Haemoproteosis and avian malaria in Columbidae and Corvidae from Iran
Abstract
Avian malaria (Plasmodium) and related genera (Haemoproteus and Leucocytozoon) are diverse and widespread parasites. Despite the extent of knowledge on avian haemosporidian parasites, information about domestic and wild bird's blood parasites is overall insufficient in Iran. Prevalence of the haemosporidian parasites’ and phylogenetic relationship of lineages are studied by using molecular and morphological results of 152 examined hosts belonging to 17 species. Molecular analysis for haemosporidian detections demonstrated overall prevalence 22.36%. Inspected hosts mostly belonging to Common Pigeons (Columba livia) parasitized by Haemoproteus spp., and Hooded Crows (Corvus cornix) and Carrion Crow (C. corone) were identified as hosting Plasmodium spp. Detected lineages COLIV03, COQUI05, LINN01, ROFI04 and SGS01 are identified as new reports from Iran. We detected no evidence of Leucocytozoon lineages, while the high prevalence of H. columbae was found in Common Pigeons. Such investigation on avian blood parasites contributes to providing new information on the prevalence, epidemiology and geographical distribution of haemosporidian parasites circulating in domestic, pets and wild birds.
1 INTRODUCTION
Avian haemosporidian intracellular parasites are transmitted by blood-sucking arthropods. Plasmodium spp. and related genera (Haemoproteus and Leucocytozoon) are diverse and widespread (Valkiūnas, 2005) leading to avian malaria, haemoproteosis and leucocytozoonosis infrequently damaging or causing mortality in birds (Valkiūnas, 2005; Valkiūnas & Iezhova, 2017). On the other hand, serious negative effects of hemoparasites on metabolism’ efficiency and avian fitness have been registered in domestic birds like poultries and pigeons (Tiwari et al., 2012) and wild birds resulting in progressive faintness and death (Dinhopl et al., 2015; Olias et al., 2011). In the last two decades using nested cytochrome b polymerase chain reaction (PCR) have uncovered greater genetic diversity of haemosporidian parasites in a wide range of birds than traditional morphologic examinations (Bensch et al., 2009; Hellgren et al., 2004; Križanauskienė et al., 2006). Different investigations have studied haemosporidian parasites detection by microscopic and molecular approaches in pet and captive birds across the world (Alley et al., 2008; Belo et al., 2009; Bensch et al., 2012; Brossy, 1992; Chagas et al., 2017; Dinhopl et al., 2011; Nakayima et al., 2019; Nourani et al., 2020a; Pacheco et al., 2011; Paperna & Martelli, 2008; Scaglione et al., 2016a; Schrenzel et al., 2003; ). Despite the extent of knowledge on the avian haemosporidian parasites, information about domestic and wild bird's blood parasites is overall insufficient in Iran (Nourani et al., 2017a; Nourani et al., 2017b, Nourani et al., 2018a; Nourani et al., 2018b; Nourani & Dinparast Djadid, 2019; Nourani et al., 2020a; Nourani et al., 2020b; Nourani et al., 2020c). To date there have been limited investigations documenting morphological and molecular results of blood intracellular parasites infection in pet and domestic birds (Chaechi-Nosrati et al., 2018; Mirzaei et al., 2016; Nematollahi et al., 2012). Haemoproteus (Hippoboscidae and Ceratopogonidae) and Plasmodium (Culicidae) parasites can be transmitted during blood meal of hematophagous dipteran (Valkiūnas, 2005) that their contribution in circulation parasites and distribution map remains unidentified in Iran (Nourani et al., 2020c). Parasites are of main hazards to bird populations; endangered or domestics (Bunbury et al., 2008) and birds may encounter multiple pathogens that contribute to the transmission of pathogens to other birds and humans (Reed et al., 2003). Thus, expanding our knowledge of distribution in novel hosts and less-studied geographical regions seems crucial (Al-Barwari & Saeed, 2012; Cray, 2011; Hunter et al., 1997; Jacob et al., 2011). Detection of parasites with a great impact on birds’ health requires more attention in designing field surveillance and performing the control programs. Here we present results from blood parasites screening in the referred pet, domestic and wild birds to the veterinary clinic during a year. Our objectives were to assess haemosporidian prevalence infections by molecular and morphological methods, to determine novel host records and/or parasitic lineages in examined birds, and to study their phylogenetic relationship with known lineages in similar hosts.
2 MATERIALS AND METHODS
2.1 Preparation of blood samples for morphological detection
Blood samples were collected of pets, domestic and wild injured birds referred to the veterinary clinic during October 2018 to September 2019 and some blood samples were collected by a veterinarian during visiting hosts in farms. Brachial vein blood was taken with a syringe (50–100 μL of whole blood). Two thin blood smears were prepared for each host, were immediately dried in the air, preserved in absolute methanol and subsequently stained with Giemsa 10% at pH 7.2 (60 minutes) for the morphological screening of haemosporidian parasites. Blood films were examined under 400× and 1,000× of the Olympus BH2 light microscope (Olympus Co, Japan) via oil immersion in 100 fields for 35–40 minutes for haemosporidian parasites gametocytes discovery (Martinsen et al., 2006; Valkiūnas, 2005; Valkiūnas & Iezhova, 2018). Blood samples were immediately stored in an anticoagulant buffer and subsequently stored at −20°C until molecular experiments. More information about the bird's health condition was registered in agreement with the history from their owners or finders (Table 1).
| Species | Common name | n | Haemosporidian parasites | Birds sampling site (n) | Other diseases diagnosis |
|---|---|---|---|---|---|
| Acridotheres tristis | Common myna | 11 | – | C | ISD (8), hematoma in neck (2), pododermatitis (2), nephropathy (1), pneumonia (1) |
| Agapornis roseicollis | Rosy-faced lovebird | 2 | – | C | Tibiotarsus fraction (1) |
| Columba livia | Common pigeon | 45 | H. columbae (COLIV03, COQUI05, HAECOL1) | C (27), F (10), S (8) | Miasis (1), eye infection (1), trichomoniasis (12), Macrorhabdus ornithogater (3), pneumonia (5), pododermatitis (1), Pseudolynchia canariensis (3) |
| Gallus gallus | Chicken | 46 | – | F | Pneumonia (6), eye infection (1), lice and mite infestation; Menopon gallinae, Dermanyssus gallinae (6), Macrorhabdus ornithogater (3), coccidiosis (1) |
| Spilopelia senegalensis | Laughing dove | 3 | – | S | – |
| Cygnus olor | Mute swan | 3 | – | F | – |
| Myiopsitta monachus | Monk parakeet | 1 | – | C | – |
| Psittacus erithacus | Grey parrot | 6 | – | C | Feather picking (2), uropygial gland infection (1) |
| Pyrrhura molinae | Green-cheeked parakeet | 2 | – | C | – |
| Nymphicus hollandicus | Cockatiel | 1 | – | C | – |
| Alectoris chukar | Chukar | 1 | – | F | – |
| Meleagris gallopavo | Wild turkey | 5 | – | F | Black head disease (Histomonas meleagridis, 2), enteritis (3) |
| Coturnix coturnix | Common quail | 7 | – | F | Pododermatitis (1), egg binding (1) |
| Corvus cornix | Hooded crow | 8 | P. relictum (SGS01), Plasmodium sp. (ROFI04) | S | Wing fracture (5), Gram-positive Bacillus in digestive system (1) |
| Corvus corone | Carrion crow | 1 | P. matutinum (LINN01) | S | – |
| Anas platyrhynchos | Mallard | 9 | – | F | Calcium deficiency (2), pneumonia (2), lice infestation (1) |
| Passer montanus | Eurasian tree sparrow | 1 | – | S | Osteomyelitis (1) |
| Total | 152 | ||||
- Number of examined hosts (n), morphological (Morph) and molecular (Mol) detection of haemosporidian parasites are summarised. Birds are brought by their host to clinic (C), visiting in the farm (F), or found in the street and referred to the clinic (S).
- ISD, iron storage disease.
2.2 Genomic DNA extraction and PCR
Genomic DNA were extracted by using DNA Isolation Kit for blood, according to the manufacturer's instructions (Yekta Tajhiz Azma, Tehran, Iran). DNA extraction was made to amplify a 478-base-pair fragment of mitochondrial DNA cytochrome b gene (cytb) by nested-PCR and sequencing method. HaemNFl/HaemNR3 (first reaction) primers were used to amplify fragments of Haemoproteus, Plasmodium and Leucocytozoon parasites. For the second PCR, the primers HaemF/HaemR2 were applied, which amplifies Haemoproteus and Plasmodium parasites and primers HaemFL/HaemR2L were used to amplify Leucocytozoon (Bensch et al., 2000; Hellgren et al., 2004). Positive PCR products of the previous investigation (Nourani et al., 2020b) and ultrapure water were utilised as positive and negative controls. Subsequently, 1% agarose gels were used for visualisation of amplified amplicons. Purification and sequencing of positive products were made by Codon Inc. (Codon Genetic Group, Tehran, Iran).
2.3 Bioinformatics and phylogenetic analysis
BioEdit v.7.1.7 was used for cleaning up, edition and alignments of raw sequences by ClustalW (Hall, 1999). The single base mutation was considered as a criterion for the novelty of detected lineages in comparison with the homologs sequences in Nucleotide BLAST analysis, accessible at NCBI and MalAvi sites (Bensch et al., 2009; Pérez-Tris & Bensch, 2005). Seventeen amplified sequences in the current study are deposited in GenBank (Accession number: MT802174-MT802190). Phylogenetic relationship of lineages was studied by Bayesian analysis that performed with two concurrent Markov Chain Monte Carlo searches of 10 million generations using MrBayes v3.2 and sampling was done for 1 of 1000 trees (Ronquist & Huelsenbeck, 2003) under the evolutionary model achieved from MODELTEST v3.7 program (Posada & Crandall, 1998). At the end of the analysis when the chains reached stationary status posterior probabilities were measured by the burn-in period of 50%. The phylogenetic consensus tree was visualised by FigTree v1.4 (Rambaut, 2012). Data analysis for prevalence and confidence interval 95% was calculated with SPSS v.16 for Windows (SPSS Inc, Chicago, IL, USA).
2.4 Identification of other diseases
For the detection of Trichomonas gallinae in pigeons, wet mounts were made by oropharyngeal swabs and subsequently were stained via Giemsa stain. The microscopic inspection of smears was accomplished under 100× magnification by CH30 light microscope (Olympus Co, Japan) (Anderson et al., 2009). Moreover, depending on the clinical signs and history of the disease, the required tests for accurate diagnoses were performed, radiographs (fractures), CT scans (respiratory infections), or through direct examinations.
3 RESULTS
3.1 Haemosporidian parasites detection and diversity of lineages
One hundred fifty-two birds belonging to 17 species were screened for the detection of haemosporidian parasites (Table 1). Gallus domesticus and Columba livia (∼60% of individuals) had the highest numbers of examined hosts. The inspection of blood smears showed 30 individuals were positive for one of the blood parasites genera, prevalence = 19.73% (95% CI 13.34, 26.14). Most of the infected birds belonged to Columbidae family. Using microscopic examinations, we discovered Haemoproteus spp. infections in 27 individuals of Columba livia (17.76%), shown in Table 1. We also recognised Plasmodium spp. infections in two individuals of Corvus cornix and one Corvus corone.
Molecular analysis results demonstrated the overall infections prevalence 22.36% (95% CI 15.67, 29.07) in examined hosts (34 individuals) that mostly belong to Columba livia (31 individuals) parasitising by Haemoproteus columbae; COLIV03 (n = 3), COQUI05 (n = 6) and HAECOL1 (n = 5) lineages. Corvid species Corvus cornix and C. corone were infected by SGS01 (P. relictum) and ROFI04, and LINN01 (P. matutinum), respectively.
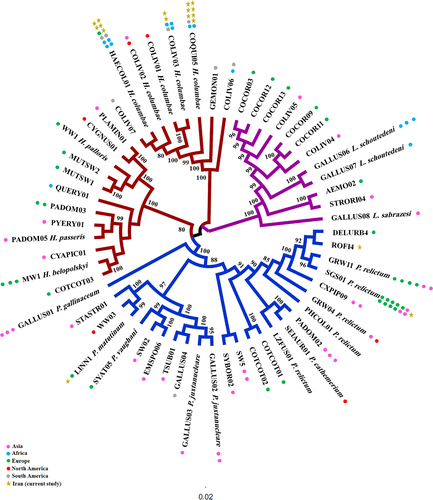
The Bayesian resultant tree constructed by using 57 retrieved sequences of Haemoproteus, Plasmodium and Leucocytozoon lineages (from NCBI and MalAvi) along with detected lineages in this study demonstrated that Leucocytozoon is positioned as the basal subclade, and Haemoproteus and Plasmodium monophyletic subclades are clustered as sister taxa (Figure 1). Returning to parasites subclade, known lineages are recorded in different continents, and any relationship among lineages and detection localities was found. Detected Haemoproteus lineages in pigeons belonged to one species (H. columbae), which are clustered in a distinct subclade, separated from other Haemoproteus lineages. Within Plasmodium, SGS01 and GRW11 (P. relictum) are more prevalent lineages, reported in different host species. The detected Plasmodium lineages in this study (ROFI4 and SGS01) are placed in different subclades, while LINN01 is placed as a sister taxa with WW03.
4 DISCUSSION
The key result of this study is the application of nested PCR sequencing along with morphological methods to identify new records of haemosporidian parasites in birds of Iran. Six Haemoproteus and Plasmodium known lineages were registered as new host records that were previously discovered in passerine and non-passerine hosts. Similar to previous researches in wild passerines across Iran, Haemoproteus was the prevalent parasitic genus among birds (Nourani et al., 2017a; Nourani et al., 2017b, Nourani et al., 2018a; Nourani et al 2018b; Nourani & Dinparast Djadid, 2019; Nourani et al., 2020a; Nourani et al., 2020b). Nested PCR assay and morphological examinations did not detect any record of Leucocytozoon spp.
Fifty percent of detected lineages in this study, COLIV03, COQUI05 and HAECOL1 have been reported in one host species (Common Pigeon), and the lineages SGS01, LINN01 and ROFI4 have been detected in various bird species, around the world. Infected hosts in this investigation were similar to preceding reports and indicate cross-infections in multiple host species of various families from different countries (Bensch et al., 2009). Haemoproteus lineage GALLUS01 was shared by two species Passer montanus (Japan) and Gallus gallus (Thailand) (Kim & Tsuda, 2010) and host shifting Plasmodium lineage SGS01 reported in Gallus gallus (Italy) and Passer montanus (Bulgaria) (Dimitrov et al., 2010; Martinez-de la Puente et al., 2015), recommending the active transmissions by suitable vectors (Chakarov et al., 2015). Furthermore, the potential significance of migration in parasitising resident birds can be deduced by multiple hosts sharing of LINN01 among migratory species Carduelis cannabina and Luscinia svecica (Nilsson et al., 2016; Svoboda et al., 2015) and residents Athene noctua and Passer montanus (Martinez-de la Puente et al., 2015). Detected lineage ROFI4, a new host record in Corvus cornix from Iran has been previously recorded in Carpodacus erythrinus from Czech Republic (Synek et al., 2013). The infections to prevalent lineages of LINN01 (reported in 14 species) and SGS01 (reported in 130 species) have been registered in MalAvi (Bensch et al., 2009). LINN01 (P. matutinum) has so far been documented in various bird hosts (MalAvi) and vectors (Culex pipiens) (Kimura et al., 2010; Martinez-de la Puente et al., 2015). For the first time, we found LINN01 in Corvidae member C. cornix. Lineage SGS01 (P. relictum) as a prevalent and widespread lineage (Zehtindjiev et al., 2008) has been recorded in more than 100 wintering and resident bird species belonging to different families around the world (MalAvi 2020) (Bensch et al., 2009; Palinauskas et al., 2008; Palinauskas et al., 2009). In this study, this lineage is recorded for the first time in a passerine bird in Iran (Corvus corone). Contrary to our study, molecular and histological examinations on Corvus corone tissues demonstrated the higher infections to Leucocytozoon (97.9%) and Haemoproteus or Plasmodium spp. (59.6%) in individuals inspected from Italy (Scaglione et al., 2016b). The higher infection by Leucocytozoon than Plasmodium and Haemoproteus spp. was registered in 85 carrion crows (89.5%) from Germany (Schmid et al., 2017).
Pigeons’ health as pets and cultural symbols or laboratory animals should be considered, which can be influenced by their growth, production and vulnerability to diseases (Dranzoa et al., 1999). In this study, the most infected host species was common pigeon, of which 17.76% were parasitised with HAECOL1 (H. columbae). To date, eight Haemoproteus, two Plasmodium and eight Leucocytozoon lineages have been detected in Columba livia, deposited in MalAvi (Bensch et al., 2009).
One-third of our samples was related to poultry, and none of them was infected with Plasmodium or Haemoproteus spp. Some species of haemosporidian parasites have a major impact on the poultry industry; high mortality up to 90% is observed in domestic chickens (Springer, 1991). Previous studies have informed absence or relative infections of haemosporidian parasites in the Phasianidae family (Castle et al., 1988; Christensen et al., 1983; Huchzermeyer, 1993; Jasim & Al-Moussawi, 2001; Laird, 1978; Mohammad et al., 2001; Otranto et al., 2010), possibly due to their specific activities avoiding encounters with vectors (Alavi et al., 2003; Loiseau et al., 2012). Nevertheless, some other investigations declared the haemosporidian infections in Phasianidae, white eared-pheasant (Crossoptilon crossoptilon) parasitised by P. juxtancleare (Murata et al., 2008), Ammoperdix griseogularis and Alectoris graeca infected by Haemoproteus and Ammoperdix griseogularis infected by Plasmodium spp. (Mohammad et al., 2001). Microscopic screening and PCR assays have been utilised to identify blood parasites in free-ranging domestic birds: chickens, ducks, turkeys and guinea fowl in Uganda. Their results demonstrated the infections to Plasmodium, Haemoproteus and Leucocytozoon, which provide information for parasite management in poultries to hinder other species (Nakayima et al., 2019). More epidemiological studies are necessary to be performed to detect haemosporidian parasites, as a prevalent infection in poultry, which prevents the spread of lineages to farm birds and other susceptible birds. The discovery of Haemoproteus and Plasmodium species and also coinfections for domestic and pet birds are also significant to prevent the potential transmission of these parasites within colonies and among wild birds in rural or urban areas (Scaglione et al., 2015).
The great chance of having different avian, both pets and captivated bird species in clinics, surely provide an opportunity for the following basic and applied investigations: systematics analysis based on morphological, molecular and morphometric tools, diagnosis of ectoparasites, endoparasites and other parasites (bacterial and viral species) mainly those that are considered as vectors of different diseases and identifying corresponding vectors (in case of VBDs) or in general zoonosis (Ryser-Degiorgis, 2013; Stallknecht, 2007). Control and surveillance of vector-borne diseases depend on the understanding of associations between pathogen, arthropod vectors and vertebrate hosts. The achieved data by developing diagnosis tools provide new prospects for surveillance, prevention and control of vector-borne diseases in the avian clinics that are mostly comprising pets, domestic birds and wild injured hosts to manage the spreading parasites among other populations. For captive animals and pets with long-term risk of diseases, epidemiologic investigations provide management practices to protect these inestimable animals (Camacho et al., 2016; Brown & O'brien, 2011; Lozano-Fuentes et al., 2011).
In this study, we reported the detection of haemosporidian parasites by using molecular and microscopic overview in pets, domestic and wild birds referred to a veterinary clinic in Iran. Forthcoming investigations will shed light on host–parasites relationships and parasites distribution that are necessary for veterinary surveillance and control of vector-borne parasites.
ACKNOWLEDGMENTS
We express our sincere appreciation to MVRG members for sharing their pearls of wisdom with us during this investigation. This research was supported by a Pasteur Institute of Iran grant to NDD. (No: 953).
ETHICS STATEMENT
This study was achieved in accordance with the guidelines and protocols permitted by the ethics committee for the care and use of animals for scientific purposes of the Pasteur Institute of Iran (No: IR.PII.REC.1395.96).
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
Amplified sequences in the current study are deposited in GenBank (Accession number: MT802174-MT802190).




