAGCACATATACTA |
| Reverse | CGCTTCACGAATTTGCGTGTCA |
RNase R assay
After extraction from cells, the RNA was incubated with RNase R. The qRT-PCR was utilized to measure circRUNX1 expression and linear RNA RUNX1 mRNA expression.
Cell counting kit 8 (CCK8) assay
A CCK8 assay kit (Dojindo) was used for examining cell viability. H1975 and PC9 cells were seeded into 96-well plates. CCK8 reagent was added into each well for 4 h. The absorbance was evaluated at 450 nm to reflect cell viability.
Colony formation assay
H1975 and PC9 cells were resuspended in a six-well plate for 2 weeks. The colonies were fixed with stained with crystal violet and photographed immediately to count the colony numbers under a microscope.
Flow cytometry
H1975 and PC9 cells were harvested and resuspended with binding buffer, and then incubated with Annexin V-FITC and propidium iodide (Biolegend). Finally, the cell apoptosis rate was analyzed using a flow cytometer and CellQuest software.
Western blot (WB) analysis
Total proteins were extracted with RIPA lysis buffer (Sangon). Protein was separated in 10% SDS-PAGE gels and transferred to PVDF membranes (Invitrogen). Subsequently, membranes were incubated with anti-Bcl-2 (1:2000, Bioss), Bax (1:1000, Bioss), anti-MMP2 (1:3000, Abcam), anti-MMP9 (1:1000, Abcam), anti-RAB3D (1:2000; Solarbio), or anti-GAPDH (1:10000, Abcam). After incubating with appropriate HRP-labeled secondary antibodies (1:20000, Abcam), protein bands were detected using ECL luminescence reagent (Sangon). Relative protein expression was quantified by ImageQuant software.
Wound healing assay
H1975 and PC9 cells were reseeded in six-well plates. After the cells had grown to 90% confluence, a 20 μl pipette tip was used to draw a line evenly on the plate. Subsequently, cells were cultured in serum-free medium for 24 h. The wound healing area at 0 h and 24 h was compared, and the wound healing rate was calculated to detect cell migration ability.
Transwell assay
The upper chamber of transwell plates, precoated with Matrigel, was used for cell invasion and noncoated was used for cell migration. In brief, H1975 and PC9 cells suspended with serum-free medium were seeded into the upper chamber. The lower chambers were covered with completed medium. After 24 h, migration and invasion cell numbers were counted under the microscope (100×).
Dual-luciferase reporter assay
The wild-type and mutant-type sequences of circRUNX1 or RAB3D 3'UTR containing miR-760 binding sites or mutation sites were cloned into pGL3 reporter vector to produce the circRUNX1 WT/MUT and RAB3D 3'UTR WT/MUT vectors. H1975 and PC9 cells were seeded in 24-well plates and were then cotransfected with the above vectors and miR-760 mimic/miR-NC. Relative luciferase activities (Firefly/Renilla) were measured by dual-luciferase assay kit (Solarbio).
LC xenograft tumors
All animal studies were approved by the Animal Ethics Committee of Central Hospital Affiliated to Shenyang Medical College. H1975 cells (1 × 106) were subcutaneously injected into BALB/c male nude mice (n = 6; Vital River Laboratories, Beijing, China) and divided into two groups. Curcumin was dissolved in dimethyl sulfoxide (DMSO, Sangon) and intraperitoneally injected into mice at 50 mg/kg every 5 days. The control group was injected with 50 mg/kg DMSO. Tumor volume was determined every week. After 30 days, the tumors were removed and weighed. An immunohistochemical (IHC) test was performed to analyze proliferation marker Ki67 positive cells in tumor tissues.
Statistical analysis
Data are presented as the mean ± SD. Differences between groups were compared by Student's t-test or analysis of variance (ANOVA). Statistical analysis was performed using GraphPad Prism software. p < 0.05 was considered statistically significant.
RESULTS
CircRUNX1 was upregulated in LC and regulated by curcumin
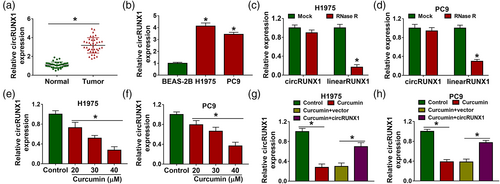
In LC tumor tissues, we discovered that circRUNX1 was higher (Figure 1a). Also, it was observed that there was elevated expression of circRUNX1 in LC cells (H1975 and PC9) (Figure 1b). CircRUNX1 could resist the RNase R digestion and showed higher stability than its linear RUNX1 (Figure 1c,d). We found that in curcumin-treated H1975 and PC9 cells, as the concentration of curcumin increased, circRUNX1 expression was significantly decreased in a concentration-dependent manner (Figure 1e,f). Since curcumin at 40 μM had a good effect, we used the concentration of 40 μM for subsequent tests. To determine the regulatory effect of curcumin on circRUNX1, circRUNX1 overexpression vector was constructed and transfected into LC cells. The results showed that overexpressed circRUNX1 effectively reversed the inhibition of curcumin on circRUNX1 expression (Figure 1g,h).
Curcumin suppressed LC cell progression by regulating circRUNX1
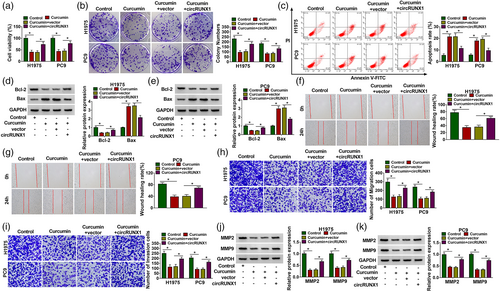
We found that curcumin inhibited the viabilities and colony numbers of LC cells, while circRUNX1 overexpression reversed this effect (Figure 2a,b). By detecting the cell apoptosis rate, we also discovered that the promotion of curcumin on H1975 and PC9 cell apoptosis could be abolished by circRUNX1 upregulation (Figure 2c). Curcumin could decrease the antiapoptosis marker Bcl-2 level and increase apoptosis marker Bax level in H1975 and PC9 cells, and overexpressed circRUNX1 also reversed these effect (Figure 2d,e). The wound healing rate of H1975 and PC9 cells could be repressed by curcumin (Figure 2f,g), and the numbers of migration and invasion cells could also be restrained by curcumin (Figure 2h,i). However, the inhibitory effect of curcumin on LC cell migration and invasion could be recovered by circRUNX1 overexpression (Figure 2f–i). Overexpressed circRUNX1 also enhanced the protein levels of metastatic markers MMP2 and MMP9, which were inhibited by curcumin (Figure 2j,k). Our data therefore suggested that curcumin regulated circRUNX1 to mediate LC progression.
CircRUNX1 served as a miR-760 sponge
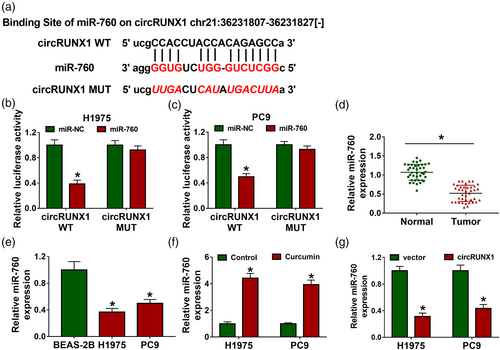
CircRNA can be used as a “miRNA sponge” to regulate downstream target expression.20, 21 To explore the mechanism of circRUNX1, StarBase version 2.0 software was used to predict the targeted miRNAs of circRUNX1, and found that circRUNX1 had a large number of targeted miRNAs. Through literature research, we selected four miRNAs with low expression in LC tissues and had an inhibition on LC progression as candidate miRNAs. By detecting the expression of each candidate miRNAs in H1975 and PC9 cells after the high expression of circRUNX1, we found that miR-760 expression was decreased most significantly (Figure S1a,b). Therefore, miR-760 was studied as the target of circRUNX1. The binding sequences are shown in Figure 3a. Subsequently, the luciferase activity of circRUNX1 WT vector could be inhibited by miR-760 overexpression (Figure 3b,c), indicating the interaction between circRUNX1 and miR-760. Moreover, a significantly low expression of miR-760 was found in LC tumor tissues and cells (Figure 3d,e). Furthermore, curcumin could notably promote miR-760 expression (Figure 3f), while circRUNX1 could inhibit miR-760 expression (Figure 3g). In addition, circRUNX1 expression was detected in H1975 and PC9 cells transfected with miR-760 mimic or miR-NC, and the results showed that miR-760 overexpression did not affect the expression of circRUNX1 (Figure S2).
MiR-760 overexpression reversed the regulation of circRUNX1 on curcumin-treated LC cell progression
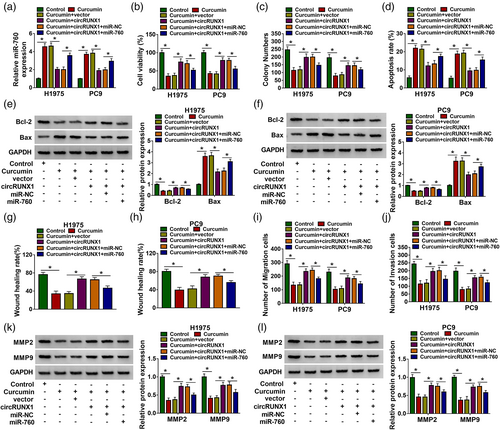
To evaluate whether miR-760 was involved in the regulation of circRUNX1 on curcumin-treated LC cell progression, circRUNX1 and miR-760 mimic were cotransfected into LC cells, followed by treating with 40 μM curcumin for 24 h. CircRUNX1 significantly inhibited the promoting effect of curcumin on miR-760 expression, which could be reversed by miR-760 mimic, suggesting that the transfection of the two was effective (Figure 4a). As shown in Figure 4b–d, the promotion effect of circRUNX1 on the viabilities and colony numbers, as well as the suppressive effect on the apoptosis rates of curcumin-treated cells, were reversed by miR-760 overexpression. MiR-760 overexpression inverted the protein level of Bcl-2 promoted by circRUNX1 and the protein level of Bax suppressed by circRUNX1 in curcumin-treated cells (Figure 4e,f). The enhancing effect of circRUNX1 on the wound healing rates and the numbers of migration and invasion in curcumin-treated cells could also be abolished by miR-760 overexpression (Figure 4g–j). Overexpressed miR-760 could reverse MMP2 and MMP9 protein levels increased by circRUNX1 in curcumin-treated H1975 and PC9 cells (Figure 4k–l). All the results indicated that circRUNX1 participated in the regulation of curcumin on LC progression by miR-760.
RAB3D was a direct target of miR-760
We also used the StarBase version 2.0 software to predict miR-760 target and RAB3D 3'UTR was found to complement to miR-760 (Figure 5a). MiR-760 overexpression reduced the luciferase activity of RAB3D 3'UTR WT vector (Figure 5b,c). Compared to negative controls, it was observed that RAB3D expression was remarkably increased in LC tumor tissues and cells at the mRNA and protein levels (Figure 5d–g). We determined that curcumin had a significant inhibitory effect on RAB3D mRNA and protein expression (Figure 5h–i). By detecting miR-760 expression, we confirmed the transfection efficiency of miR-760 mimic (Figure 5j). Subsequently, in H1975 and PC9 cells transfected with miR-760 mimic, the mRNA and protein levels of RAB3D were markedly inhibited by miR-760 overexpression (Figure 5k,l). RAB3D expression in H1975 and PC9 cells cotransfected with circRUNX1 overexpression vector and miR-760 mimic was also detected. As shown in Figure 5m,n, overexpressed circRUNX1 obviously enhanced RAB3D mRNA and protein expression, but miR-760 mimic could reverse this effect.
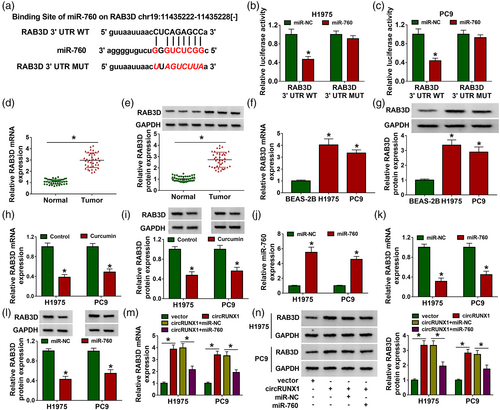
Silencing RAB3D reversed the promotion of miR-760 inhibitor on curcumin-treated LC cell progression
In addition, si-RAB3D was used to reduce RAB3D expression in H1975 and PC9 cells (Figure S3). To determine whether miR-760 participated in curcumin-treated LC cell progression by targeting RAB3D, anti-miR-760 and si-RAB3D were cotransfected into curcumin-treated LC cells. By measuring RAB3D protein expression, we found that miR-760 inhibitor could promote the RAB3D expression inhibited by curcumin, but this effect could be eliminated by si-RAB3D, indicating a successful transfection (Figure 6a). Further experiments revealed that miR-760 inhibitor could enhance cell viabilities and colony numbers, and repressed apoptosis rates in curcumin-treated H1975 and PC9 cells, while RAB3D knockdown could reverse these effects (Figure 6b–d). The promotion of miR-760 inhibitor on Bcl-2 protein level and the inhibition on Bax protein level in curcumin-treated H1975 and PC9 cells could be inverted by silencing RAB3D (Figure 6e,f). Downregulated RAB3D also reversed the increasing effect of miR-760 inhibitor on the wound healing rates, the numbers of migration and invasion cells, as well as MMP2 and MMP9 protein levels in curcumin-treated H1975 and PC9 cells (Figure 6g–l).
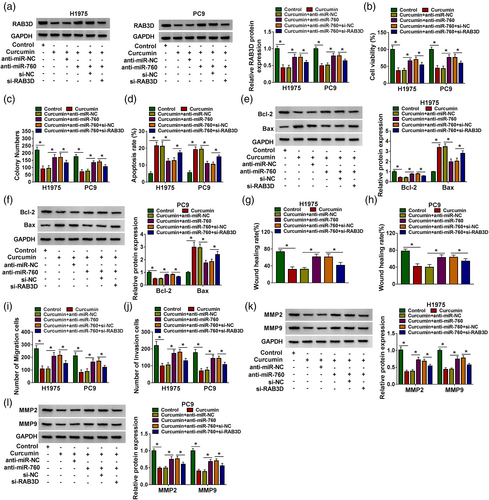
Curcumin inhibited LC tumor growth by regulating the circRUNX1/miR-760/RAB3D axis in vivo
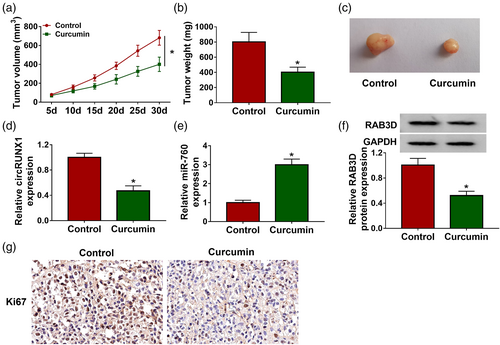
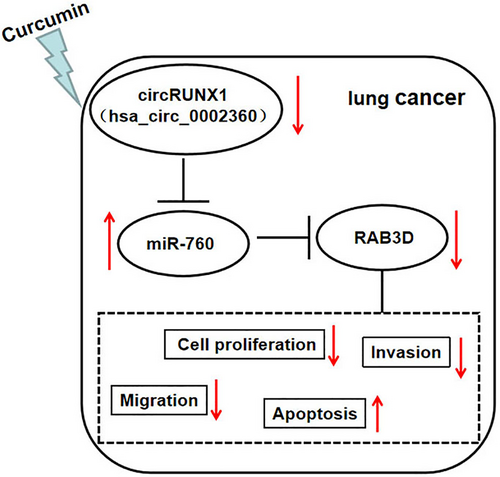
To further confirm the regulatory effect of curcumin on LC progression, we constructed LC xenograft tumors to conduct in vivo experiments. After 30 days of curcumin injection, we found that the volume of xenograft tumors in the curcumin treatment group was lower (Figure 7a). Also, tumor weight in the curcumin-treated group was significantly reduced (Figure 7b). Figure 7c shows a schematic diagram of the size of the two groups of tumors. Subsequently, we detected circRUNX1, miR-760 and RAB3D expression in the two groups of tumor tissues. The results showed that the circRUNX1 expression and RAB3D protein level were markedly decreased, while miR-760 expression was increased in the curcumin treatment group (Figure 7d–f). IHC results showed that Ki67 positive cells were markedly reduced in the curcumin treatment group (Figure 7g). In conclusion, we confirmed that curcumin inhibited LC progression through regulating the circRUNX1/miR-760/RAB3D axis (Figure 8).
DISCUSSION
Curcumin, as a natural phytochemical, is attracting more and more attention. The anticancer role of curcumin has previously been confirmed in hepatocellular,22 breast23 and pancreatic cancers.24 Here, we reveal the anticancer effect of curcumin in LC progression. Curcumin hindered proliferation, metastasis, increased the apoptosis of LC cells, and restrained LC tumor growth. Our results are consistent with previous studies,11, 12 which indicate that curcumin is an effective substance to suppress LC progression.
CircRUNX1 is a newly discovered functional circRNA. Research has suggested that circRUNX1 is overexpressed in colorectal cancer (CRC) and can act as a cancer-promoting factor to promote CRC proliferation and metastasis.25 Consistent with previous research results,19 high expression of circRUNX1 was confirmed in LC in our study Surprisingly, circRUNX1 expression was found to be negatively regulated by curcumin. Functional tests confirmed that overexpressed circRUNX1 reversed the inhibitory effect of curcumin on LC progression, showing that curcumin might regulate the progression of LC by inhibiting circRUNX1 expression. These findings are of great significance for molecular targeted therapy in LC.
CircRUNX1 has been shown to sponge miR-245-5p to regulate IGF1 expression in CRC.25 To elucidate circRUNX1 mechanism, we used bioinformatic analysis and experimental verification and identified that circRUNX1 could serve as a sponge for miR-760. MiR-760 has been found to be significantly downregulated in many cancers, including CRC,26 gastric cancer,27 and cervical cancer.28 In LC, miR-760 was considered to hinder LC cell proliferation and metastasis.29, 30 In addition, it had been stated that radiation therapy could inhibit LC progression by increasing miR-760 expression.31 Similarly, our study revealed that miR-760 was underexpressed in LC, and its expression was promoted by curcumin. Further experiments showed that miR-760 reversed the promoting effect of circRUNX1 on curcumin-treated LC cell progression, which confirmed that miR-760 was the downstream miRNA of circRUNX1 to participate curcumin-mediated LC progression.
RAB3D is a member of the Rab3 subfamily and has been found to be upregulated in several types of cancers. For example, RAB3D has been shown to promote the malignant progression of CRC,32 and its knockdown inhibited esophageal squamous cell carcinoma progression.33 Li et al. found that the overexpression of RAB3D could promote LC cell proliferation and metastasis.34 Here, we discovered that RAB3D could interact with miR-760, and its expression was negatively regulated by curcumin and miR-760. Moreover, RAB3D expression was also positively regulated by circRUNX1. The reversal effect of RAB3D silencing on miR-760 inhibitor illuminated that RAB3D was targeted by miR-760 and it participated in curcumin regulating LC progression.
In conclusion, our study showed that curcumin plays an anticancer role in the progression of LC, which is mainly realized by circRUNX1/miR-760/RAB3D axis. Our study revealed for the first time that curcumin inhibits LC cancer progression by regulating the circRNA network, which not only provides more evidence for the anticancer role of curcumin, but also provides a new molecular target for LC treatment.
AUTHOR CONTRIBUTIONS
HC supervised and managed the study. XW conducted the experiments and drafted the manuscript. NL and SL collected, analyzed the data and contributed the methodology. GL edited the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




