Scaling-up RADseq methods for large datasets of non-invasive samples: Lessons for library construction and data preprocessing
Abstract
Genetic non-invasive sampling (gNIS) is a critical tool for population genetics studies, supporting conservation efforts while imposing minimal impacts on wildlife. However, gNIS often presents variable levels of DNA degradation and non-endogenous contamination, which can incur considerable processing costs. Furthermore, the use of restriction-site-associated DNA sequencing methods (RADseq) for assessing thousands of genetic markers introduces the challenge of obtaining large sets of shared loci with similar coverage across multiple individuals. Here, we present an approach to handling large-scale gNIS-based datasets using data from the spotted hyena population inhabiting the Ngorongoro Crater in Tanzania. We generated 3RADseq data for more than a thousand individuals, mostly from faecal mucus samples collected non-invasively and varying in DNA degradation and contamination level. Using small-scale sequencing, we screened samples for endogenous DNA content, removed highly contaminated samples, confirmed overlap fragment length between libraries, and balanced individual representation in a sequencing pool. We evaluated the impact of (1) DNA degradation and contamination of non-invasive samples, (2) PCR duplicates and (3) different SNP filters on genotype accuracy based on Mendelian error estimated for parent–offspring trio datasets. Our results showed that when balanced for sequencing depth, contaminated samples presented similar genotype error rates to those of non-contaminated samples. We also showed that PCR duplicates and different SNP filters impact genotype accuracy. In summary, we showed the potential of using gNIS for large-scale genetic monitoring based on SNPs and demonstrated how to improve control over library preparation by using a weighted re-pooling strategy that considers the endogenous DNA content.
1 INTRODUCTION
Suitable wildlife conservation management requires baseline population data, which include accurate demographic and life-history information, reliable population genetic parameter estimates and an understanding of the factors that shape population dynamics, adaptation and evolution (Boakes et al., 2016; Willi & Hoffmann, 2009). The unprecedented declines in global biodiversity driven by human-mediated environmental disturbance (Maclean & Wilson, 2011) emphasize the need for genetic monitoring to mitigate impacts on wildlife (Carroll et al., 2018).
Genetic non-invasive sampling (gNIS) is a cost-effective sampling approach that allows for genetic studies of large numbers of free-ranging animals without the need to capture or observe them (Waits & Paetkau, 2005). Thus, gNIS provides an opportunity to gather population data and develop monitoring programs, especially for rare and threatened taxa for which compliance with ethical standards and regulations precludes other types of sampling (Zemanova, 2020). DNA sources for gNIS include eggshell membranes (Hu & Wu, 2008), egg chorion debris (Storer et al., 2019), feathers (Miño & Del Lama, 2009), mucus swabs (Lieber et al., 2013), hair (De Barba et al., 2010), faeces (Schultz et al., 2018) and others. Despite the wealth of sample types and logistical and economic benefits to gNIS, there are caveats of non-invasive samples that must be taken into consideration in population genetic analysis. DNA extracted from non-invasive samples is often degraded, which has downstream effects on genotyping accuracy: missing data, allele dropout, fewer loci and variant calls and erroneous estimates of allele frequencies (Graham et al., 2015; Schultz et al., 2022; Taberlet et al., 1999; Valière et al., 2007). Furthermore, certain gNIS DNA sources (e.g. mucus, faeces) are often contaminated with non-endogenous DNA, which increases the costs necessary to obtain appropriate sequencing coverage per individual (Hernandez-Rodriguez et al., 2018; Perry et al., 2010).
In addition to the number of samples, population genetic inference power relies on the number of genetic markers analysed (Davey et al., 2011). Advances in high-throughput sequencing technologies have allowed for data acquisition of increasingly large numbers of samples and loci at improved cost–benefit ratios, especially when referring to methods covering a reduced representation of the genome. Methods based on genome digestion using restriction enzymes (RADseq) have commonly been chosen due to their lack of need for a reference genome, and their flexibility in controlling the number of loci sequenced (Baird et al., 2008). However, artefacts generated during library construction can introduce bias into downstream analyses if not carefully treated (Mastretta-Yanes et al., 2015). For example, PCR duplicates, which stem from a random allele at a given locus being amplified more than the other allele, result in a spurious inflation of homozygosity and a false increase in variant call confidence (Andrews et al., 2016; Flanagan & Jones, 2018). Discrepant fragment-size range among libraries and variable coverage across loci and individuals in a given dataset strongly influence the number of SNPs and genotyping confidence (DaCosta & Sorenson, 2014; Driller et al., 2021). Furthermore, different bioinformatic processing approaches for loci building and SNP calling, as well as SNP filtering strategies, can strongly impact results, potentially creating false patterns and leading to incorrect biological interpretations (O'Leary et al., 2018). Moreover, any genomic method requiring individual pooling presents the challenge of obtaining an even read-depth distribution across individuals, which is not straightforward (Maroso et al., 2018), especially in large datasets. Read-depth inconsistency can impact the missing rate among individuals and genotyping accuracy, ultimately impacting the estimation of population genomic parameters (Davey et al., 2013). All of these challenges in library construction and data analysis highlight the need to develop new strategies to overcome these issues.
Here, we present a novel strategy for the improved handling of large-scale gNIS-based datasets using non-invasive samples collected from spotted hyenas (Crocuta crocuta) inhabiting the Ngorongoro Crater in Tanzania. We generated 3RADseq data (Bayona-Vásquez et al., 2019) for 1142 individuals, mostly from faecal mucus samples collected non-invasively and varying in DNA degradation and non-endogenous contamination level. We demonstrate how to control library preparation and efficiently reduce costs by using a weighted re-pooling strategy that considers the endogenous DNA content. Using small-scale sequencing to obtain a low number of reads per individual, we screened samples for endogenous DNA content, filtered out highly contaminated samples, confirmed overlap fragment length between libraries and balanced individual representation in a sequencing pool. Furthermore, we take advantage of a special characteristic of our dataset, which is composed of several related individuals, to identify parent–offspring trios and measure genotype error rate based on patterns of Mendelian inheritance. Thus, we evaluate the impact of (1) DNA degradation and non-endogenous contamination of non-invasive samples, (2) PCR duplicates and (3) different SNP filters on genotype accuracy based on trio datasets.
2 MATERIALS AND METHODS
2.1 Sampling
Samples were collected from spotted hyenas inhabiting the Ngorongoro Crater in Tanzania between April 1996 and December 2020, as part of the long-term monitoring of behaviour and life history of all individually known spotted hyenas in this population (https://hyena-project.com/, Davidian et al., 2016). Non-invasive samples were collected from faecal mucus (N = 573), blood on faeces (N = 2) or grass (N = 1), hair (N = 17) or tissues from dead animals: muscle (N = 9), skin (N = 5), liver (N = 1), ear (N = 1), and heart (N = 1). Mucus covering the hyena faeces, which contains intestinal epithelial cells, was collected immediately after defaecation to ensure reliable individual assignment. Hair samples were opportunistically collected when cubs approached the monitoring vehicle. Skin biopsy samples (N = 532) were also analysed. They were collected using a Telinject GUT 50 dart gun fitted with a biopsy needle designed for spotted hyenas. Instruments used to collect genetic samples were sterilized and then held in the flame of a cigarette lighter before use. Genetic material was either stored and transported at or below −70°C, or stored in dimethyl sulfoxide (DMSO) salt solution or ethanol at 5°C. DNA extractions were performed using QIAamp DNA kit, Machery and Nagel Tissue and Soil kit, Qiagen Dneasy tissue kit or Chelex extraction method following the manufacturer's instructions.
The quality of the DNA extracts was evaluated by running samples on an agarose gel electrophoresis. The level of DNA degradation was assessed by visualizing the fragment length distribution (or smear). We observed a high variation in DNA quality, especially among mucus samples (Figure S1).
We applied the method described in the next sections for 1142 samples, of which 761 were sequenced at high coverage (i.e. at least 20×). For the scope of this work, we present a detailed analysis of a subset of 284 high-coverage individuals, which form 158 parent–offspring trios.
2.2 Library construction
We used the method 3RADseq following the protocols described by Bayona-Vásquez et al., 2019 with a modification in one primer to include unique template molecule tags (Hoffberg et al., 2016). We chose the 3RADseq method because of two main features: (1) the use of the third enzyme increases the adapter ligation efficiency by cutting adapter dimers and making them available for further sample DNA ligation, which allows for the use of low quantity and quality DNA samples (Bayona-Vásquez et al., 2019); and (2) the inclusion of the iTru5-8N primer, which incorporates 8 degenerate bases in the P5 adapter and permits the identification and removal of PCR duplicates during preprocessing of the sequence data.
We simulated the digestion of the Crocuta crocuta genome (GenBank accession number GCA_008692635.1) with different restriction enzyme combinations in order to choose a set of enzymes that would provide us with approximately 25,000 loci. We used a dedicated Python script (RAD_digestion_v2.0.py) that performs in silico digestion based on selected restriction enzymes (in ddRAD or 3RAD mode) and generates double-digested DNA sequences and their length distribution. We selected the combination of EcoRI, XbaI and NheI, which allowed us to sequence around 23,500 loci in a range between 380 and 460 bp. We started the library preparation with an initial DNA amount per sample of 20, 50, 70 and 100 ng, depending on the available material, in a volume of 10 μL. The three restriction enzymes (10 units of each enzyme), NEB 1× CutSmart Buffer and a unique combination of dual-internal barcodes (5 μM) were added to each sample separately for the DNA digestion reaction. After 2 h at 37°C, we added DNA ligase and ATP to the reaction and exposed it to multiple temperature cycles to promote ligation (22°C) followed by digestion (37°C), ending with 80°C for 20 min. After the digestion/ligation reaction, a maximum of 132 samples with the same initial amount of DNA were pooled and cleaned with 0.8X CleanPCR magnetic beads (GC biotech). We decided to pool a maximum of 132 samples, as the higher the number of samples pooled, the more difficult it is to achieve balance between individuals. The final volume of the digestion/ligation product consisted of 20 μL per individual, of which we used 2 μL for the first pooling. We distributed the samples to different pools according to the sample type (generalized as biopsy and faecal mucus pools, aiming to put together samples with similar quality) and DNA input available (20, 50, 70 or 100 ng), resulting in 14 libraries total (library composition can be found in Table S1 and a methodological scheme is displayed in Figure S2). Importantly, all the pipetting steps for normalization of the DNA concentration, DNA digestion, adapter ligation and pooling were performed with the Beckman Coulter Biomek i7 automated Workstation. While not a requirement for implementing this method, a robot is useful to optimize these steps.
Following sample pooling and cleaning, we performed fragment size selection for each library with the Blue Pippin using a 1.5% cassette (Sage Science). After standardization, we set the size-selection range configured in the BluePippin software to 480–640 bp in order to obtain a fragment-size range of 390–450 bp. The difference in the configured and obtained size range refers to the adapter size (80 bp) and recurrent deviation error (tending to narrower and shorter size ranges) we often observe in our BluePippin equipment.
The 40 μL size-selected product was split into two aliquots of 20 μL, and each replicate then underwent a single cycle PCR to incorporate the iTru5-8N primer, followed by an indexing PCR using P5 outer and P7 primers. We used P7 primers with unique indexes to differentiate libraries and combine them for the sequencing run. The combination of different external P7 indexes and internal P5 and P7 barcodes allowed us to pool a high number of samples and libraries (Peterson et al., 2012). The number of indexing PCR cycles varied according to the number of samples pooled and the DNA concentration of each library. Pools with 60–240 ng DNA underwent 12 PCR cycles, pools with 240–750 ng DNA underwent 10 PCR cycles and pools with >750 ng DNA underwent 8 PCR cycles (Figure S2); this served to minimize the formation of PCR duplicates while achieving pools with at least 80 ng of DNA for the sequencing run. Note that the PCR reactions were done after the size selection. This is a strategy to overcome small fragment preferential amplification that can bias subsequent processing steps (DaCosta & Sorenson, 2014).
The final libraries were checked with an Agilent Tapestation using High Sensitivity DNA ScreenTape and sequenced using 300 bp paired-end reads on a MiSeq platform (Illumina), aiming to obtain 2000 reads per individual. This first small-scale sequencing aiming to obtain a low number of reads per individual is hereby referred to as a spike-in run. These data can be obtained cost-efficiently by running spike-ins in shared sequencing runs or using nano or microsequencing kits. The sequencing output is used to check the balance between the individuals, the length distribution of each library and the individual contamination level. This approach is described below in the section ‘Spike-in strategy’.
2.3 Preprocessing analysis
The 2 × 300 bp reads are submitted to a dedicated preprocessing pipeline automatized in a Snakemake workflow, as described in Figure S3A. The first step involves phiX control library cleaning, as phiX is included in the sequencing run to increase sequence diversity and improve base reading quality. The raw data are mapped to the Enterobacteria phage phiX174 reference genome (NC_001422.1) using Bowtie2 (Langmead & Salzberg, 2012) with default parameters, and the unmapped reads are saved for the following processing steps. Next, we demultiplexed reads per individual using the software Flexbar (Roehr et al., 2017). PCR duplicates were then filtered from each individual, based on recognition of identical reads with the same iTru-8 N index sequence, using a custom Python script (filterPCRdups_CM.py) that outputs only one copy of each unique DNA molecule. After this step, we concatenated the two replicates of each library and merged forward and reverse reads using PEAR v.0.9.11 (Zhang et al., 2014). At this point, the Snakemake pipeline generates a series of plots with the fragment length distribution of each individual. Then, we perform an in silico digestion, in order to recognize and filter out undigested and chimeric sequences. Next, we check restriction sites using a custom Python script (checkRestrictionSites.py), keeping only reads with correct sequences at both ends in the output. Finally, quality filtering is performed using the software Trimmomatic (Bolger et al., 2014), keeping only reads with a Phred score higher than 30.
2.4 Spike-in strategy
The filtered spike-in reads are produced for three main objectives. First, they allow us to check the balance between the individuals by estimating the proportion of reads belonging to each individual in the library. Based on this proportion, we calculate the new volumes of the digestion/ligation product (of which 18 μL remain) that will be re-pooled for second library preparation (Figure 1).
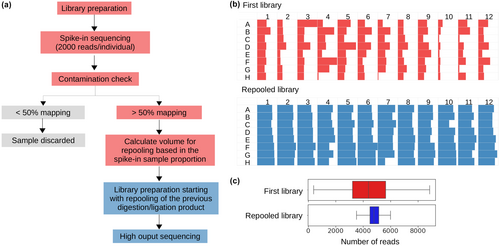
Second, we use the filtered spike-in reads to check the length distribution of each library. Long Illumina reads (300 bp) allow us to sequence loci in their entirety, as RADseq libraries typically consist of fragments between 200 and 600 bp. We merge the forward and reverse reads and analyse the length distribution to confirm the overlap between different libraries, ensuring that we have the same set of loci across all individuals.
Third, filtered spike-in reads allow us to check for contamination. We mapped reads to the C. crocuta genome (GenBank accession number GCA_008692635.1) using Bowtie2 (default parameters). Unmapped reads were submitted to Blastn analysis implemented in NCBI's BLAST+ package (v.2.8.1+) with a maximum e-value of 10−20 for taxon identification. We created a custom database by adding the C. crocuta and Hyaena hyaena genomes to the NT NCBI database. Blastn results were visualized in MEGAN v.6.19.2 (Huson et al., 2011). The percentage of reads mapping to the C. crocuta genome was thereafter considered to be the endogenous hyena DNA content. Individuals with less than 50% endogenous DNA content were discarded, due to the high effort (in terms of sequencing coverage and associated costs) needed to obtain good quality data from these samples.
Once we define the samples that will continue in the analysis, we calculate the volumes for re-pooling considering the number of mapped reads from each sample. This accounts for the proportion of non-endogenous reads per sample, as well as varying sample proportions, in order to obtain a balanced library (i.e. samples with fewer reads after filtering and mapping are included in greater volumes in the re-pool). After weighted re-pooling, library preparation steps are repeated as described before. Adjustments in size selection can be done at this point to reach the target range with the desired number of loci. The re-pooled library is submitted again for a spike-in run to confirm the final library balance and finally submitted for a high-output paired-end (150 bp) sequencing run on two lanes of the NovaSeq S4 platform (Illumina), aiming to obtain at least 30× coverage per individual. We combined 3RADseq libraries with 10% of whole genome sequencing (WGS) libraries to increase the sequence diversity in all sequencing cycles, especially in the first cycles where all reads share the same restriction site.
NovaSeq reads are preprocessed following the pipeline described before, except for the phiX filter (because no phiX was added to the high-output sequencing run) and the merging of reads (Figure S3B). As we now have shorter reads (150 bp), only short fragments (<270 bp) overlap (with minimum overlap length of 30 bp), thus merging allows us to filter out these short fragments and continue analysing the unassembled forward and reverse reads. Additionally, we used the software Cutadapt (Martin, 2011) to perform adapter trimming, aiming to filter out any remaining adapter sequences in the 3′-sequence ends.
Preprocessed reads were mapped to the C. crocuta genome using Bowtie2 with default parameters and the options “-no-mixed” and “-no-discordant” to ensure unique paired-end alignments. We excluded the mitogenome and sex scaffolds using Samtools (Danecek et al., 2021). Considering that the genome used as reference is at scaffold level, we used RADsex (Feron et al., 2021) to recognize scaffolds presenting sex-biased markers. We checked the distribution of fragment lengths per sample by extracting the fragment size from the resultant sam file using an awk command. This strategy allows us to check the variation between the set of loci selected for individuals pooled in different libraries, which may often have slightly different fragment length ranges (Driller et al., 2021).
2.5 SNP calling
The Stacks reference-based pipeline v2.61 (Catchen et al., 2011; Rochette et al., 2019) was used to call variant sites and perform population filters, allowing only loci genotyped in at least 60% of individuals within at least one population to be retained in the final dataset (N = 761). This selection was achieved using the population program with parameters p = 1 and r = .6. We subsampled the number of reads for individuals with greater than 2.5 million reads (equivalent to roughly 60× coverage per site), in order to obtain even coverage among individuals.
Further variant filtering was done using VCFtools (Danecek et al., 2011) using constrained (minimum depth: 20, maximum depth: 100, missing rate: 0.96, minimum allele frequency: 0.05, unlinked SNPs) and permissive (minimum depth: 5 or 10, maximum depth: 100 or 110, missing rate: 0.8, minimum allele frequency: 0.01) thresholds (Table S2). Each dataset was used for different downstream analyses, as described below.
2.6 Relatedness analysis
The constrained set of SNPs was used to assign parentage among individuals. Pairwise relatedness values were calculated with the software Related (Pew et al., 2015), and parent–offspring trio relationships were identified with the software Colony (Jones & Wang, 2010). We identified 158 trios in our dataset, comprising 284 individuals, which were used to estimate genotype accuracy in subsequent analyses. We confirmed trio identification by comparing our results with trios previously identified based on microsatellite data (Höner et al., 2010).
2.7 DNA degradation and non-endogenous contamination analysis
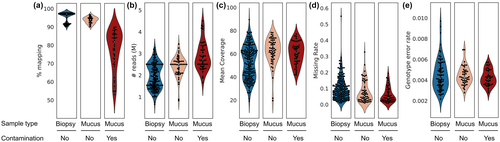
We compared the performance of contaminated and non-contaminated samples by comparing the percentage of mapping to the C. crocuta genome, number of sequenced reads, mean coverage per individual, mean missing rate per individual and genotype error rate (see the next section for more details). We considered samples with mapping percentages between 50% and 90% as contaminated samples. This maximum threshold was chosen based on the lower mapping limit of skin biopsy samples, which are considered as truly non-contaminated samples. Contaminated samples included 48 faecal mucus and one blood on faeces, so we considered this group as a composite of mucus samples for simplicity. Non-contaminated samples included skin biopsy (N = 175), faecal mucus (N = 56), liver (N = 1), and muscle (N = 3) samples. Since faecal mucus samples commonly presented high levels of DNA degradation (Figure S1), we also compared the performance of non-contaminated faecal mucus (>90% mapping) and biopsy samples. Liver, muscle and skin biopsy samples were grouped as biopsy samples, totalling 179 samples.
2.8 Genotype accuracy estimation based on parent–offspring trio datasets
We estimated genotype accuracy using parent–offspring trio datasets, assuming correctly specified familial relationships. We identified genotype errors as any variant calls inconsistent with the rules of Mendelian inheritance, assuming that de novo mutation rates (~10−8 per bp) are several orders of magnitude smaller than calling and genotyping error rates (often 10−2–10−5) (Kómár & Kural, 2018). Using a dedicated python script (GenotypeAccuracyEstimation.py), we classified each variable position in a VCF file as missing data, correct or incorrect genotype calling for each trio based on the offspring genotype assuming that the parents' genotypes were correct. Although genotype classification is defined based on the offspring genotype, the Mendelian error might have occurred in either the offspring or one or both parents. We calculated the genotype error rate as the relationship between the number of correct and incorrect sites, discarding missing data. The script also removes the incorrect and missing genotypes and outputs a filtered VCF file.
- Faecal mucus samples are more likely to be degraded, as observed when DNA samples were submitted to gel electrophoresis analysis (Figure S1). We hypothesize that these samples contain DNA damage and breaks that prevent some loci from being sequenced and lead to erroneous SNP calling, thus the genotype error rate is higher for faecal mucus samples in comparison with biopsy samples. We also tested the effect of high non-endogenous contamination levels by comparing the genotype error rate between contaminated (<90% mapping) and non-contaminated (>90% mapping) faecal mucus samples. We considered exclusively the offspring sample type, as trios sharing the same sample type were rare (e.g. only three trios had DNA extracted exclusively from faecal mucus). The genotype error rate comparison between contaminated faecal mucus samples (N = 43) and non-contaminated biopsy (N = 75) and faecal mucus (N = 40) samples was done using the dataset ‘High coverage SNP-filtered’ (Table S3).
- We hypothesize that the presence of PCR duplicates will lead to an increase in the genotype error rate, as it spuriously increases homozygosity. To test this hypothesis, we compared the PCR duplicates filtered and unfiltered datasets. The only difference between the datasets is the presence/absence of the PCR duplicates filter during preprocessing. We grouped samples according to the number of PCR cycles used during library preparation, as this directly impacts PCR duplicate levels. The subgroups included 100, 39 and 19 trios with 8, 10 and 12 PCR cycles, respectively. We also conducted this analysis including only samples with the same sampling type (biopsy) and DNA input concentration (100 ng), in order to eliminate confounding effects that can also impact the genotype accuracy and PCR duplicate rate. We also evaluated the effect of coverage on genotype accuracy by simulating datasets with low coverage. We subsampled only the offspring individuals, as the error classification was based on the offspring genotype assuming that the parents' genotypes were correct. We used Samtools (-s option) to subsample the bam files to ~13× coverage and ran the SNP calling with a Stacks reference-based pipeline. The summary statistics of the low and high coverage, and PCR duplicates filtered and unfiltered datasets are presented in Table S3.
- The effect of SNP filters on genotype accuracy was evaluated after each SNP filtering step separately and after combining all filtering steps. The rationale for the different filters (minimum depth, maximum depth, missing rate, minimum allele frequency) and the chosen thresholds (10, 110, 0.8, 0.05, respectively) are described in Table S2.
We predicted that DNA degradation, PCR duplicates and the lack of SNP filters would result in reduced genotype accuracy and reduced precision of genetic measures, especially at the individual level. Statistical analyses were performed using a linear mixed model to study the fixed effects of coverage, number of PCR cycles and PCR duplicates filter on the (log) genotype error rate while accounting for the dependence between observations stemming from the same samples using a random intercept. We considered all possible two-way interactions as well as the triple interaction among all three predictor variables. The p-values were computed using the R package lmerTest v3.1–3 (Kuznetsova et al., 2017) using the so-called “Satterthwaite” method to compute the denominator degrees of freedom after fitting the model using the lme4 R package v1.1–34 (Bates et al., 2015). Prediction and 95% confidence intervals were computed using the R package spaMM v4.3.20 (Rousset & Ferdy, 2014) using the setting “predVar” to compute the intervals. The model was fitted by maximum likelihood in both lme4 and spaMM and produced identical estimates. We used the parametric one-way analysis of variance (ANOVA) with post hoc t-test with Holm correction to compare the genotype error rate and missing rate for samples grouped by DNA source and contamination level (non-contaminated biopsy, non-contaminated mucus and contaminated mucus). The same statistical test was used to compare the percentage of PCR duplicates between groups of samples that underwent 8, 10 or 12 PCR cycles and datasets treated with different SNP filters. Pearson coefficient and its respective p-value were calculated for correlation analysis. These tests were performed using SciPy v1.10.1 (Virtanen et al., 2020) or scikit-postdocs (Terpilowski, 2019) Python packages and a two-tailed p-value less than .05 was considered statistically significant.
3 RESULTS
3.1 Obtaining homogeneous 3RADseq libraries across loci and individuals
Our strategy of weighted re-pooling of individuals based on the number of reads obtained in the spike-in sequencing successfully homogenized the final library. Figure 1 shows the number of reads per individual displayed in a 96-well plate before (blue) and after (red) re-pooling procedure (Figure 1b) and the decrease in the variance (Figure 1c).
Our approach also allowed us to confirm the overlap in the fragment length distribution among all libraries, by sequencing 2 × 300 bp reads, merging paired-end reads and comparing the length distribution of different libraries. Thus, we avoided sequencing libraries that did not share the same set of loci.
The spike-in data also proved to be a powerful resource for dealing with non-endogenous contaminated samples. Of the 1142 samples analysed, 238 had contamination levels higher than 50% and were discarded after the first spike-in data analysis. Obtaining good coverage for these individuals would have required at least double the number of reads compared to non-contaminated samples, which we judged to be prohibitively expensive with regard to sequencing costs. In-depth Blast analysis of a subset of 10 individuals using the spike-in data showed that the vast majority of contamination derived from Bacteria (83.7%), followed by Platyhelminthes (8.9%) (Figure S4). Blast hits for Hyaenidae and Bovidae families were also found and represented 2.84% and 4.52% of the hits, respectively. We also analysed the high-output data of another 10 random individuals using the unmapped reads (subsampled to 50,000 reads) and a similar contamination content was observed in the Blast results, showing 69.7%, 0.86%, 28.6% and 0.39% of Bacteria, Platyhelminthes, Hyaenidae and Bovidae hits, respectively. Among the remaining samples, 761 samples were sequenced with high coverage, and 143 samples presented read numbers after spike-in that were too low to allow for subsequent balancing (i.e. >18 μL of sample would have been required), and thus were not included in the final library pool.
The percentage of reads mapping to the C. crocuta genome for the spike-in data was overall highly correlated (r = .98) to the high-output data (Figure S5A) and showed that even a low number of reads (~1000) is informative enough to quantify the per-sample contamination level (Figure S5B). Out of the 14 libraries, two libraries presenting read numbers below 1000 showed high levels of variance in the mapping difference (calculated as the percentage of mapping for the high-output data minus the percentage of mapping for the spike-in data). Another three libraries presented a higher mapping percentage for the spike-in data than for the high-output data (>2%—Figure S5C). No association with mapping quality, DNA source, PCR duplicate rate or coverage was observed (data not shown).
The Snakemake pipeline streamlined the preprocessing filtering steps for the analysis of the spike-in data. For the final high-output dataset, the number of reads after each preprocessing step is shown in Figure S3C. The two filtering steps responsible for the greatest reduction in read number were PCR duplicates (29.4%) and restriction site filters (26.9%). In the latter filter, reads cut by the third restriction enzyme (NheI) represented 68% of the removed reads. These fragments represent remnants of NheI-EcoRI combinations that were intended to be digested by the third enzyme. The 3RAD protocol was designed in such a way that the restriction site for the third enzyme is recreated any time that NheI fragments are ligated to the adapters, with the expectation that the enzyme would cut these fragments (Bayona-Vásquez et al., 2019). However, the substantial amount of the NheI fragments points to the partial inefficiency of this process, which is a drawback of the 3RAD protocol that needs to be taken into consideration.
3.2 The effect of non-endogenous contamination and DNA degradation on the missing rate and genotype accuracy
Samples classified as contaminated (N = 49) according to the mapping percentage to the C. crocuta genome (<90%—Figure 2a) yielded a higher number of reads (Figure 2b) as a result of our weighted re-pooling approach, which takes into consideration the number of mapped reads. Thus, the coverage of contaminated and non-contaminated samples was similar (Figure 2c), despite the difference in the mapping percentage. Due to the observed DNA degradation of non-invasive samples, we compared the missing rate and genotype accuracy between contaminated and non-contaminated biopsy and faecal mucus samples. No significant difference was observed among the three groups for both parameters (one-way ANOVA: F2,155 = 2.69, p = .071 for the missing rate comparison and F2,155 = 0.68, p = .51 for the genotype accuracy comparison) (Figure 2d,e).
3.3 The impact of PCR duplicates, number of PCR cycles and coverage on genotype accuracy
The percentage of PCR duplicates varied according to the number of PCR cycles, showing an average of 13.9%, 14.5% and 31.3% for 8, 10 and 12 PCR cycles, respectively (Figure 3a). The results of the one-way ANOVA test indicated a statistically significant difference among the three groups (F2,1648 = 2386, p < .0001), revealing an increase of 4.5% from 8 to 10 PCR cycles (post-hoc t-test with Holm correction, p = .0023) and a substantial increase of 115.7% from 10 to 12 PCR cycles (p < .0001). Predictably, higher coverage per locus was observed for the dataset with no PCR-duplicates filter (27.67 ± 6.02) in relation to the PCR-duplicates filtered dataset (25.84 ± 5.05). PCR duplicates are also expected to result in larger variance in the locus coverage (Andrews et al., 2016; Flanagan & Jones, 2018). Our results supported this hypothesis, as the PCR-duplicates unfiltered dataset had higher variance in coverage (1022.6) than the PCR-duplicates filtered dataset (752.3).
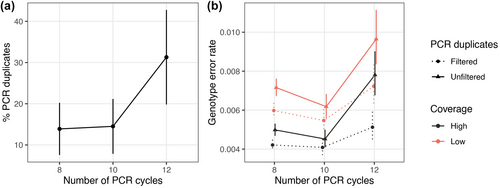
Genotype error rate calculated for the datasets after SNP filters (Table S3) was influenced by the number of PCR cycles, the presence or absence of PCR-duplicates filtering, the coverage and interactions between these variables (Linear mixed model: LRT χ2 = 527, df = 11, p < .0001; Figure 3b, Table S4). The overall effect (accounting for the main term and interactions) was clear for each experimental variable (PCR cycles: χ2 = 57.7, df = 8, p < .0001; PCR-duplicates filtering: χ2 = 187, df = 6, p < .0001; coverage: χ2 = 422, df = 6, p < .0001). The only interaction reaching significance was the two-way interaction between the number of PCR cycles and the PCR-duplicates filtering (F2,474 = 14.7, p < 0.0001), implying that the effect of PCR-duplicates filtering on the genotype error rate varied with the number of PCR cycles. At 12 PCR cycles, the effect of filtering was stronger than at 8 or 10 PCR cycles, irrespective of the coverage. Despite the presence of the interaction, the treatment that presented the lowest genotype error rate was the one with high coverage and filtered PCR duplicates, irrespective of the number of PCR cycles (Figure 3b). Similarly, the treatment with low coverage and unfiltered PCR duplicates showed the highest genotyping error rate. Intriguingly, samples that underwent 10 PCR cycles exhibited the least amount of genotyping error, whereas those subjected to 8 PCR cycles presented a slight increase in errors, and those with 12 PCR cycles exhibited a large increase in genotype error rates. The slightly higher genotype error rate observed for 8 PCR cycles compared to 10 PCR cycles may be attributed to the presence of outliers (Figure S6) and lower coverage in the 8 PCR cycles group (12.3× and 13.3× for 8 and 10 PCR cycles, respectively, for the low coverage PCR-duplicates unfiltered dataset), as partially confirmed when outliers were excluded (Figure S7). Within the datasets, an overall weak correlation between the genotype error rate and coverage (minimum 9× for the low-coverage datasets) was observed (Figure S6).
Considering that both DNA input mass and number of PCR cycles can potentially impact the PCR duplicate rate and genotype accuracy (Rochette et al., 2023), we conducted an analysis including only samples with the same sampling type (biopsy) and DNA input amount (100 ng). Despite the low sample size (14, 4 and 5 samples for 8, 10 and 12 PCR cycles, respectively), a significant increase in PCR duplicates (F2,266 = 5177, p < .0001) and genotype error rates was observed when 12 PCR cycles were performed compared to 8 and 10 PCR cycles for all datasets, irrespective to PCR-duplicates filtering or coverage (Linear mixed model: LRT χ2 = 94.61, df = 11, p < .0001, Figure S8).
3.4 The impact of minor allele frequency, minimum and maximum coverage and missing rate on genotype accuracy
Despite our efforts to obtain a similar fragment range across libraries, the upper limit differed across 3RADseq libraries (Figure 4a). The coverage per locus for the Stacks output showed a large variation across the fragment length distribution and the presence of outliers, including the two loci that appear as peaks at 326 and 417 bp fragment lengths (Figure 4b). They likely represent repetitive collapsed paralogous loci, which should be removed from the final dataset to avoid genetic estimations based on non-orthologous loci. Thus, we decided to perform further SNP filtering aimed at removing potential paralogues and dropout alleles, spurious variant calls due to low coverage, and variant calls present in low frequency and with a high missing rate.
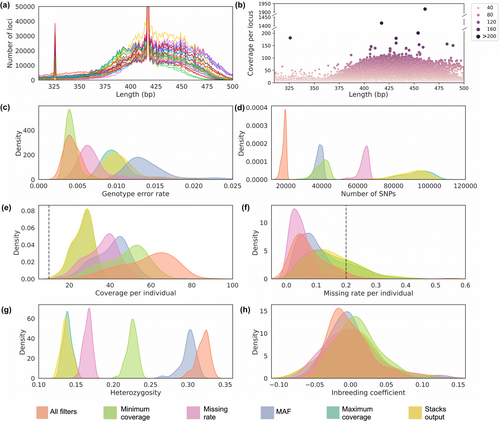
We compared the effect of each SNP filter independently (datasets named according to the unique filter) and all filters combined (‘All filters’ dataset) in relation to the dataset with only Stacks baseline filters (‘Stacks output’ dataset). In general, applying different SNP filters resulted in a decrease in the number of SNPs (Figure 4d) and missing rate per individual (Figure 4f), and in an increase in the coverage per individual (Figure 4e) and in the heterozygosity (Figure 4g). An exception was the dataset ‘Maximum coverage’, which had little impact on the estimations compared to the ‘Stacks output’ dataset, as it removed only 62 out of 107,256 SNPs. Low variation was observed in the inbreeding coefficient among the datasets (Figure 4h).
The genotype error rate varied between 1.4%, when only the MAF filter was applied, to 0.4%, when all filters were applied. Filtering SNPs with low coverage (‘Minimum coverage’ dataset), high missing rate (‘Missing rate’ dataset), or combining all filters (‘All filters’ dataset) led to a statistically significant decrease in the genotype error rate (p < .01, Figure 4c). Surprisingly, filtering by MAF led to an increase in genotype error rate in comparison with the baseline-filtered dataset (‘Stacks output’) (p < .01). A similar increase in genotype error rate was observed when MAF threshold was reduced to 0.01 (data not shown). Despite that, the absolute number of incorrect genotypes decreased in the MAF dataset in relation to the baseline-filtered dataset, suggesting that the increase in the genotype error rate is associated with the exclusion of true heterozygous variants by the MAF filter. Among the SNP filters, removing variant sites with coverage lower than 10× caused the greatest decrease in the genotype error rate, leading to similar error rates between the ‘All filters’ and ‘Minimum coverage’ datasets (p = .12).
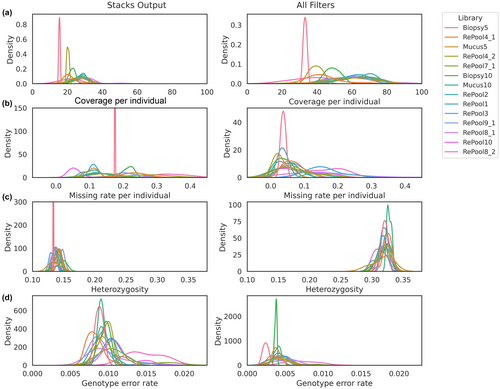
Aiming to explore the variation observed in the summary statistics and genotype error rate within each dataset, we investigated the consistency among individuals pooled in different libraries. Both datasets, before (‘Stacks output’) and after (‘All filters’) applying enhanced SNP filters, presented a large variation in the mean coverage, missing rate and genotype error rate among libraries (Figure 5). Heterozygosity estimations were similar among libraries (Figure 5c).
4 DISCUSSION
We present here an innovative approach to handle large RADseq datasets, with particular emphasis on its value for the processing of non-invasively collected samples that are commonly used and highly valuable for conservation but that are often contaminated. Based on our findings, we present a series of recommendations for RADseq library preparation and data preprocessing to overcome technical or biological artefacts that can bias downstream analyses.
4.1 Optimizing RADseq experiments with small-scale sequencing
Small-scale sequencing (spike-in) proved to be a powerful resource to control library preparation by providing preliminary information on the endogenous DNA content of each sample and the balance among individuals in a given pool. The weighted re-pooling strategy considering endogenous DNA content allowed for a guaranteed set of shared loci with similar coverage across multiple individuals with variable DNA quality.
Datasets with uneven sample representation and excessive (>100×) or poor (<10×) individual coverage present a challenge for accurate locus building and variant calling (Christiansen et al., 2021). It has been common practice until now to balance libraries using DNA quantification to control for the quantity of input DNA used in the DNA digestion reaction of RADseq protocols (Peterson et al., 2012). However, the equimolar and equal-volume pooling strategy has ultimately been shown to be inadequate to achieve even sample representation. The combination of samples at different quality/concentrations has been attributed as the main influencing variable, rather than single sample quality (Maroso et al., 2018). Solutions to this issue of individual unbalance have inherent caveats; e.g., excluding low-coverage individuals reduces inference power through lower sample numbers, and increasing sequencing effort to improve individual coverage incurs increased project costs. Larger sample sets further complicate the capacity to control for coverage distribution across individuals. Our approach overcomes this issue by first preparing a library with a small volume of the DNA digestion/adapter ligation product, and then using the same reaction for re-pooling based on the filtered and mapped spike-in read numbers.
RADseq methods rely on consistent loci selection across libraries (Andrews et al., 2016). Size-selection methods, such as manual or automated gel-cutting techniques or magnetic beads, often result in deviations in the desired fragment length range selected (Heffelfinger et al., 2014), deviations which may not be recognized by fragment analysers. We sequenced long reads (2 × 300 bp) to obtain the full locus sequence and check the length distribution of different libraries, confirming loci selection overlap among libraries and adjusting the selected range, if needed.
Based on our results, we recommend a minimum of 1000 reads per individual for spike-in sequencing to obtain a reasonable estimate of contamination level, balance among samples and length distribution per library. The financial burden of small-scale sequencing is easily outweighed by the cost savings due to more effective and reliable high-throughput sequencing, which is often the greatest expense in a given project. Combining test libraries from different projects in order to share a high-output sequencing run can greatly reduce costs.
4.2 RADseq applied to non-invasive samples
Working with gNIS brings additional challenges to library building and the preprocessing of data. Non-invasive samples are often characterized by low levels of input DNA and high levels of non-endogenous contamination, often resulting in lower genotype accuracy, increased allelic dropout and fewer variant loci and SNPs (Valière et al., 2007). Some of these pitfalls may derive from the lower number of reads produced from highly degraded samples (Graham et al., 2015). Our approach, built from multiple existing 3RADseq protocols (Bayona-Vásquez et al., 2019; Hoffberg et al., 2016), allows us to address the challenge of working with low-quality and -quantity DNA by: (1) increasing the efficiency of adapter ligation through the use of a third enzyme and concomitant digestion and adapter ligation reactions; and (2) running small-scale sequencing (spike-in) to control individual representation and compensate for non-endogenous DNA content. Despite these benefits, the addition of a third enzyme also can incur additional costs, as misleading adapter ligation to reads cut by the third enzyme was shown to occur at a relatively high frequency (18.2% of the sequenced reads, in our dataset). Additional sequencing effort can compensate for this issue.
Furthermore, we show the importance of taking into account endogenous DNA content for the pooling of non-invasive samples. Our results, combined with previous evidence (Hernandez-Rodriguez et al., 2018), indicate that weighted pooling can minimize the loss in representation of low endogenous DNA content samples in relation to samples with higher endogenous content in the pool.
This is the first time, to our knowledge, that faecal mucus samples from hyenas were tested as a potential source of DNA for SNP-based genetic studies. Given their low error rate and binary nature, SNPs are preferable to other genetic markers (e.g. microsatellites) when using non-invasive or degraded samples (Morin & Mccarthy, 2007). We demonstrated that the missing rate and genotype accuracy of faecal mucus samples (with mapping percentage higher than 50%) is comparable to biopsy samples, thus establishing faecal mucus as a promising source of gNIS. The level of DNA degradation and bacterial contamination of faecal mucus samples varied widely, which is likely associated with factors such as the diet and the length of time between the last consumption of water or fresh meat and defaecation (unpublished data). We emphasize that all samples were collected immediately after defaecation and our findings might not be valid for dry samples. The transferability of our findings to other studies using gNIS will rely on the extent of DNA degradation and non-endogenous contamination present in samples. It is crucial to note that additional research is imperative to thoroughly assess the accuracy of genotypes in any given study that employs gNIS, taking into account potential downstream implications. That said, our results provide compelling evidence to support the use of non-invasively sampled DNA to derive SNP-based data as a valuable tool for genetic monitoring of wild populations.
4.3 Improving genotype accuracy
Our results show that minimizing PCR cycles and optimizing SNP filters can significantly improve genotype accuracy. Regarding the first, the genotype error rate was higher in the PCR duplicates unfiltered than in the filtered dataset. PCR duplicates are a perennial concern in the production of RADseq libraries, particularly when a greater number of PCR cycles are used (Díaz-Arce & Rodríguez-Ezpeleta, 2019; Flanagan & Jones, 2018). PCR duplicates impact population genomics analyses by spuriously increasing homozygosity, making PCR errors appear to be true alleles and resulting in false confidence in downstream variant calls as a result of the increased read coverage (Andrews et al., 2016; Casbon et al., 2011; Schweyen et al., 2014).
Several studies have shown that PCR duplicate rate is associated with the number of PCR cycles (Ebbert et al., 2016; Flanagan & Jones, 2018). Our results support this hypothesis, as we observed higher PCR duplicate rate (and genotyping error rate) for libraries that underwent 12 PCR cycles in comparison with 8 and 10 PCR cycles. In contrast, Rochette et al. (2023) argue that PCR duplicate rate is primarily determined by the library complexity and previous studies did not control for the amount of starting material in the protocol, such that either the number of PCR cycles or the DNA input mass could be responsible for the observed differences. We tested this hypothesis by comparing samples with the same initial DNA input but submitted to different PCR cycles, which resulted in similar results: a positive correlation between the number of PCR cycles and the PCR duplicate rate (and genotyping error rate) (Figure S8). Thus, we confirmed the pivotal role of PCR cycles in determining the duplicate rate.
Despite the use of single-molecule tagging to identify and remove PCR duplicates, and strict SNP filtering (to remove variant sites with too low or too high coverage, with high missing rate and low in frequency), our results showed that the genotype error rate increased with an increasing number of PCR cycles (Figure 3). Such results emphasize the need for RADseq-users to plan library preparation with as few PCR cycles as possible. This can be achieved by increasing the library composition, via, e.g., pooling more individuals and/or using greater quantities of DNA input material per individual.
Sequencing coverage plays an important role in the genotype error rate, as low coverage increases stochasticity and reduces accuracy of the variant calling (Fountain et al., 2016). Our results corroborate this, as the low-coverage datasets (offspring subsampled to ~13×) presented a higher genotype error rate than the high-coverage (~56×) datasets. PCR duplicates impact genotype accuracy by causing overdispersion of the allelic ratios observed at heterozygous sites, leading to a bias against heterozygotes (Rochette et al., 2023). The high coverage observed in our dataset (~56×) likely permitted the heterozygote likelihood to be significantly larger than the homozygous likelihood, resulting in a lower genotype error rate than the low coverage dataset.
We showed the importance of applying posterior SNP filters, which resulted in a decrease of 60% in the genotype error rate (from 0.01 with no filters applied to 0.004 when all filters were applied). We highlight here the importance of filtering by minimum coverage, considering a study-appropriate minimum threshold (in our case, ≥10), as this parameter had the greatest impact on the genotype error rate in our analysis. Filtering by maximum coverage was also important to remove potentially collapsed paralogues that passed Stacks SNP caller requirements. In order to set minimum and maximum coverage thresholds, it is necessary to use other tools (e.g. VCFtools), as Stacks does not provide this option. We note that we did not test the impact of different SNP filter thresholds on the genotype error rate. Indeed, SNP filter thresholds should be tested for appropriate settings (Nazareno & Knowles, 2021) and chosen according to the studied taxon, specific research goals and dataset features (Díaz-Arce & Rodríguez-Ezpeleta, 2019). For example, studies focused on linkage disequilibrium should aim for higher coverage than a study based on allele frequencies (Pool et al., 2010).
Several technical factors can bias the summary statistics of a RADseq dataset (DaCosta & Sorenson, 2014). We point out that inconsistency among individuals pooled in different libraries can, in part, explain the variation observed within each dataset. While variation in the mean coverage and missing rate among individuals from different libraries is to be expected, we also found a surprising pattern in the genotype error rate associated with libraries even after applying SNP-filters (Figure 5d). This is probably connected to the coverage and missing rate results. This emphasizes the need for RADseq users to consider the bias that can be introduced by individual libraries when designing library pools.
Genotyping errors can have serious consequences on downstream analysis, including overestimates of inbreeding, impact on the resolution of tree topologies and erroneous demographic and population structure inferences (Martín-Hernanz et al., 2019; Mastretta-Yanes et al., 2015; Pool et al., 2010). The innovative bioinformatic pipeline provided here is a powerful tool for estimating genotype accuracy based on trio datasets and filtering a VCF file for Mendelian errors. A limitation of our pipeline is that it does not incorporate allele frequency in the population to assign errors (Douglas et al., 2002). We also only take into account Mendelian compatible errors (i.e. errors that produce genotypes that are consistent with Mendelian inheritance among relatives), which might represent only 61% of existing errors (Geller & Ziegler, 2002; Pompanon et al., 2005). Regardless, we recommend checking and filtering for Mendelian errors when there is the potential for related individuals to exist within a dataset, as some errors remain even after further SNP filters are applied.
In summary, we showed the potential of using gNIS for large-scale genetic monitoring based on SNPs and demonstrated how to improve control over library preparation by using a weighted re-pooling strategy that considers the endogenous DNA content. We found that PCR duplicates lead to an increase in the genotype error rate, especially when the number of PCR cycles is as high as 12 cycles and the coverage is low, even after bioinformatically removing PCR duplicates with single-molecule tagging methods. Finally, we demonstrated the impact of SNP filters and library variation patterns on genotype accuracy and summary statistics, concluding with recommendations on how to avoid associated biases in SNP calling.
AUTHOR CONTRIBUTIONS
L.S.A. and C.J.M. conceived the work and designed the research. L.S.A., S.S. and S.M. conducted the wet lab experiments. L.S.A. performed the bioinformatic analysis. L.S.A. and J.K.S. wrote the scripts. J.A.C. and L.S.A. drafted the manuscript with input from all authors. O.P.H. provided the samples and revised the manuscript. All authors read and approved the manuscript.
ACKNOWLEDGEMENTS
This work was supported by the European Research Council (ERC) grant ERC-2020-ADG – project number 101020503 and partially funded by the German Federal Ministry of Education and Research (BMBF, Förderkennzeichen 033W034A). The publication of this article was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - project number 491292795. We thank Loeske Kruuk for facilitating and funding this work, Ivan Belo Fernandes for his contributions to Python scripting, Alexandre Courtiol for his contributions to the statistical analysis, Bettina Wachter, Philemon Naman, Arjun Dheer and Eve Davidian for sample collection and Stephan Karl and Dagmar Thierer for DNA extraction and concentration measurement. Sample collection and export were permitted by the Vice President's office of the United Republic of Tanzania (Ref. No. BA 78/130/01/42) and the Tanzania Commission for Science and Technology (Permit No. 2021-380-NA-1990). Open Access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
BENEFIT-SHARING STATEMENT
Benefits from this research accrue from the sharing of our data and results on public databases as described above.
Open Research
DATA AVAILABILITY STATEMENT
The raw sequencing data are deposited in the Genbank with the NCBI BioProject accession no. PRJNA951614. The preprocessing Snakemake pipeline is available at https://git.imp.fu-berlin.de/begendiv/radseq-preprocessing-pipeline, the script used for the linear model analysis and the SNP data is available at https://git.imp.fu-berlin.de/begendiv/hyenasproject, and the ‘Genotype Accuracy based on trios’ script is available at https://git.imp.fu-berlin.de/begendiv/genotype-accuracy-estimation-based-on-trios.




