Parallel comparison and combining effect of radiomic and emerging genomic data for prognostic stratification of non-small cell lung carcinoma patients
Abstract
Background
A single institution retrospective analysis of 124 non-small cell lung carcinoma (NSCLC) patients was performed to identify whether disease-free survival (DFS) achieves incremental values when radiomic and genomic data are combined with clinical information.
Methods
Using the least absolute shrinkage and selection operator (LASSO) Cox regression method, radiomic and genetic features were reduced in number for selection of the most useful prognostic feature. We created four models using only baseline clinical data, clinical data with selected genetic features, clinical data with selected radiomic features, and clinical data with selected genetic and radiomic features together. Multivariate Cox proportional hazards analysis was performed to determine predictors of DFS. Receiver operating characteristic (ROC) calculation was made to compare the discriminative performance for DFS prediction by four constructed models at the five-year time point.
Results
On precontrast scan, improved discrimination performance was obtained in a merging of selected radiomics and genetics (AUC = 0.8638), compared with clinical data only (AUC = 0.7990), selected genetic features (AUC = 0.8497), and selected radiomic features (AUC = 0.8355). On post-contrast scan, discrimination performance was improved (AUC = 0.8672) compared with the clinical variables (AUC = 0.7913), and selected genetic features (AUC = 0.8376) and selected radiomic features (AUC = 0.8399) were considered.
Conclusions
The combination of selected radiomic and genomic features improved stratification of NSCLC patients upon survival. Thus, integrating clinicopathologic model with radiomic and genomic features may lead to improved prognostic accuracy compared to conventional clinicopathological data alone.
Key points
Significant findings of the study
Receiver operating characteristic (ROC) calculation was made to compare the discriminative performance for disease-free survival (DFS). The discriminative performance for DFS was better when combining radiomic and genetic features compared to clinical data only, selected genetic features, and selected radiomic features.
What this study adds
The combination of selected radiomic and genomic features improved stratification of NSCLC patients upon survival. Thus, integrating a clinicopathological model with radiomic and genomic features may lead to improved prognostic accuracy compared to conventional clinicopathological data alone.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide, with non-small cell lung carcinoma (NSCLC) accounting for 85% of cases.1 However, the widespread survival time that exists even after complete resection of NSCLC at the same stage indicates the vital importance of personalized medicine.2, 3 Improvements in survival estimation have largely been made as a result of advances in biological and genomic technologies that enable the integration of survival-related biological or genetic signatures.4, 5 However, the difficulty of obtaining comprehensive information on heterogeneous tumors is a limitation of such invasive methods.6, 7 Notwithstanding, enabled by the possibilities of large-scale high-throughput next-generation sequencing (NGS) technologies along with the computational resources and tools to store, process and analyze the data, more powerful methods for the characterization of individual patients and tumor types became available.8
While substantial progress like NGS has been made in genetic information, new areas of attention have been created in the medical imaging field. Radiomics is a field of medical study that aims to extract a large amount of quantitative features from medical images using data-characterization algorithms.9-12 Radiomics enables noninvasive profiling of tumor heterogeneity by extracting high-throughput quantitative descriptors from routinely gained computed tomography (CT) studies.9, 10, 13 Recent advances in radiomics have provided insights into personalized medicine in oncologic practice in the areas of tumor detection, subtype classification, and response assessment to treatment.13-15
Despite recent developments in Genomics, there is still a need for surgical procurement of tissue. Radiographic imaging is routine in clinical practice, but is currently based on histopathology. If specific imaging traits and gene expression patterns that can predict underlying cellular pathophysiology can be correlated with each other through radiomics, radiologic data can be used as a molecular surrogate marker to monitor diagnosis, prognosis, and possibly gene-expression-associated treatment response of various human cancers.16
The aim of this study was to analyze disease-free survival (DFS) in patients with NSCLC through radiomic signature and to compare the results of radiomics with those of traditional staging systems or genetic analysis to determine if an incremental value could be obtained when they are combined.
Methods
Patient population
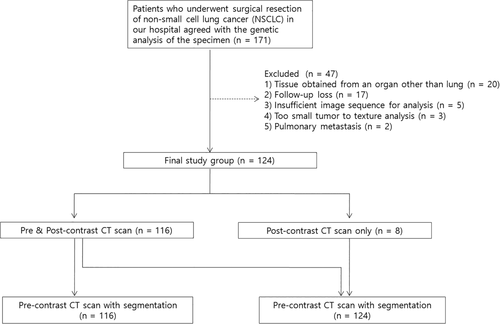
The Institutional Review Board approved this retrospective study which was exempt from the need to acquire informed consent. From May 2002 to January 2015, 171 patients with NSCLC had a surgical resection and agreed to genetic analysis of the specimen in our institution. After the review of medical records including imaging and pathologic studies, a total of 124 patients were included in this study (Fig 1). The exclusion of 47 patients was because genetically analyzed specimens had not been obtained from the lung (n = 20), patient was lost to follow-up (n = 17), insufficient image sequence for analysis (n = 5), tumor too small to texture analysis (n = 3) and metastatic pulmonary nodules (n = 2). In our study, radiomics were all obtained by three-dimensional (3D) texture analysis, so it required at least three axial scan slices. Therefore, three cases whose axial scans were only one and two slices were included in the exclusion criteria.
Research data collected through electronic medical records and clinical characteristics at the time of the diagnostic work-up were evaluated. Age, sex, smoking status, and American Joint Committee on Cancer (AJCC) stage were recorded. Pathology include histologic type and grade were recorded.
Follow-up
The end point of this study was DFS, which was defined as the time from the surgical resection date until either the date of relapse (event), which refers to tumor recurrence within. or immediately adjacent to, the treated field, mediastinal relapse, distant relapse, or death, or until the date that the patient was last known to be free of relapse (censored). The minimum follow-up period to ascertain DFS was 18 months after surgical resection; while the maximum follow-up time was 175 months (median, 36 months). Following our institution's follow-up protocol, the patient was followed-up postoperatively by chest CT every 6–12 months for the first two years and annually thereafter.
Imaging and texture analysis
Chest CT scan was used to assess the imaging characteristics of each lesion. Dedicated chest CT images were obtained with several multidetector CTs including eight-, 16-, or 64-detector row CT scanners. Among 124 patients, 116 CT examinations consisted of pre- and post-contrast scan, while eight CT examinations consisted of post-contrast scan only. CT images were obtained with the following parameters: detector collimation was 1.25 or 0.625 mm, the tube peak potential energies ranged from 80 to 140 kVp, tube current ranged from 150 to 200 mA, and reconstruction interval ranged from 1 to 2.5 mm. All patients underwent chest CT at full inspiration through breath hold to minimize the effect of the tumor motion due to breathing. Chest CT scanning was obtained 90 seconds after the administration of contrast material. A total of 1.5 mL/kg (bodyweight) iomeron 300 (iomeprol, 300 mg iodine/mL; Bracco; Milan, Italy) was injected at an infusion rate of 3 mL/second using a power injector (MCT Plus; Medrad; Pittsburgh, PA, USA). All CT images were displayed at standard mediastinal (window width, 400 HU; window level, 20 HU) and lung (window width, 1500 HU; window level, −700 HU) window settings. In-plane resolution varied from 0.49 to 0.88 mm with a mean and standard deviation (SD) of 0.7 and 0.07, respectively. The mean slice thickness of images was 2.33 (range: 1–5 mm) and the SD was 0.98. Chest CT data were interfaced directly with a picture archiving and communication system (PACS) (Path-Speed or Centricity 2.0; GE Healthcare, Mt. Prospect, IL, USA), which displayed all image data on two monitors (1536 × 2048 matrix, eight-bit viewable grayscale, 60-foot-lambert luminescence).
For quantitative CT analyses, our in-house software was used for lesion segmentation. Tumors were segmented by drawing a region of interest (ROI) with a semiautomatic approach using a MRIcro (version 1.40, Chris Rorden, University of Nottingham, UK) that traced the edge of the tumor to include the largest area along all axial slice CT images until the entire tumor was covered, containing the lesion displayed in the lung window setting.
Segmentations were performed on precontrast CT scans (n = 116) and post-contrast CT scans (n = 124) each. Two radiologists carried out additional manual corrections to eliminate bronchovascular bundles and ground glass opacity boundary. After the nodule had been segmented, radiomic features were automatically calculated and extracted.
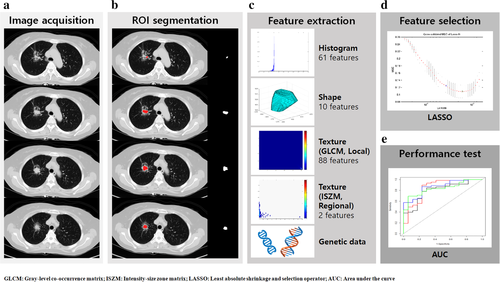
We calculated a total of 161 features of raw imaging over the given ROI using a combination of open-source (Pyradiomics) and in-house MATLAB code (MATLAB 2017b, Mathworks Inc., MA, USA). Our MATLAB code was used for features that were not calculated in pyradiomics.13 The features could be classified into four categories of histogram-based, shape-based, gray level co-occurrence matrix (GLCM)-based, and intensity size zone matrix (ISZM)-based. All of the features were verified for stability by image biomarker standardization initiative (IBSI).13, 17 A detailed definition of the features adopted is given in Table S1. The workflow of radiomic feature extraction is illustrated in Figure 2.
 ) Clinical, (
) Clinical, ( ) Clinical + SGI, (
) Clinical + SGI, ( ) Clinical + radiomics, (
) Clinical + radiomics, ( ) Clinical + radiomics + SGI.
) Clinical + radiomics + SGI.Genetic analysis
We obtained genomic variant information by using CancerSCAN, a targeted sequencing platform designed by Samsung Medical Center. This customized platform provides the researchers and clinicians the flexibility to include selected genes in the literature required. Single nucleotide variation (SNV), small indels, copy number variation, and gene fusion were detected using both existing18 and custom algorithms.19 Samples were typically sequenced without paired normal tissue. Genomic deoxyribonucleic acid (DNA) was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue or fresh tissue using a QIAamp DNA mini kit (Qiagen, Valencia, CA, USA) and Promega Maxwell 16 CSC DNA FFPE kit or QIAamp DNA after the pathologist's diagnosis and tumor content were examined. DNA concentration and purity were determined using a Nanodrop 8000 UV-Vis spectrometer (Thermo Scientific, Waltham, MA, USA) and a Qubit 2.0 fluorescence meter (Life Technologies, Grand Island, NY, USA). The degree of DNA degradation was measured using a 200 TapeStation Instrument (Agilent Technologies, Santa Clara, Calif., USA) and real-time PCR (Agilent Technologies, Santa Clara, CA, USA). Genomic DNA was shared using Covaris S220 (Covaris, Woburn, Massachusetts, USA). We used SureSelect-XT Reagent Kit, HSQ (Agilent Technologies, Santa Clara, CA, USA) to capture the target and create a bidirectional sequencing library using barcodes. After checking the quality of the library, sequencing was performed on a HiSeq 2500 with a 100 bp reading (Illumina, San Diego, California, USA). When sequencing is complete, the analytics workflow has started automatically. Pair-ended reads were aligned to the human reference genome (hg19) using BWA-MEM (v.0.7.5). SAMTOOLS (v0.1.18), GATK (v3.1–1), and Picard (v1.93) were used for reading alignment, local realignment and duplicate read removal, respectively. The baseline quality score was recalibrated using the GATK BaseRecalibrator based on known single nucleotide polymorphisms (SNPs) and indels of dbSNP138. SNV was identified using a combination of the two algorithms MuTect20 (v1.1.4) and LoFreq21 (v0.6.1). We applied a custom-designed filtering step based on a regression model trained on the low variant allele frequency (VAF) variance of the normal sample to improve its accuracy.22 Pindel was used to search insertions and deletions.23 To calculate copy number variations, the read depth per exon was divided by the estimated normal reads per exon using in-house references. Gene fusion of the target site was identified using an in-house algorithm. A detailed analysis workflow has been described in a previous report.22 An open SNP database such as the Exome Aggregation Consortium (EXID),,19 Korean Reference Genome (KRG), and Korean Variant Archive (KOVA)24 was used to differentiate somatic mutations from germline mutations. VAF of at least 0.1% in any of the SNP databases were considered as germ-line variants. For the remaining variants, a manually constructed internal database containing variants frequently found in the entire cohort was used for additional variant filtering. The variant was annotated by ANNOVAR.25
Mutation data was condensed into multiple binary values for each gene to represent functional effects. (i) Loss of function (LoF) mutation: Damaging mutations including frame-shift insertions and deletions and stop gain mutations; (ii) S1 mutation: Mutations that have been implicated in cancer development or therapy; (iii) missense (MS) mutation: Mutations in the coding region causing amino acid change other than S1 mutations; (iv) amplification (Amp): Gene amplifications with estimated copies ≥4; (v) deletion (Del): One or two copy gene deletions; and (vi) gene fusion (Fusion): Gene fusions inferred from the sequenced DNA reads that were specially designed to detect gene fusions in the CancerSCAN platform. Only driver gene fusions were included.
Statistical analysis
Continuous variables were presented as means and standard deviations and categorical variables as numbers and percentages.
We created four models using only clinical data, clinical data with selected genetic features, clinical data with selected radiomic features, and clinical data with selected genetic and radiomic features together. We performed least absolute shrinkage and selection operator (LASSO) to remove redundancy within the radiomic and genetic information by selecting the most prognostic characteristics in radiomic and genetic analysis.18, 26 LASSO is a method of selecting a few suitable features using L1-norm regularization and the method is frequently used for the feature selection in radiomics.27, 28 We performed multivariate Cox proportional hazards analysis to determine predictors of DFS. The variables investigated included demographics, pathological characteristics, and selected radiomic and genetic features.
Next, leave-one-out cross validation tests were performed to test the validity of our prediction model. We divided the data set into equal subgroups where the number of folds was equal to the number of instances in the data set. Subsequently, to compare the discrimination performance for DFS prediction by four constructed models at the five-year time point, the receiver operating characteristic (ROC) analysis with area under the curve (AUC) calculation was performed. Statistical analyzes were performed using SAS v9.4 (SAS Institute Inc., Cary, NC) and R 3.4.0 (Vienna, Austria; http://www.R-project.org/). P-values less than 0.05 indicated statistical significance.
Using 39 randomly selected patients, the reliability and reproducibility of tumor segmentation and feature extraction was attained by comparing the intraclass correlation coefficient (ICC)29 values between both radiologists.
Results
Patient group characteristics
Table 1 shows baseline clinicopathologic data such as age, sex, smoking status, AJCC stage, histologic type and histologic grade. A model using only clinical factors was designed with these variables. A total of 62 women (50%) and 62 (50%) men (median age, 56.6 years) were enrolled. We analyzed a total of 124 measurable lesions from 124 patients. There were 77 (62.1%) patients who were never-smokers, and 47 (37.9%) were ever-smokers. Among 124 lesions, 114 (91.9%) were adenocarcinomas, five were squamous cell carcinomas, two were pleomorphic carcinoma, two were large cell neuroendocrine carcinoma, and one was adenosquamous cell carcinoma. As of the last follow-up, 59 patients (34.4%) had experienced a confirmed disease relapse. The mean DFS was 27.66 months, and the median DFS was 23 months.
| Characteristics | No. of patients (n = 124) |
|---|---|
| Age at diagnosis (year) | 56.6 ± 11.9 |
| Sex | |
| Female | 62 (50) |
| Male | 62 (50) |
| Smoking history | |
| Never | 77 (62.1) |
| Ever | 47 (37.9) |
| Pathology | |
| Adenocarcinoma | 114 (91.9) |
| Squamous cell carcinoma | 5 (4.0) |
| Pleomorphic carcinoma | 2 (1.6) |
| Large cell neuroendocrine carcinoma | 2 (1.6) |
| Adenosquamous cell carcinoma | 1 (0.7) |
| Histologic grade | |
| Well-differentiated | 3 (2.4) |
| Moderately-differentiated | 89 (71.8) |
| Poorly-differentiated | 32 (25.8) |
| AJCC stage at diagnosis | |
| I | 53 (42.7) |
| II | 25 (20.2) |
| III | 35 (28.2) |
| IV | 11 (8.9) |
| Follow-up time (months) | |
| Median (quartile) | 36 ± (28.6–58.9) |
| Maximum | 175 |
Feature selection and diagnostic performance of constructed models
Using the LASSO method, radiomic and genetic features were reduced in number. On precontrast CT scan, two radiomic features (IMC1_val_out and IMC1_val_delta_sub2) and six genetic features (ALK_Fusion, ATRX_Amp, MET_MS, MLH1_MS, MTOR_MS, and STK11_MS) were selected. On post-contrast CT scan, 12 radiomic features (Kurtosis_val_in, mean_delta, Skewness_delta, Sphericity_val, Contrast_glcm_val_delta_sub2, Entropy_glcm_val_in, Energy_glcm_val_delta_sub2, IMC1_val_delta, Max_probability_val_in_sub2, Max_probability_val_out_sub2, Variance_glcm_val_delta, and Variance_glcm_val_delta_sub2) and seven genetic features (ALK_Fusion, ATRX_Amp, MET_MS, MLH1_MS, MTOR_MS, NF1_LoF, and STK11_MS) were selected. Radiomic and genetic features obtained from pre- and post-contrast scans were arranged and are shown in Tables S1 and S2.
Tables 2 and 3 summarize the survival prognosis performance of the three models. Using radiometric data in a precontrast CT scan, when selected radiomic and genetic features were integrated with clinical variables, the discrimination performance improved (AUC = 0.8638) compared with consideration of only clinical variables (AUC = 0.7990), selected genomic features incorporated with clinicopathological data (AUC = 0.8497), and selected radiomic features incorporated with clinicopathological data (AUC = 0.8355). When selected radiographic and genetic features were integrated with clinical variables using radiomic data on post-contrast CT scan, the discrimination performance improved (AUC = 0.8672) compared with consideration of only clinical variables (AUC = 0.7913), selected genomic features incorporated with clinicopathological data (AUC = 0.8376), and selected radiomic features incorporated with clinicopathological data (AUC = 0.8599).
| Model | AUC | P-value |
|---|---|---|
| Clinical data only | 0.7990 | Reference |
| Clinical + genetic data | 0.8497 | 0.1923 |
| Clinical + radiomic data | 0.8355 | 0.2793 |
| Clinical + genetic + radiomic data | 0.8638 | 0.0523 |
| Model | AUC | P-value |
|---|---|---|
| Clinical data only | 0.7913 | Reference |
| Clinical + genetic data | 0.8376 | 0.1929 |
| Clinical + radiomic data | 0.8599 | 0.1994 |
| Clinical + genetic + radiomic data | 0.8672 | 0.2092 |
Reliability and reproducibility of tumor segmentation and feature extraction
The ICC values ranged from 0.484 to 1.000 with a mean value of 0.929, representing a higher reliability level of agreement.
Discussion
Recently, several researchers have succeeded in extracting radiomic features in the oncology field by computing enormous amounts of quantitative variables from conventional medical images.30-32 Although previous studies have reported the prognostic value of some radiomic or genomic features, the question remains whether these new radiomic and genomic features have any incremental value above the current clinicopathologic model. Identifying the predictive power and clinical relevance of the numerous radiomic and genomic features for lung cancer survival remains an important issue.33, 34
In this study, we compared four models to evaluate selected radiomic and genomic feature increments along with clinicopathologic data using: (i) only clinicopathological data; (ii) selected genomic features incorporated with clinicopathological data; (iii) selected radiomic features incorporated with clinicopathological data; and (iv) selected radiomic and genomic features incorporated with clinicopathological data. The merging of selected radiomic and genomic features improved stratification in lung cancer patients upon survival. Thus, our results show that integration of the current clinicopathologic model with radiomic and genomic features may lead to improved prognostic accuracy compared to conventional clinicopathological data alone.
We employed a cancer panel sequencing platform to explore relationships between cancer genomic features and radiomic characteristics. There have been many debates on using whole-genome sequencing (WGS) versus whole-exome sequencing (WES) versus gene panels.35, 36 For clinical sequencing, we previously reported that many hotspot variants have a low VAF variant,22 which make it difficult to detect variants from a clinical sample at a standard coverage of WGS or WES platform (100–200x). The CancerSCAN panel sequencing platform aims at 1000x coverage to detect low VAF variants such as 2% VAF. This capability enables the detection of variants that only a small subclone contains, even in a low purity tumor sample. Thus our data represents near complete variant profiles in the panel genes, which other studies who employed WGS or WES would have missed.
Most lung adenocarcinoma genomic expression appears as a mixed-subtype such as polyclonal composition of two or more different pathologic subtypes.37, 38 Recent NGS and bioinformatic studies have studied different types of tumor heterogeneity including intratumor heterogeneity in which heterogeneity between tumor cells exists due to the presence of multiple subclones within the tumor in addition to interpatient or intertumor heterogeneity.39, 40 Based on this concept, recently published large databases characterizing the molecular features of human tumors are attempting to change the determination of each cancer type from the conventional histopathological classification to a new classification based on genetic identity.41-44 Such changes could better explain the symptoms of partial tumor response to treatment, the emergence of drug-resistant malignant cells, or the dissemination of metastatic cells with the emergence of genomically distinct minor subclones of malignant cells more practically.45-47 Several observations have demonstrated that the presence of minor subclones affects tumor progression.48-50
However, performing multiple tumor biopsies to precisely determine the clonal composition of a tumor is not a simple or practical solution in the context of clinical research. In addition, the results obtained with a single tumor-biopsy sample may also underestimate the overall genomic landscape. The error rate of cancer histopathology can be as high as 23%, and it is estimated that even the most clinically relevant solid tumors become very heterogeneous at phenotypic, physiologic, and genomic levels over time.11 Phylogenetic reconstruction has been reported to show a branching evolutionary tumor growth in which almost 70% of all somatic mutations were not detected across all tumor regions.51 In comparison, the medical image has the advantage of a quantified comprehensive macroscopic picture in estimating the entire tumor plus surrounding tissue relatively safely and less inconveniently for patients.
Radiogenomics focuses on defining relationships between radiomic phenotypes and genomic information to interconnect macroscopic imaging and subcellular characteristics to as oncologic diagnosis changes from traditional tissue-based approaches to molecular stratification.52 Although it is still poorly understood, many studies have attempted to find radiographic tumor phenotypes using radiomics and to find correlations with specific genetic expressions, and some have been successful.53-55 Radiomics may function as complementary “virtual biopsy” information that can be monitored more frequently than the invasive method during an entire course of cancer treatment, but it will still be difficult to completely replace conventional biopsy, which allows more detailed genomic analysis.
Of the 14 radiomics selected in our study, 10 were GLCM-based features, one was shape, and three were histogram-based features, and we conclude that the results reflect intratumoral heterogeneity well. GLCM texture features contains information about the positions of pixels having similar gray level values. They describe the high-order statistical spatial distributions of the voxel intensities characterizing heterogeneity with spatial information within a tumor or ROI. Therefore, GLCM features extracted from the raw image can act as an effective texture descriptor to reflect lesion heterogeneity. Representative features extracted from GLCM included correlation, cluster, contrast, energy, and entropy.
In addition, in our study, the features that were finally selected through LASSO were generally partial ROI associated features (in, out, and subsampled) than features related to whole ROI. In a study conducted on breast cancer, Wu et al.56 reported that the subregional radiomic analysis method had an advantage in quantifying the tumor subregion which was more correlated with the tumor growth or aggressiveness. Similarly, in our research, most of the radiomic features finally selected were partial ROI related features correlated with subregion microenvironment, and we can assume that these features were accompanied with risk group stratification.
We focused on CT-derived radiomic features in this study. CT is the most popular and routinely performed modality in the lung cancer field, and CT-derived radiomic features are practical for clinical application. We included both precontrast-enhanced CT scan and enhanced CT scan in this study. Theoretically, precontrast-enhanced CT-derived features are indicative of cellularity of the tumor and contrast-enhanced CT-derived features are indicative of vascularity of the tumor, reflecting both characteristics may represent tumor heterogeneity and complexity more accurately. As we utilized both precontrast scan CT-derived features and post-contrast CT scan-derived features, it broadens the applicability of our study, given the different imaging protocols of each hospital.
Despite the advantages of the radiomic approach to lung cancer prognostication, our study had some limitations. First, there was a small number of subjects included in our study, and they were collected from a single institute. The second limitation is that our study was retrospectively performed and based on previously acquired gene-expression data. Therefore, our findings will need to be further characterized and validated in future studies by patient groups receiving the same treatment to generate gene-expression data prospectively.
Our results indicate that integrating the current clinicopathologic model with selected radiomic and genomic features may lead to improved prognostic accuracy compared to conventional clinicopathologic data alone.
Acknowledgments
The authors would like to offer our special thanks to Insuk Sohn, PhD (Statistics and Data Center, Research Institute for Future Medicine, Samsung Medical Center, Seoul, Korea) for his assistance with the statistics used in this report. This research was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), which was funded by the Ministry of Health & Welfare (HI17C0086) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP; Ministry of Science, ICT & Future Planning) (No. NRF-2016R1A2B4013046 and NRF-2017M2A2A7A02018568). The funders had no role in the design of the study, data collection, data analysis, interpretation, or the writing of this report.
Disclosure
The authors declare that they have no conflicts of interests.