A case of lung adenocarcinoma with a novel CD74-ROS1 fusion variant identified by comprehensive genomic profiling that responded to crizotinib and entrectinib
Abstract
ROS1 rearrangements are found in 1–2% of patients with non-small-cell lung cancer. The detection of the rearrangements is crucial since clinically effective molecular targeted drugs are available for them. We present a case of lung adenocarcinoma with a previously unknown ROS1-CD74 fusion variant, CD74 exon 3 fused to ROS1 exon 34, which was not detected by a conventional RT-PCR-based test for ROS1 fusion gene detection but identified by hybrid capture-based next-generation sequencing. This tumor responded to crizotinib initially and to entrectinib after relapse with brain metastasis, indicating the oncogenic activity of this novel fusion variant.
INTRODUCTION
ROS1 rearrangements are found in 1–2% of patients with non-small-cell lung cancer1 and so far 24 partner genes for the rearrangements have been identified.2, 3 We need to identify these rearrangements since patients with the rearrangements gain significant benefits from molecular targeted drugs such as crizotinib,4 ceritinib,5 lorlatinib,6 and entrectinib.7
Reverse transcription polymerase chain reaction (RT-PCR) using specific primers has been used for the detection of ROS1 rearrangements. The commercial OncoGuide®AmoyDx® (Amoy Diagnostics, Xiamen, China) can detect already-known 15 ROS1 fusion variants by RT-PCR. On the other hand, next-generation sequencing (NGS) has become a useful tool in clinical settings for comprehensive analysis of cancers to identify druggable targets. Relatively rare ROS1 fusions have been identified by NGS.8, 9 Here, we present a case of lung adenocarcinoma with a novel breakpoint of CD74-ROS1 fusion gene which was not detected by the PCR-based OncoGuide®AmoyDx® but was identified by the NGS-based FoundationOne® CDx (Foundation Medicine, Cambridge, MA, USA).
CASE REPORT
A 38-year-old female never-smoker presented with enlargement of cardiac silhouette on a chest x-ray, and a chest computed tomography (CT) scan showed a small lung nodule in the left upper lobe inferior lingular segment, bilateral enlargement of mediastinal lymph nodes, and significant pericardial effusion (Figure 1(a)). A transbronchial tumor biopsy and additional examinations revealed clinical stage IVA (T1aN3M1a) lung adenocarcinoma. EGFR mutations and ALK translocation were not detected from the biopsy specimens. After partial response by five cycles of first-line chemotherapy with cisplatin, pemetrexed, and bevacizumab, maintenance chemotherapy with pemetrexed and bevacizumab was started. Complete response was obtained after two cycles of the maintenance therapy.
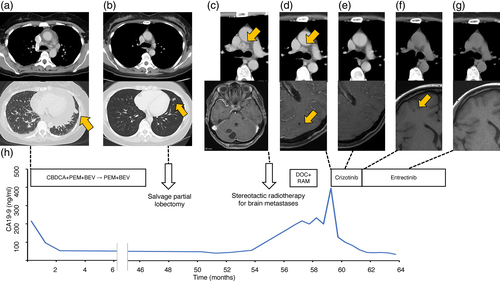
Four years after the first diagnosis, when 43 cycles of the maintenance chemotherapy were completed, a chest CT scan showed a small shadow at the primary lesion (Figure 1(b)). Salvage partial lobectomy was performed and viable cancer cells were confirmed in the resected specimens. ROS1 fusion transcripts were not detected from the specimens by using the OncoGuide®AmoyDx® kit. They were also negative for BRAF mutations.
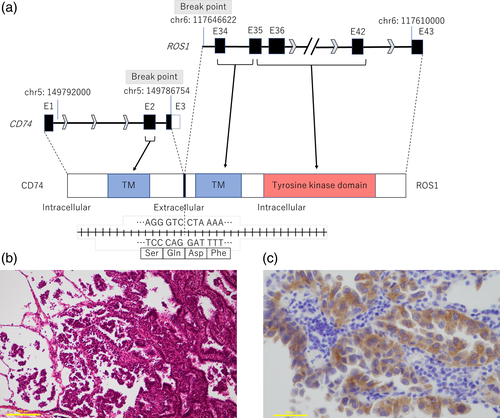
Nine months after the operation, her lung cancer relapsed with bilateral mediastinal and hilar lymphadenopathy, multiple bone metastases, and multiple brain metastases (Figure 1(c)). Several symptomatic brain metastases were treated with stereotactic irradiation and second-line chemotherapy with docetaxel and ramucirumab was started. Meanwhile, we undertook NGS analysis of the surgical specimens with FoundationOne®CDx and this revealed a novel CD74-ROS1 rearrangement. This novel fusion variant C3R34 was generated by the fusion of exon 3 of CD74 on chromosome 5q to exon 34 of ROS1 on chromosome 6q (Figure 2(a)). When we examined ROS1 protein expression by immunohistochemistry (IHC) staining with anti-ROS1 (D4D6®) rabbit monoclonal antibody (Cell Signaling Technology, Danvers, MA), cancer cells in the surgical specimens were strongly stained with this antibody (Figure 2(b),(c)). Based on these results, she was treated with oral crizotinib since at this point new asymptomatic brain metastases had appeared despite shrinkage of the enlarged mediastinal and hilar lymph nodes (Figure 1(d)). After 1 month, a chest CT scan showed further shrinkage of the mediastinal and hilar lymph nodes, and a head magnetic resonance imaging (MRI) showed disappearance of untreated brain metastases (Figure 1(e)). However, 2 months after administration of crizotinib, another asymptomatic small brain metastasis had appeared while lymph node metastases and bone metastases remained stable (Figure 1(f)). Next, we switched crizotinib to entrectinib. After 1 month, a head MRI showed disappearance of the new brain metastasis again (Figure 1(g)). Carbohydrate antigen 19–9 shown in Figure 1(h) was considered to reflect her disease activity.
DISCUSSION
In this case, we identified a novel CD-74-ROS1 fusion variant C3R34 in a patient with lung adenocarcinoma. We were able to detect this fusion variant by NGS after a negative result of testing for ROS1 rearrangements with a PCR-based method. While known breakpoints of CD74-ROS1 fusion gene are exon 6 of CD74 and exon 34 of ROS1 (C6R34), exon 6 of CD74 and exon 32 of ROS1 (C6R32), exon 7 of CD74 and exon 32 of ROS1 (C7R32), to the best of our knowledge C3R34 has not been previously reported.
ROS1 encodes a receptor tyrosine kinase that is a single-pass transmembrane protein with an intracellular C-terminal tyrosine kinase domain and a large extracellular N-terminal domain.10 In detail, exons 1–34 encode an extracellular domain, exons 34 and 35 encode a transmembrane domain, and exons 35–42 encode an intracellular tyrosine kinase domain. It has been reported that expression of ROS1 fusions leads to oncogenic transformation both in vitro and in vivo,11, 12 and constitutive kinase activation is thought to be a main part of their oncogenic activities.13 However, neither the normal function of native ROS1 with its unidentified ligand nor mechanisms of dysregulated kinase activity by rearranged ROS1 is still unclear. The CD74 gene encodes the type II integral membrane glycoprotein CD74, which is a single-pass transmembrane protein with an intracellular N-terminal domain and works as a surface-binding receptor for migration inhibitory factor (MIF), pro-inflammatory cytokine.14 Among nine exons of the CD74 gene, exons 1 and 2 encode intracellular and transmembrane domains while exons 3–8 encode an extracellular domain.15 Therefore, this C3R34 fusion protein is supposed to contain the tyrosine kinase and transmembrane domains of ROS1, and the intracellular and transmembrane domains of CD74. Based on the predicted structure of the fusion protein, we speculated that it had potential oncogenic activity for which targeted therapies might be effective. In fact, we were able to confirm that crizotinib and entrectinib were both effective against this tumor, although crizotinib was less effective on brain metastasis. Since the site of tumor progression after starting crizotinib was only the brain, we speculated that early resistance to crizotinib in this case was probably due to its poor penetration through the blood–brain barrier.16-19
Importantly, we detected this fusion variant by applying hybrid capture-based NGS. While PCR-based approaches for clinical use such as OncoGuide®AmoyDx® and amplicon sequencing-based NGS systems such as Oncomine® Dx Target Test can detect only known fusion variants, FoundationOne®CDx can detect unknown variants of the CD74-ROS1 fusion gene when the breakpoints are either within introns 6 to 8 of CD74 or within introns 31 to 35 of ROS1, which enabled us to identify the new CD74-ROS1 fusion variant C3R34 in this case. Clinicians should be aware of the differences in testing systems for the detection of gene rearrangements.
Despite the identification of this novel fusion variant, whether this is actionable by ROS1-targeted therapy remains to be seen. In this regard, we observed overexpression of ROS1 protein by IHC staining of the tumor tissue. IHC analyses using the anti-ROS1 rabbit monoclonal antibody (D4D6®) have shown promise for accurate identification of ROS1-rearranged cancers.20 Taken together with the prediction of the fusion gene structure described above, we considered that this case was treatable by the ROS1-targeted drugs. Indeed, we could observe the efficacy of crizotinib and entrectinib in this case.
In summary, we present a case of lung adenocarcinoma with a novel CD74-ROS1 fusion variant C3R34 which was not detected by conventional PCR-based testing but identified by hybrid capture-based NGS, confirming an important role for the diagnostic application of NGS in precision medicine.
CONFLICT OT INTEREST
Dr. Hashiguchi reports personal fees from AstraZeneca, outside the submitted work. Dr. Sato reports personal fees from Chugai Pharmaceutical, personal fees from Bristol Myers Squibb, and personal fees from Boehringer Ingelheim, outside the submitted work. Dr. Kato reports grants and personal fees from Abbvie, grants and personal fees from Amgen, grants and personal fees from AstraZeneca, grants and personal fees from Bristol Myers Squibb, grants and personal fees from Chugai, grants and personal fees from Eli Lilly, grants and personal fees from Merck Biopharma, grants and personal fees from MSD, grants and personal fees from Novartis, grants and personal fees from Ono, grants and personal fees from Pfizer, grants and personal fees from Taiho, personal fees from Boehringer Ingelheim, personal fees from Daiichi-Sankyo, personal fees from Nippon Kayaku, personal fees from Takeda, and grants from Regeneron, outside the submitted work. Dr. Nakahara reports personal fees from MSD, personal fees from Ono Pharmaceutical, personal fees from Chugai Pharmaceutical, grants and personal fees from Eli Lilly, grants and personal fees from Bristol-Myers Squibb, personal fees from Boehringer Ingelheim, and grants from Takeda Pharmaceutical Company Limited, outside the submitted work. Dr. Shiomi reports personal fees from Boehringer Ingelheim, personal fees from Bristol Myers Squibb, personal fees from Chugai Pharmaceutical, personal fees from Pfizer, and personal fees from Taiho, outside the submitted work. The remaining authors declare no conflict of interest.




