Elucidation of ligand binding and dimerization of NADPH:protochlorophyllide (Pchlide) oxidoreductase from pea (Pisum sativum L.) by structural analysis and simulations
Funding information: Carl Tryggers Foundation, Grant/Award Numbers: CTS 15:34, CTS 17:32; Magyar Tudományos Akadémia, Grant/Award Number: Bolyai János Research Scholarship; Ministerio de Ciencia e Innovación, Grant/Award Number: PID2019-106370RB-I00; Országos Tudományos Kutatási Alapprogramok, Grant/Award Number: OTKA FK 124748
Abstract
NADPH:protochlorophyllide (Pchlide) oxidoreductase (POR) is a key enzyme of chlorophyll biosynthesis in angiosperms. It is one of few known photoenzymes, which catalyzes the light-activated trans-reduction of the C17-C18 double bond of Pchlide's porphyrin ring. Due to the light requirement, dark-grown angiosperms cannot synthesize chlorophyll. No crystal structure of POR is available, so to improve understanding of the protein's three-dimensional structure, its dimerization, and binding of ligands (both the cofactor NADPH and substrate Pchlide), we computationally investigated the sequence and structural relationships among homologous proteins identified through database searches. The results indicate that α4 and α7 helices of monomers form the interface of POR dimers. On the basis of conserved residues, we predicted 11 functionally important amino acids that play important roles in POR binding to NADPH. Structural comparison of available crystal structures revealed that they participate in formation of binding pockets that accommodate the Pchlide ligand, and that five atoms of the closed tetrapyrrole are involved in non-bonding interactions. However, we detected no clear pattern in the physico-chemical characteristics of the amino acids they interact with. Thus, we hypothesize that interactions of these atoms in the Pchlide porphyrin ring are important to hold the ligand within the POR binding site. Analysis of Pchlide binding in POR by molecular docking and PELE simulations revealed that the orientation of the nicotinamide group is important for Pchlide binding. These findings highlight the complexity of interactions of porphyrin-containing ligands with proteins, and we suggest that fit-inducing processes play important roles in POR-Pchlide interactions.
1 INTRODUCTION
Chlorophyll biosynthesis is crucial for photosynthesis, and thus photoautotrophic growth of land plants. It is subject to complex regulation, involving various anterograde and retrograde signaling pathways and several feedback mechanisms.1, 2 One of the key regulatory steps is the penultimate step in chlorophyll formation, the reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide). Two nonhomologous enzymes with completely different structures and reaction mechanisms have evolved that catalyze this process.3 Dark operative (or light-independent) Pchlide oxidoreductase (DPOR) putatively evolved from a nitrogenase-like enzyme of methanogenic archaea under anoxygenic conditions.3 This is a multimeric, oxygen-sensitive enzyme composed of three different subunits encoded by the plastid genome. The other enzyme is nuclear-encoded and oxygen-insensitive, but requires light for its activity and is thus called light-dependent NADPH:Pchlide oxidoreductase (LPOR, hereafter simply POR, for convenience).2, 4 Anoxygenic photosynthetic bacteria contain only DPOR, many organisms (most green algae, bryophytes, pteridophytes and gymnosperms) contain both types of enzymes, but angiosperms and some other organisms have lost DPOR during evolution and thus the ability to produce chlorophyll in the dark.3, 5
Without light-induced developmental signals and chlorophyll, chloroplast formation is also inhibited in dark-germinated angiosperm seedlings. Instead, proplastids become etioplasts, containing three-dimensional networks of tubular membrane structures called prolamellar bodies (PLBs).2, 6 PLBs are also found in plants grown under natural light conditions, for example, in fruit pericarps and leaf buds.2, 6, 7 The lipid composition of PLBs favors formation of the highly regular and curved network of PLB membranes, but is not the crucial factor.6, 8, 9 Key identified factors include the presence and supramolecular organization (dimerization or oligomerization) of ternary complexes of POR with Pchlide and NADPH.2, 10-17 However, the mechanisms involved in POR's interactions with PLB lipids and carotenoids that stimulate formation of the highly regular network of PLB membranes are poorly understood as no high-resolution structure of POR is available. Information about its structure is limited to relative amounts of secondary structure elements, experimentally determined by circular dichroism,18 and predictions of secondary and tertiary structures obtained using various bioinformatic tools.18-23
Several in vivo and in vitro washing experiments and mutagenesis experiments have been performed in attempts to elucidate the strength of POR's membrane association. The results show that its association with thylakoid or etioplast inner membranes requires NADPH and ATP.24, 25 Furthermore, charged amino acids, cysteine (Cys) residues and the C-terminal are important for membrane association.20, 26 In addition, various salts,24, 25, 27, 28 detergents27, 29 and proteases24, 25, 30 have been used in washing experiments. These studies have shown that POR is firmly attached to PLBs and PTs, but more loosely attached to thylakoids, indicating that POR's affinity for the membranes may decline during greening.31
The difficulties of releasing POR from PLBs and PTs using salts and detergents led to the conclusion that POR must be an integral membrane protein.27, 28 Integral membrane proteins are generally classed as integral transmembrane proteins with one or more regions spanning the membrane or integral monotopic membrane proteins permanently attached to one side of a membrane.32 However, the suggestion that POR is an integral transmembrane protein was subsequently withdrawn33 in the light of its deduced amino acid sequence.34, 35
Today, numerous sequences of POR from diverse plant groups have been characterized.4, 36, 37 Two differently regulated POR isoforms, designated PORA and PORB, have been characterized in barley (Hordeum vulgare),38 Arabidopsis thaliana,39 wheat (Triticum aestivum)40 and rice (Oryza sativa).41 PORA accumulates in dark-grown tissues and is active during the first hours of greening while PORB is active throughout the life of the plant. A third isozyme, PORC, has only been found, to date, in light-grown A. thaliana42 and may be involved in regulation of oxidative stress.43 POR sequences generally share high sequence similarity and have high contents of basic and hydrophobic amino acids.5 However, no evidence of hydrophobic segments long enough to span a membrane have been detected in hydropathy plots,44-46 which have been persistently cited since their publication. However, these plots were based exclusively on searches of sequences for a sufficiently long hydrophobic stretch of residues to traverse a membrane based on the hydrophobicity scale.47 Refinements of the methodology have included consideration of statistical and structural information about known transmembrane helices, such as the presence of specific sequence patterns and motifs, the packing of helix bundles, and properties of inter-helical residue interactions. Methods considering this kind of information are referred to as advanced transmembrane methods and generally perform better than simple hydrophobicity scale-based methods.48
Some early experiments involving attempts to isolate and characterize photoactive POR complexes showed that they may be associated with carotenoids, particularly zeaxanthin and violaxanthin,13 or carotenoids may play important roles in their membrane association.49 In vitro reconstitution experiments have also shown the importance of various membrane lipids in the proper organization of the photoactive complexes,50 which as yet have not been reconstituted without lipids, and thus membranes. Similarly, data on lipid biosynthesis mutants have shown that membrane lipids are crucial for proper POR activity, which requires its oligomerization as well as membrane association.51, 52
As already stated, despite its importance in chlorophyll biosynthesis, and thus plant metabolism, as well as its highly interesting photochemical catalytic activity, there is still no available crystal structure of POR and we do not fully understand its structure and oligomerization. Therefore, as reported here, we performed a detailed bioinformatic analysis of POR, focusing on its potential dimerization and binding of Pchlide. The modeling and simulations indicate that α4 and α7 helices of the POR protein form the dimer interface, and that Pchlide binds close to the pro-S hydrogen of the nicotinamide ring of NADPH and Tyr191, which is involved in the catalytic mechanism.
2 MATERIALS AND METHODS
2.1 Sequence and structural analysis
The amino acid sequence of the POR protein of pea (Pisum sativum L.) was retrieved from the Uniprot database (Q01289). Its domain pattern was analyzed using the protein families (PFAM)53 and Prosite54 databases. PSI-Blast55 was used to search the POR sequence against the PDB database56 to identify homologous proteins with crystal structures, which were superimposed using the PDBefold server.57 The root means square deviation (RMSD) between the homologs was calculated based on pairwise comparison using PDBefold. Structure-based sequence alignment was applied to identify regions of these proteins that were structurally conserved, despite low sequence identity. We downloaded additional POR sequences from different organisms. The accession numbers of these sequences are as follows: Arabidopsis thaliana PORA (Q42536), Arabidopsis thaliana PORB (P21218), Arabidopsis thaliana PORC (O48741), Pisum sativum (pea) POR (Q01289), Hordeum vulgare (barley) PORA (P13653), Hordeum vulgare (barley) PORB (Q42850), Triticum aestivum (wheat) PORA (Q41578), Avena sativa (oat) POR (P15905), Cucumis sativus (cucumber) PORA (Q41249), Chlamydomonas reinhardtii POR (Q39617), Marchantia paleacea (liverwort) POR (O80333), Daucus carota (carrot) POR (Q9SDT1), Plectonema boryanum POR (O66148), Synechocystis sp. strain PCC 6803 POR (Q59987). The POR sequence was then aligned with the multiple sequence alignment of these templates using the MAFFT program.58
2.2 Homology model
Homologs of POR were identified by a BLAST search of the PDB database using the retrieved pea sequence as candidate templates for model building. Nine with crystal structures were identified that bind NADPH (PBD_ID: 3WXB, 2JAH, 3P19, 3CSD, 3SJU, 1XU9, 3O26, 1WMA, and 1N5D), and the crystallographically determined structure of chicken carbonyl reductase (3WXB) was selected as a template, as it has high resolution (1.98 Å), bound cognate ligand (NADPH), dimeric form, and sequence identity. An optimized model of POR was built using Modeller v. 9.1959 with the sequence alignment described above. Models of both monomeric and dimeric forms of the protein were generated. Ten models were constructed and the best, according to Modeller's atomic statistical potential function, was selected. Multiple templates were used to model the long insertions of amino acids in POR. The model was validated using a Ramachandran plot, VERIFY3D60 and ProSA-web.61
2.3 Molecular Docking
Molecular docking of NADPH with POR was applied to study its mode of interaction with its POR binding site using GOLD (Genetic Optimization for Ligand Docking) v. 5.38. This is a widely used protein-ligand docking program,62 which applies a genetic algorithm for protein-ligand docking that allows full ligand and partial protein flexibility. The homology model of POR with added hydrogen atoms was used as the receptor, the NADPH ligand model was corrected with the Auto-Edit Ligand option, and 200 conformers were generated using the Conformation Generation module of Mercury Software.63 The NADPH active site was identified and reconfirmed by superposition and multiple structure-based sequence alignments of POR with the nine selected NADPH-bound crystal structures (described above). Based on the alignment, the binding site of NADPH was mapped onto the POR protein and used as input for GOLD calculation. Default genetic algorithm (GA) settings were used for docking, generating 10 poses for each of the 200 conformers. Early termination of the GA runs was allowed when the root mean square deviations (RMSD values) of the top three GA solutions were < 1.5 Å. The best pose of the docked ligand was selected based on GOLD score and calculated RMSD between the reference NADPH and crystal structure of 3WXB. The RMSD between the different docked conformation of NADPH was calculated using the GOLD software. The docking was submitted to the Albiorix cluster available at the University of Gothenburg (http://albiorix.bioenv.gu.se/).
2.4 PELE simulations
While the NADPH binding site can be inferred from homologs, Pchlide binding information is not available. Thus, to map Pchlide's binding to POR we used Protein Energy Landscape Exploration (PELE) Monte Carlo (MC) molecular simulation software.64 PELE has been developed for analyzing protein-ligand interactions, through functions such as binding site search and refinement, ligand migration, and fragment growth.65-67 Each PELE MC step consists of a ligand perturbation based on a (random) translation and rotation, followed by a backbone protein perturbation using a low frequency subset of normal modes, side chain prediction and, finally, a global minimization. The resulting structure is accepted or rejected by applying a Metropolis criterion, based on energy calculations generated using a OPLS2005 force field with an implicit solvent, as explained in detail by.68 In the applied approach, which we call the AdaptivePELE enhanced sampling method,66 several iterations are run, consisting of a short PELE simulation, then the output is clustered and some of the clusters are selected to start the next epoch. All accepted MC steps are clustered with the leader algorithm using the ligand RMSD (after protein superposition). The following initial structures are then selected using criteria deemed appropriate, for example, the clusters' populations or value of some metric such as solvent-accessible surface area (SASA) or interaction energy. AdaptivePELE can be an order of magnitude faster than PELE simulations, allowing us to map complex unbiased binding mechanisms in less than an hour.
Two PELE simulations were performed. First a global simulation, in which the ligand was allowed to explore the entire protein surface by using large translations and rotations, as well as 20 different initial structures randomly distributed at the enzyme surface. In the second simulation local sampling was applied, with reduced ligand translations and rotations together with application of a steering vector (translation direction) for two consecutive MC steps. The aim of the second simulation was to refine the binding site, by allowing the ligand to enter inner cavities.
3 RESULTS AND DISCUSSION
3.1 Sequence search
POR is a member of the short-chain dehydrogenase/reductase (SDR) superfamily of proteins. Accordingly, a Blast sequence homology search against the PDB database showed POR to be highly similar to other members of this family, which have a highly conserved nucleotide-binding Rossmann fold,69 despite sharing very low sequence identity.70, 71 However, based on the Blast search results, we selected two crystal structures: carbonyl reductase from chicken fatty liver (PDB_ID: 3WXB)72 and salutaridine reductase from Papaver somniferum (opium poppy, PDB_ID: 3O26)73 from the hits, based on total sequence coverage and presence of a bound NADPH cofactor.
3.2 Structural alignment of POR-homologous SDR proteins
POR belongs to the SDR family that includes enzymes that utilize both NADPH and NADH as cofactors. However, the enzymes have small structural differences and residue preferences within their cofactor binding motif that may account for the differences in specificities for these two cofactors.74, 75 Presence of a basic amino acid in the glycine-rich motif immediately preceding the second conserved glycine has been reported in NADP(H)-specific enzymes.71 Other studies indicate that POR of a Synechocystis species can bind to NADPH, but not NADH.76, 77 Thus, a key task was to identify structural homologs bound with NADPH as a cofactor to find residues that play important roles in its binding in the homologs and, potentially, POR.
Using the selected template, 3WXB, a Blast search against the PDB database (using the DALI server) identified more than 1000 structures with Z-scores between 42.3 and 12. From this set we selected SDR protein structures that met two criteria: possession of the adh_short domain (PFAM ID: PF00106) and binding of the NADPH cofactor (Figure 1A). SDR protein family members have structural differences that depend on whether they bind to NAD(H) or NADP(H).71 POR has two conserved regions: a consensus Gly-X-X-X-Gly-X motif associated with cofactor binding78 and a region that has been strictly conserved through evolution in the SDR family with a Tyr-X-X-X-Lys sequence that is essential for enzyme activity.79 However, in the set of nine structures considered as candidate templates here, Arg, Ser and Lys precede the second conserved Gly. The presence of the Ser residue before the second Gly of the Gly motif confers preference for NADP(H) binding in the human 17β-hydroxysteroid dehydrogenase type 1 enzyme.80
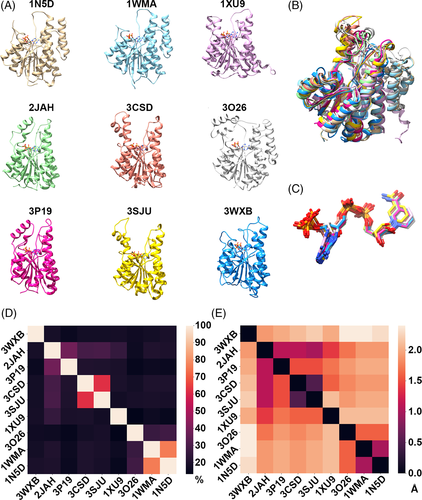
The pairwise sequence identity and RMSD of these nine proteins range from 13.2% to 76% and 0.5 Å to 2.8 Å, respectively (Figure 1D and E, Table S1). Structural superposition of these structures shows that they have a well-conserved Rossmann fold, despite its very low sequence identity (Figure 1B). Each of these proteins has a α/β doubly wound structure, in which seven parallel β strands that form a β sheet are sandwiched between two arrays of three α helices. Similarly, the bound conformation of NADPH within pockets of these protein structures was found to have an RMSD less than 0.7 Å (Figure 1C). Further, we analyzed the interaction between each of the nine protein structures and the bound NADPH. Aligned residues of the proteins at positions corresponding to Asn15, Arg16, Ile18, Arg41, Arg47, Val68, Asn96, Thr178, Lys182, Val211, Thr213 in 3WXB were observed to form hydrogen bonds with the NADPH cofactor in all nine structures (Figure 2A, B). Thus, the bound conformation of NADPH is observed to be well conserved among these proteins, despite low overall sequence identity (Figure 2C). The hydrogen bond interaction involving these residues (Asn15, Arg16, Ile18, Arg41, Glu67, Val68, Asn96, Tyr178, Lys182, Val211 and Thr213) is conserved in all cases except for amino acids Glu67 and Asn96 in 3SJU, and Arg16 in 1XU9, 3O26, 1WMA and 1N5D. Thus, these residues are identified as critical for binding to NADPH. In addition, 10 atoms of the ligand (O7N, O2D, O3D, O1N, O2N, O3B, O1X, O2X, O3X, and N1A according to standard labelling; reference) form hydrogen bonds in all nine structures (Figure S1). We observed that the adenine group is captured near the loop connecting β3 and α3 via interactions with Glu67 and Val68 (in 3WXB and corresponding residues in the other proteins), while the ribose phosphate interacts with Asn15 and Arg41 located on the loop connecting β1 and α1, immediately after β3. The nicotinamide moiety appears to interact with Val211 and Thr213 in the loop after β6.
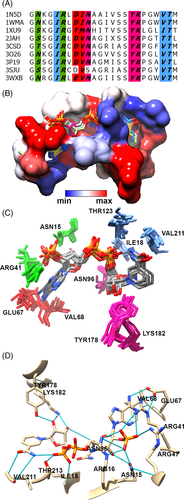
We further aligned 14 POR sequences from different organisms with the nine sequences associated with the nine crystal structures, then mapped the NADPH binding site onto the POR sequences (Figure S2). We found that eight of the 24 residues identified in the binding site of NADPH of co-crystal structures were identical to residues at corresponding positions in the POR enzymes. Of these eight residues, four form hydrogen bonds with ribose groups of the NADPH cofactor. We infer from results of this analysis that these residues play an important role in the geometric alignment of the NADPH cofactor within active sites of these SDR enzymes.
The 3D structure of POR was predicted using Modeller software v. 9.19 and the NADPH bound co-crystal structure of carbonyl reductase from chicken fatty liver (PDB_ID:3WXB) as template. 3WXB was used for this purpose because both POR and 3WXB proteins have the short-chain dehydrogenase domain and the crystal structure of dimeric 3WXB bound with NADPH provides insights into the spatial arrangement of the active site residues and two monomeric subunits. However, the sequence identity between the target and template is low (21.19%) so we applied profile-based alignment to improve the alignment, as follows. First, the nine crystal structures and set of 14 POR sequences were subjected to structure-based sequence alignment using PDBeFold and sequence alignment using Clustal Omega. Both alignments were then combined using the Merge program provided by the MAFFT server, and the merged alignment was then used as input for modeling POR by the Modeller software. The most noticeable differences from the sequence alignment between POR and the template are the two long amino acid insertions, one 38 amino acid long (between β5 and α7) and the other 22 amino acid long (between β6 and α8) were observed in the central region of POR.
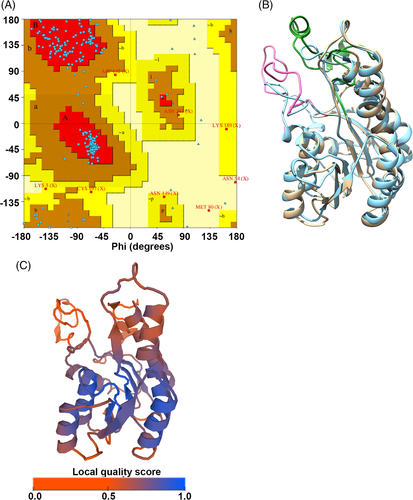
We modeled POR by first modeling the POR monomer (residues 84-371) then the dimer using both one template (3WXB) and a combination of multiple templates to cover the long amino acid insertions. The quality and accuracy of the model was then assessed using a Ramachandran plot, Verify3D and ProSA. In the Ramachandran plot of the POR monomer about 85.9, 13 and 0.8% of the residues are in the most favored, generously allowed and disallowed regions, respectively (Figure 3A), according to criteria presented by Reference (Date). Verify 3D indicated that about 77.78% of the modeled structure is compatible with its amino acid sequence and the Z score for the model calculated using ProSA is −5.87. Following this validation, the modeled POR structure was superposed onto the template 3WXB, resulting in an RMSD value of 1.34 Å (Figure 3B). Thus, the overall topology of the modeled structure is similar to the template structure except for the two long insertions. The quality of the model evaluated using the qualitative model energy analysis (QMEANS),81 suggests that the predicted model is overall good and has a high quality score in the core of the protein structure, whereas the two long loops (mentioned above) has lower quality, which is expected for such low identity (Figure 3C). The overall analysis confirms that the model generated is reliable and can be used for further analysis.
Most SDR enzymes reportedly occur predominantly in dimeric and tetrameric forms in nature and the two long α-helices are involved in oligomerization.70 Oligomerization of POR in an experimental reaction mixture was recently reported,82 and dimerization or oligomerization of POR proteins isolated from in vivo systems has been observed both with no pretreatment13 and following use of protein cross-linkers.17 The interface between two monomers of a protein in a dimer or oligomer is considered to be conserved if it tends to overlap strongly in similar protein structures.83
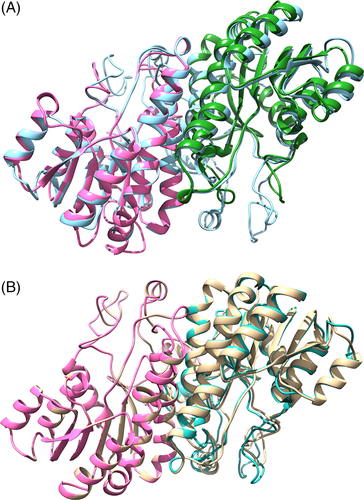
In the 3WXB template, the two monomers interact with each other through α4 and α5 helices of both monomers. Most of the residues involved in dimerization of the template are conserved in POR, according to the sequence alignment. Thus, we modeled the dimeric form of POR using 3WXB in its dimeric form as the template (Figure 4A). To further confirm the dimerization of POR, we also docked the two monomers using the MZDOCK server. The top three predictions of MZDOCK had similar dimeric interfaces to that of the modeled POR dimer (Figure 4B), corroborating the inference that α4 and α7 helices of POR form the dimer interface between pairs of POR monomers.
3.3 Docking of the cofactor NADPH in the POR binding site
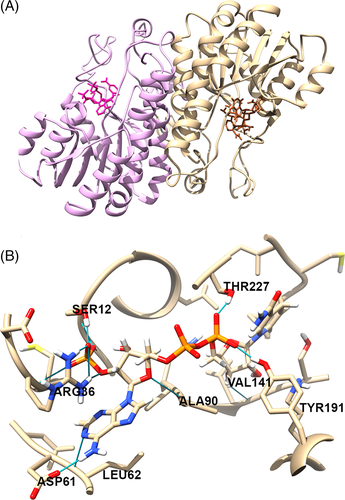
Molecular docking was applied to further elucidate the binding of NADPH in the active site of both chains of POR dimers. A large number of conformers were generated for NADPH using CCDC Mercury software to identify the most stable NADPH binding pose, and the identified NADPH-binding residues in POR (Table 1) were used to guide the POR-NADPH docking analysis. The best docking pose of NADPH in both chains of POR was identified based on GOLD scores and RMSD values between the docked pose and template NADPH, following superposition (Figure 5). The GOLD scores for NADPH in the A and B monomeric chains of POR were 389 and 393 kJ/mol respectively. The NADPH docked in silico in the POR binding site forms twelve hydrogen bonds with Ser12, Arg36, Asp61, Leu62, Ala90, Val141, Tyr191, and Thr227. Of these twelve H-bonding residues, six (Ser12, Arg36, Asp61, Leu62, Tyr191 and Thr227) are the ones conserved with respect to the 3WXB crystal structure.
| PDB_ID | Arg (16) | Glu (67) | Asn (96) |
|---|---|---|---|
| 3SJU | x | - | - |
| 1XU9 | - | x | x |
| 3O26 | - | x | x |
| 1WMA | - | x | x |
| 1N5D | - | x | x |
3.4 Structural similarity of protochlorophyllide (Pchlide) with other molecules
Pchlide is a precursor of chlorophyll, and is structurally similar to chlorophyll but lacks the phytol side chain of chlorophyll and has one more double bond, between C17 and C18 of the porphyrin ring. Pchlide is the major pigment accumulating in dark-grown plants,2, 6 and the substrate of POR, which catalyzes reduction of the C17-C18 double bond of Pchlide to form Chlide, an essential regulatory step in chlorophyll biosynthesis.84 Many studies have shown that the POR-NADPH-Pchlide ternary complex accumulates in vivo when plants are germinating in the absence of light.5, 37, 85, 86
The availability of numerous structures of protein complexes in the PDB database enables comparison of the binding pockets of proteins with different structures that bind similar ligands.87-89 Crystal structures of the light-independent (dark-operative) Pchlide oxidoreductase (DPOR) enzymes bound with Pchlide from Rhodobacter capsulatus (PDB_ID:3AEK) and Prochlorococcus marinus (PDB_ID: 2YNM) have been reported.90, 91 Pchlide binds between the interface of protein subunits A, B and D in the 3AEK structure, and protein subunits C and D in the 2YNM structure. Superposing the two structures 3AEK (A, B) and 2YNM (C, D) revealed high structural similarity (RMSD 1.6 Å), together with high conservation in the binding site and conformation of Pchlide (Table 2).
| 3AEK | 2YNM | Property | |||||
|---|---|---|---|---|---|---|---|
| Chain.ID | A. A. | Type | Chain.ID | A. A. | Type | Source | Same AA |
| B.274 | Asp | ACC | D.45 | Met | ACC | ||
| C.25 | Phe | PII | C.16 | Phe | PII | s | * |
| C.29 | Thr | ALI | C.20 | Thr | ALI | s | * |
| C.33 | Trp | PII | C.24 | Trp | PII | s | * |
| C.53 | His | DAC | C.48 | Ser | DAC | ||
| C.54 | Leu | ALI | C.45 | Leu | ALI | s | * |
| C.57 | Ala | ACC | C.48 | Ser | ACC | ||
| C.58 | Ala | ACC | C.49 | Ala | ACC | b | * |
| C.58 | Ala | ALI | C.49 | Ala | ALI | s | * |
| C.150 | Phe | PII | C.141 | Phe | PII | s | * |
| C.372 | Leu | DON | C.363 | Met | DON | ||
| C.372 | Leu | ALI | C.363 | Met | ALI | ||
| C.387 | Trp | PII | C.378 | Trp | PII | s | * |
| C.389 | Ile | ALI | C.380 | Ile | ALI | s | * |
| D.38 | Tyr | PII | D.38 | Tyr | PII | s | * |
| D.41 | Leu | PII | D.41 | Leu | PII | b | * |
| D.41 | Leu | ALI | D.41 | Leu | ALI | s | * |
| D.42 | Leu | ALI | D.42 | Leu | ALI | s | * |
| D.45 | Met | ALI | D.45 | Met | ALI | s | * |
| D.164 | Leu | ALI | D.177 | Leu | ALI | s | * |
| D.379 | Val | ALI | D.395 | Val | ALI | s | * |
- Note: * identical amino acids between the two structures.
- Abbreviations: A.A., amino acids; ACC, hydrogen bond acceptor; ALI, aliphatic hydrophobic property; DAC, hydrogen bond donor and acceptor; DON, hydrogen bond donor; PII, aromatic property (pi contacts); B, amino acid backbone, buried; s, amino acid side chain, exposed.
To identify the Pchlide binding pocket in POR based on pocket similarity, we performed a chemical structure search using the Pchlide structure as a query against the PDB database with a 90% similarity cut-off. We searched for pockets that accommodate chemically similar or identical ligands, and identified Pchlide (PMR, according to the PDB 3-letter code) bound to the DPOR enzymes and three other chemical structures: chlorophyll b (CHL), chlorophyll a (CLA) and bacteriochlorophyll g (GB0) that share some structural similarity with Pchlide in their closed tetrapyrrole structure (Figure 6A). We picked and compared five crystal structures binding to these four ligands (PMR, CHL, CLA and GB0), and found that most proteins that bind to these structurally similar porphyrin-containing ligands have different folds. The analysis further confirmed that these molecules are capable of binding to pockets with different geometries (Figure 6B), reorienting themselves in different positions within the active sites. Comparison of the binding sites in the five crystal structures showed that four atoms in the porphyrin ring had non-bonded interactions in all the five structures. However, we observed no conservation of the amino acids (Figure S3) involved in interactions with these four atoms (Table 3). Comparing the protein-ligand interactions from five different crystal structures, both 3AEK bound with Pchlide and 5L8R bound with CLA, were observed to have only one hydrogen bonded interaction with O2A and OBD atoms of the ligand, respectively. The porphyrin binding pockets in all these protein structures are hydrophobic as calculated using the Kyte-Doolittle scale47 (Figure 6C).
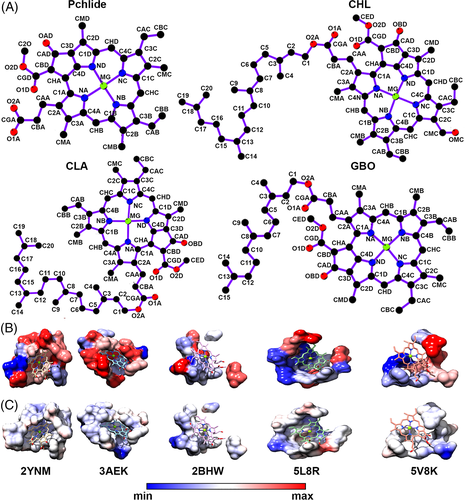
| 2BHW | 2YNM | 3AEK | 5L8R | 5V8K | |
|---|---|---|---|---|---|
| CHB | Leu25A | Met45D His394D | Met45D | Glu1841 Tyr1801 | Trp540A His537A |
| CMD | Tyr44A | Trp378C | Trp387C | Phe1651 | Thr518A Leu605A |
| NB | Leu25A, Tyr24A | Leu41D Ser48C | Ala57C | Glu1841 | His537A |
| NC | Tyr24A | Met363C | Ile389C Ala58C | Glu1841 AegG851 | His537A |
To summarize, analysis of the overall ligand atom-amino acid interactions in the 14 structures show that frequencies of these non-bonded interactions in the five (A-E) rings of the CBB, CBC, CMD, CMA, and O2D ligands were 64, 78, 71, 64, and 57%, respectively (Table 4), strongly suggesting that the interaction of these atoms of the porphyrin ring within the ligand binding pocket are important. Overall, our analysis suggests that these ligands are capable of binding to proteins with diverse folds and active sites, through both the ligands and proteins undergoing adaptive conformational changes that enable them to accommodate their partners in binding sites with surprisingly different shapes. The findings suggest that we cannot easily predict the Pchlide binding site in pea POR, which does not share fold similarity with any of these 14 protein structures. Thus, it will be interesting to explore Pchlide's mode of binding in the undetermined binding site of POR.
| Sub-structure | PDB_ID | C1A | C2A | C3A | C4A | CAA | CBA | CGA | CMA | NA | O1A | O2A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ring A | 1JB0 | ✓ | - | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| 1Q90 | - | - | - | - | - | - | - | - | - | ✓ | - | |
| 2BHW | ✓ | - | - | ✓ | ✓ | ✓ | - | - | ✓ | - | - | |
| 2C9E | - | - | - | - | - | - | - | - | - | - | ✓ | |
| 2DRE | - | - | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| 2YNM | - | ✓ | ✓ | - | ✓ | - | - | ✓ | - | - | - | |
| 3AEK | - | - | - | - | - | ✓ | ✓ | ✓ | - | - | ✓ | |
| 3IIS | - | - | - | - | - | - | - | ✓ | - | - | - | |
| 4OGQ | - | - | - | - | ✓ | - | - | ✓ | - | ✓ | - | |
| 4RI2 | - | - | ✓ | - | - | - | - | ✓ | - | - | - | |
| 5L8R | - | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| 5V8K | - | - | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | ✓ | - | |
| PDB_ID | C1B | C2B | C3B | C4B | CAB | CBB | CMB | NB | ||||
| Ring B | 1JB0 | ✓ | - | - | - | - | - | - | ✓ | |||
| 1PPR | - | - | - | - | - | ✓ | - | - | ||||
| 1Q90 | - | ✓ | ✓ | - | ✓ | ✓ | ✓ | - | ||||
| 2BHW | ✓ | - | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| 2C9E | - | - | - | - | - | ✓ | - | - | ||||
| 2DRE | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| 2YNM | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| 3AEK | - | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| 3IIS | - | - | - | ✓ | - | - | - | - | ||||
| 4OGQ | ✓ | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | ||||
| 4RI2 | - | - | - | - | ✓ | ✓ | ✓ | - | ||||
| 5L8R | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| 5V8K | ✓ | - | - | - | - | - | - | ✓ | ||||
| PDB_ID | C1C | C2C | C3C | C4C | CAC | CBC | NC | |||||
| Ring C | 1JB0 | - | ✓ | - | - | ✓ | ✓ | ✓ | ||||
| 1PPR | ✓ | ✓ | ✓ | ✓ | - | ✓ | ✓ | |||||
| 1Q90 | ✓ | ✓ | ✓ | - | - | ✓ | - | |||||
| 2BHW | ✓ | ✓ | - | ✓ | - | ✓ | ✓ | |||||
| 2C9E | ✓ | ✓ | - | ✓ | - | ✓ | ✓ | |||||
| 2DRE | - | - | - | ✓ | ✓ | ✓ | ✓ | |||||
| 2X20 | ✓ | ✓ | ✓ | ✓ | - | ✓ | ✓ | |||||
| 2YNM | ✓ | ✓ | - | - | ✓ | ✓ | ✓ | |||||
| 3AEK | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||
| 3IIS | ✓ | ✓ | ✓ | ✓ | - | ✓ | ✓ | |||||
| 4OGQ | ✓ | ✓ | ✓ | - | - | ✓ | - | |||||
| 4RI2 | - | - | - | ✓ | ✓ | - | ✓ | |||||
| 5L8R | - | - | - | ✓ | ✓ | ✓ | ✓ | |||||
| 5V8K | - | - | - | - | ✓ | - | ✓ | |||||
| PDB_ID | C1D | C2D | C3D | C4D | CMD | ND | ||||||
| Ring D | 1JB0 | ✓ | ✓ | - | ✓ | ✓ | ✓ | |||||
| 1PPR | - | - | - | - | ✓ | - | ||||||
| 2BHW | ✓ | - | - | ✓ | ✓ | ✓ | ||||||
| 2C9E | ✓ | - | - | - | ✓ | - | ||||||
| 2DRE | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||
| 2X20 | ✓ | - | - | - | ✓ | - | ||||||
| 2YNM | - | ✓ | ✓ | - | ✓ | - | ||||||
| 3AEK | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||
| 3IIS | ✓ | - | - | - | ✓ | - | ||||||
| 4OGQ | ✓ | ✓ | ✓ | ✓ | - | ✓ | ||||||
| 4RI2 | - | - | - | - | ✓ | - | ||||||
| 5L8R | - | - | - | - | ✓ | ✓ | ||||||
| 5V8K | - | - | - | ✓ | ✓ | ✓ | ||||||
| PDB_ID | C2O | C3D | C4D | CAD | CBD | CGD | CHA | O1D | O2D | OAD | ||
| Ring E | 1JB0 | - | - | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | - | |
| 1Q90 | - | - | - | - | - | - | - | ✓ | ✓ | - | ||
| 2BHW | - | - | ✓ | - | ✓ | - | - | - | - | - | ||
| 2DRE | - | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | - | - | ||
| 2YNM | ✓ | ✓ | - | ✓ | - | - | - | - | ✓ | ✓ | ||
| 3AEK | ✓ | ✓ | ✓ | ✓ | ✓ | - | - | ✓ | ✓ | ✓ | ||
| 4OGQ | - | ✓ | ✓ | - | - | - | - | ✓ | ✓ | - | ||
| 5L8R | - | - | - | ✓ | ✓ | ✓ | - | ✓ | ✓ | - | ||
| 5V8K | - | - | ✓ | ✓ | ✓ | - | - | - | ✓ | - |
3.5 Short-chain dehydrogenase protein bound with substrate
To further elucidate the binding of the Pchlide substrate with POR, we searched the available X-ray crystal structures of short-chain dehydrogenases/reductases with structural homology to POR and identified the crystal structure of enoyl-ACP-reductase (INHA) protein from Mycobacterium tuberculosis (PDB_ID: 1BVR) in complex with NAD+ and a C16-fatty-acyl substrate.92 Comparison of the crystal structure of INHA and the modeled structure of dimeric POR revealed very high structural similarity between the two proteins (RMSD of Cα: 3 Å). Comparing the secondary structures close to the substrate-binding region in INHA to that of the POR model, we observed a large insertion and significant structural differences. However, we mapped five amino acids (Pro94, Ser143, Gly229, Leu230 and Arg232) binding to the substrate in INHA to the POR protein based on the POR and INHA structural alignment. In the surroundings of this predicted binding site there are two large structurally flexible exposed loop regions (Gly142-Ala188 and Pro222-Glu256) between β5-α5 and β6-α7, which we hypothesize favor the adaptation of POR to its Pchlide binding partner and facilitate the binding process.
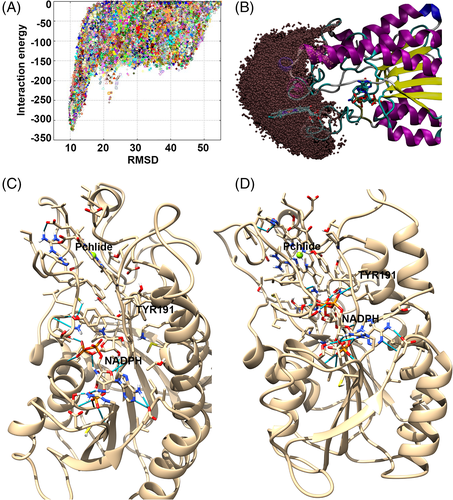
To explore the Pchlide binding mechanism in POR and corroborate the Pchlide binding site, we performed several PELE simulations in which the ligand was allowed to freely explore (and enter) the POR surface. PELE has proven ability to portray porphyrin diffusion, active site searching and binding.93 First, we performed a global search in which the substrate was allowed to explore the entire protein surface. As can be seen in Figure 7A, this procedure returned one main surface binding mode, from which we started a finer local search by exploring a space around up to 20 Å from the initial position (which the ligand's center of mass could not leave, Figure 7B). PELE clearly identified a binding pocket where two poses, differing only in the level of pocket penetration, were selected based on population analysis. The second most populated pose (designated A, Figure 7C), shows a slightly more solvent-exposed Pchlide than the most populated pose (B, which was also best in terms of POR-Pchlide interaction energy according to the PELE simulations), in which the substrate is more deeply bound (Figure 7D). According to earlier studies, the mechanism of Pchlide reduction involves several steps, including: light absorption, subsequent hydride transfer from the pro-S face of the nicotinamide ring of NADPH to C17 of the Pchlide molecule, and finally the transfer of a proton from Tyr280 to C18 of Pchlide.75, 94 To further clarify which of the two poses represents the substrate Pchlide binding pocket in POR most accurately, we compared the positioning of Pchlide and NADPH molecules in the POR complex. In the bound conformation of the Pchlide molecule in pose B (Figure 7D) the C17-C18 bond of Pchlide is close to the pro-S hydrogen from the nicotinamide ring of the NADPH cofactor and Tyr191, which plays an important role in proton transfer to C18 of Pchlide in the catalytic mechanism. In contrast, in pose A the pro-S hydrogen of the NADPH cofactor is far from the C16-C17 bond of the Pchlide molecule. Thus, we predict that the deep docking position of Pchlide in the POR structure is closer to the real substrate binding pocket.
4 CONCLUSION
In this study, the structure of POR protein from pea was predicted in its monomeric and dimeric form. Alpha helices 4 and 7 form the interface between the two POR monomeric structures. Comparison of the NADPH binding site and its conformation within the active site of other NADPH binding proteins showed that it is well conserved, despite low sequence identity. Analysis of the binding sites of tetrapyrrole-containing molecules revealed high variability in the amino acids involved and active site conformation. In addition, exploration of the Pchlide binding mechanism in PELE simulations yielded two binding poses (designated A and B). In pose B the C17-C18 bond of Pchlide is close to the pro-S hydrogen from the nicotinamide ring of the NADPH and Tyr191, which is involved in proton transfer.
ACKNOWLEDGEMENT
he authors are grateful for the following providers of financial support: the Carl Tryggers Foundation (grants CTS 15:34, CTS 17:32 for Hassan Sameer, Aronsson Henrik), the Hungarian Academy of Sciences (a Bolyai János Research Scholarship for Solymosi Katalin), Hungarian Scientific Research Fund (grant OTKA FK 124748 for Solymosi Katalin), and the Spanish Ministry of Science and Innovation (grant PID2019-106370RB-I00 for Guallar Victor). The authors declare that there is no conflict of interest regarding the publication of this article.
AUTHORS CONTRIBUTIONS
Conceived and designed the experiments: Hassan Sameer, Aronsson Henrik. Performed the experiments: Hassan Sameer, Guallar Victor. Analyzed the data: Hassan Sameer, Guallar Victor, Solymosi Katalin, Aronsson Henrik. Wrote the paper: Hassan Sameer, Guallar Victor, Solymosi Katalin, Aronsson Henrik. All authors read and approved the final manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




