Selenomethionine incorporation in proteins expressed in Lactococcus lactis
Abstract
Lactococcus lactis is a promising host for (membrane) protein overproduction. Here, we describe a protocol for incorporation of selenomethionine (SeMet) into proteins expressed in L. lactis. Incorporation efficiencies of SeMet in the membrane protein complex OpuA (an ABC transporter) and the soluble protein OppA, both from L. lactis, were monitored by mass spectrometry. Both proteins incorporated SeMet with high efficiencies (>90%), which greatly extends the usefulness of the expression host L. lactis for X-ray crystallography purposes. The crystal structure of ligand-free OppA was determined at 2.4 Å resolution by a semiautomatic approach using selenium single-wavelength anomalous diffraction phasing.
Introduction
Membrane protein structures are under-represented in the PDB and account for less than 1% of the database entries. A major bottleneck in studying membrane proteins is the difficulty to produce the molecules in their native state in sufficient quantities. It is often worthwhile testing more than one expression host because proteins that express poorly in one host may be produced in large amounts in other organisms. Over the past decade, the gram-positive bacterium Lactococcus lactis has been developed as a robust expression host for (membrane) proteins.1-4 Protocols for cloning, expression, and quality control of membrane proteins in L. lactis are available.1, 2 The organism has proven to be particularly suitable for overexpression of complex, multisubunit proteins (such as ABC and TRAP transporters) and mammalian membrane proteins, often when E. coli failed to deliver.1, 4-8 To fully exploit the potential of the host for production of proteins to be used in crystallographic studies, we have developed a protocol for incorporation of selenomethionine (SeMet) into expressed proteins, which can be used for experimental phasing. Two proteins from L. lactis were tested for SeMet incorporation: the oligopeptide-binding protein OppA without lipid anchor (termed OppA*) was produced in the cytoplasm of L. lactis,9 and the ABC transporter for glycine betaine (OpuA), consisting of the two subunits OpuAA and OpuABC, was produced in the membrane.10 Using mass spectrometry and X-ray crystallography, we show that L. lactis can effectively incorporate SeMet in overexpressed proteins and produce the amounts sufficient for crystallization and structure determination.
Results
Selenomethionine incorporation
L. lactis NZ9000 cells containing the expression plasmids were cultivated in chemically defined medium (CDM) to an OD600 of 1.5, after which the medium was changed to CDM containing SeMet instead of methionine, and expression was induced. OpuA was purified as previously described,11 and details of OppA* purification are presented in the Experimental Procedures section.
The extent of SeMet incorporation in OppA* and OpuA was measured by mass spectrometry. Purified proteins with and without incorporated SeMet were digested with trypsin, and peptides were analyzed using MALDI-TOF MS. Selenium has six different naturally occurring isotopes (74Se 76Se 77Se 78Se 80Se 82Se), five of which have an abundance of more than 5%, making Se-Met-containing peptides readily distinguishable from peptides containing methionine [32S (95%) and 34S (4.3%)] on the basis of their m/z value and the distribution of isotope masses [Fig. 1(A)]. Peptides were matched to the amino acid sequence of each protein, resulting in the identification of tryptic peptides covering 56, 42, and 29% of the protein sequences of OppA*, OpuAA, and OpuABC, respectively. For OppA* the identified peptides contained 9 of the total 14 (seleno)methionines, and for the OpuA complex 18 of 38. The ratio of peak areas from SeMet-containing and wild-type peptides was calculated to estimate the incorporation efficiency. OppA* and OpuAA/OpuABC had 93 and 94% SeMet incorporated, respectively.
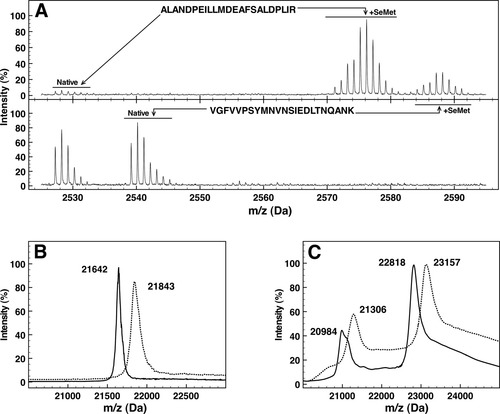
Selenomethionine incorporation in OppA* and OpuA. A: Mass spectra of the tryptic peptides ALANDPEILLMDEAFSALDPLIR and VGFVVPSYMNVNSIEDLTNQANK from OppA* containing SeMet (upper panel) or methionine (lower panel). B, C: Mass spectra of intact OppA* (panel B) and OpuAA/OpuABC (panel C). Continuous lines represent methionine-containing proteins, dotted lines represent proteins with SeMet incorporated. In panel B, the triply charged peaks of OppA* are shown. In panel C, the triply charged peaks of OpuAA (m/z values of 20,984 and 21,306) and the doubly charged peaks of OpuABC (m/z 22,818 and 23,157) are shown.
MALDI mass spectrometry was also used to determine the masses of the intact SeMet-substituted and wild-type proteins. The mass of OppA* increased from 64,923 to 65,526 Da on SeMet incorporation, that of OpuAA from 45,634 to 46,312 Da, and the mass of OpuABC from 62,949 to 63,915 Da, indicating the incorporation efficiencies of 92, 96, and 89%, respectively [Fig. 1(B,C)].
Crystallization and structure determination
OppA* was stripped of any copurified peptide ligands by guanidium chloride treatment, which generates ligand-free OppA*,9 and the protein was crystallized by vapor diffusion as detailed in the Experimental Procedures section. SeMet-substituted OppA* yielded crystals belonging to space group P21, with 1 protein molecule in the asymmetric unit and a solvent content of 42%. Single-wavelength anomalous diffraction (SAD) data were collected at the K-absorption edge of selenium on beamline BM16 at the ESRF, Grenoble. The data were of sufficient quality to allow the structure to be built in a semiautomated manner by SAD phasing, using the Auto-Rickshaw server12 (see Fig. 2). Thirteen of the 14 possible selenium sites were identified and used for phasing, confirming the high efficiency of SeMet incorporation determined by mass spectrometry. Relevant crystallographic statistics are given in Table I. The refined model contains the continuous polypeptide trace from Asn18 to Ala576. Residues 1–17 and 577–590 could not be built because of weak or missing electron density. Similar to other bacterial substrate-binding proteins,13 OppA* consists of two domains with α/β folds connected by a hinge. OppA* is in an open conformation with the two domains separated from each other, leaving the binding site, which does not contain ligand, accessible to the solvent (see Fig. 3). The biological relevance of the structure will be discussed elsewhere (Berntsson et al., in preparation).

Experimental electron density and model of OppA*. A: Stereoview of the anomalous difference Fourier map superimposed on a backbone trace of OppA*. Selenomethionine residues are highlighted in a balls-and-sticks representation (orange). The Se anomalous difference Fourier map, calculated between 61 and 2.4 Å and contoured at 5σ, is shown in blue. Thirteen of a total of 14 SeMet were visible in the map, SeMet at position 1 was in a disordered region of the protein. Peak heights are as follows: MSE-178, 36.3σ; MSE-530, 35.5σ; MSE-62, 32.7σ; MSE-45, 32.6σ; MSE-555, 22.0σ; MSE-457, 21.3σ, MSE-562, 20.2σ; MSE-448, 19.8σ, MSE-182, 17.8σ; MSE-278, 17.5σ; MSE-305, 17.3σ; MSE-300, 14.3σ; MSE-542, 11.7σ. No noise peaks were visible at a 5σ cutoff. B: Electron density (2Fo-Fc map contoured at 1.5σ) showing part of OppA*, visualizing the good quality of the experimentally derived phases. The protein is shown in a stick representation.
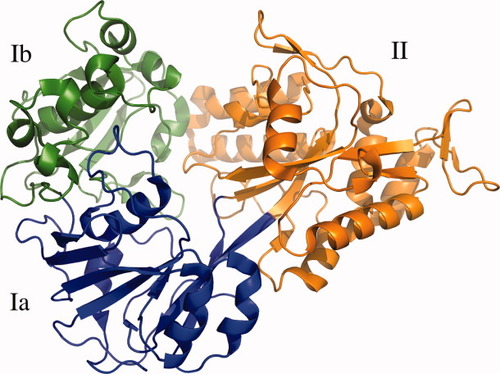
Ribbon representation of OppA* in an open unliganded conformation. The refined model contains a continuous polypeptide trace from Asn18 to Ala576. The two α/β domains are made up of residues 18–300 and 543–572 (domain I, green and blue), and residues 301–572 (domain II, orange). Domain I has two subdomains: domain Ia (residues 18–82, 220–300, and 543–573, blue) and domain Ib (residues 83–219, green). The hinge region corresponds to the segments that link domain Ia and domain II, i.e., the blue to orange transitions.
| Data collection | |
|---|---|
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 39.6, 122.3, 59.4 |
| a, b, g (°) | 90, 104, 90 |
| Wavelength (Å) | 0.979 |
| Resolution range (Å) | 61–2.4 |
| Rsym | 0.079 (0.178) |
| I/σ (I) | 6.1 (4.0) |
| Completeness (%) | 99.6 (100.0) |
| Redundancy | 6.0 |
| SAD phasing | |
| Resolution (Å) | 19.8–2.38 |
| Cullis R-factor | 0.67 |
| FOMacentric | 0.272 |
| Refinement | |
| Resolution (Å) | 61.2-2.4 |
| Number of reflections | 20965 |
| Rwork/Rfree | 0.203/0.257 |
| No. atoms | |
| Protein | 4355 |
| Water | 290 |
| Average B-factors (Å2) | |
| Protein | 30.048 |
| Waters | 37.7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 1.362 |
- a The number in parentheses corresponds to the highest resolution shell.
Discussion
SeMet incorporation in proteins expressed in L. lactis expands the utility of the organism as protein production host. The first expression host used for SeMet incorporation was Escherichia coli,14 but in the last decade, SeMet has been incorporated into proteins produced in Baculovirus,15, 16Saccharomyces cerevisiae,17Pichia pastoris,18, 19 and mammalian cells.20, 21 The SeMet incorporation efficiency of L. lactis is comparable to that of E. coli and superior to that of the eukaryotic expression systems.14L. lactis has proven to be a well-suited host for (eukaryotic) membrane protein overexpression.1, 4-8 The possibility to efficiently incorporate SeMet into those proteins makes the organism a complete protein production host for crystallography purposes.
Experimental Procedures
Selenomethionine incorporation
For inducible production of OppA* in L. lactis NZ9000,22 the oppA* gene9 was inserted into vector pNZcLIC, using ligation-independent cloning.1 Production of the ABC transporter OpuA in L. lactis NZ9000 from plasmid pNZOpuAhis was done as previously described.11 A 10-mL preculture was grown overnight at 30°C in M17 medium (Oxoid), supplemented with glucose (0.5%) and chloramphenicol (5 μg/mL). The preculture was used to inoculate 1 L of CDM composed of 10 mL vitamin mix, 10 mL metal mix, 10 mL base mix, 50 mL amino acid mix, 1 mL MnSO4 × H2O, 0.25 g cysteine–HCl, and 919 mL basic medium (see Table II for the exact composition of the CDM), and containing 5 μg/mL chloramphenicol and 1% glucose. The culture was grown in a 2-L bioreactor (Applikon Biotechnology) at 30°C, pH 6.5, stirred at a speed of 200 rpm, until the OD600 reached a value of 1.5. The cells were spun down in sterile centrifuge tubes for 10 min, 10,000g, 30°C, washed with 150 mL buffer (50 mM KPi pH 7.0, preheated to 30°C), spun down again, and subsequently resuspended to the same OD600 in 1 L CDM, with SeMet (0.84 mM) instead of methionine, and containing 5 μg/mL chloramphenicol plus 1% glucose. The culture was put back in the bioreactor and incubated for 20 min before inducing the expression with 0.05% (v/v) of culture supernatant of the nisin A producing strain NZ9700.22 The cells were grown until the OD600 reached a value of 3–5 and then harvested. Cells were resuspended to an OD600 of ∼200 in 50 mM Tris-HCl pH 7.8, 300 mM NaCl (buffer A), frozen in liquid nitrogen, and stored at −80°C.
| Amino acid mix, 20× solution (adjusted to pH 7.0, filter sterilized, heated to 50°C) | mg/L | Vitamin mix, 100× solution (adjusted to pH 7.0, filter sterilized) | mg/L |
|---|---|---|---|
| L-Alanine | 237.5 | Nicotinic acid | 200 |
| L-Glutamine | 390 | Thiamine dichloride | 100 |
| L-Asparagine | 350 | Riboflavin | 100 |
| L-Arginine | 125 | Ca-(D+)pantothenate | 100 |
| L-Lysine | 437.5 | K-p-Aminobenzoate | 1000 |
| L-Isoleucine | 212.5 | D-Biotin | 1000 |
| L-Methionine | 125 | Folic acid | 100 |
| L-Phenylalanine | 275 | Vitamin B12 | 100 |
| L-Serine | 337.5 | Orotic acid | 500 |
| L-Threonine | 225 | 2-Deoxythymidine | 500 |
| L-Tryptophan | 50 | Inosine | 500 |
| L-Valine | 325 | DL-6,8 Thioctic acid | 250 |
| Glycine | 175 | Pyridoxamine dichloride | 500 |
| L-Histidine | 150 | Pyridoxal-chloride | 200 |
| L-Leucine | 475 | Base mix, 100× solution (in 0.2M NaOH) | g/L |
| L-Proline | 675 | Adenine | 1 |
| L-Aspartate | 0 | Uracil | 1 |
| Metal mix, 100× solution (in H2O) | g/L | Xanthine | 1 |
| MgCl2 × 6H2O | 20 | Guanine | 1 |
| CaCl2 × 2H2O | 5 | Basic medium (set pH to 6.4 with HCl, autoclaved) | g/L |
| FeCl2 × 4H2O | 0.5 | Tyrosine (in boiling H2O) | 0.29 |
| ZnSO4 × 7H2O | 0.5 | KH2PO4 | 2.5 |
| CoCl2 × 6H2O | 0.3 | K2HPO4 | 3 |
| CuSO4 × 5H2O | 0.02 | (NH4)3-citrate | 0.6 |
| Separately | Na-Acetate | 1 | |
| MnSO4 × H2O | 28 | ||
| Per liter CDM | |||
| Amino acid mix | 50 mL | ||
| Vitamin mix | 10 mL | ||
| Metal mix | 10 mL | ||
| Base mix | 10 mL | ||
| MnSO4 × H2O | 1 mL | ||
| Basic medium | 919 mL |
- a The Vitamin mix, Amino acid mix, and the Base mix can be stored at −20°C. The Metal mix has to be made fresh. The Basic medium should be autoclaved and stored in the dark.
Purification of OppA*
Frozen cells were thawed at room temperature and diluted twofold with buffer A. Subsequently, 100 μg/mL deoxyribonuclease type I, 10 mM MgSO4, and 0.1 mM phenylmethanesulfonyl fluoride were added. The cells were broken by two consecutive passes in a cell disruptor (Constant Systems) at 39,000 psi, 5°C. Unbroken cells and cell debris were removed by ultracentrifugation, 267,000g, 4°C for 75 min. The supernatant was collected (resulting in ∼50 mL lysate per liter culture), and 0.5 mL of Ni2+-sepharose resin (Amersham Biosciences) was added per 50 mL of lysate. The mixture was incubated for 1 h at 4°C in buffer A supplemented with 15 mM imidazole and 10% (v/v) glycerol. Subsequently, the resin was poured into a disposable column (BioRad) and washed with buffer A containing 50 mM imidazole, for 20 column volumes (CV). Bound peptides were removed from OppA* by partially unfolding the protein while bound to the resin. The following wash steps were performed (all in buffer A with 15 mM imidazol): 40 CV with 2M guanidine–HCl (GndHCl), 4 CV with 1.5M GndHCl, 4 CV with 1M GndHCl, 4 CV with 0.5M GndHCl, and finally 8 CV with 0M GndHCl. The protein was eluted with 20 mM Na-MES, pH 6.0, 300 mM NaCl, and 500 mM imidazole, pH 6.0, 2 CV.
Purified SeMet-OppA* was concentrated to 0.5 mL in spin concentrators with a 30 kDa cutoff (Vivaspin with PES membrane from Sartorius) and further purified on a Superdex 200 10/300 GL size exclusion column (Amersham Biosciences) in 20 mM Na-MES, pH 6.0, 150 mM NaCl. Fractions containing OppA were pooled, concentrated 10-fold, and diluted such that the final buffer composition was 10 mM Na-MES, pH 6.0, 10 mM NaCl, and finally concentrated again to 11 mg/mL of protein. The final yield was about 1 mg SeMet-OppA* per liter culture, which is comparable to that of OppA* produced in L. lactis grown in normal CDM.
Purification of OpuA
OpuA was purified as described.11 For SeMet-OpuA, the final yield was 50 μg of pure protein from 10 mg of total membrane protein, which is similar to the yields of OpuA produced in L. lactis grown in normal CDM.
Mass spectrometry analysis of SeMet-substituted proteins
Ten micrograms of purified protein was subjected to digestion for 18 h with 125 ng porcine trypsin (MS-grade, Promega, Leiden, The Netherlands) in 200 mM triethylammonium bicarbonate, pH 8.0, 10% acetonitrile. The peptide digests were mixed 1:1 v/v with a solution of α-cyano-4-hydroxycinnamic acid matrix (5 mg/mL in 50% ACN and 0.1% TFA, LaserBio Labs), spotted directly onto stainless steel MALDI targets, and analyzed using either a 4700 or 4800 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems). For the digested peptide samples, the MALDI-TOF/TOF was operated in reflector positive ionization mode in the m/z range 700–4000. MS peak-lists were extracted by GPS Explorer Software, version 3.5 (Applied Biosystems), applying default parameters. A list of theoretical tryptic peptides, obtained with the program GPMAW, version 7.10sr2 (Lighthouse), allowing for one missed cleavage, was used to interpret the MS spectra, estimating an average increase in m/z of 47 Da for each SeMet.
For mass spectrometry analysis of the intact OppA*, the protein was mixed 1:1 v/v with a solution of α-cyano-4-hydroxycinnamic acid matrix (5 mg/mL in 50% ACN and 0.1% TFA, LaserBio Labs), spotted onto stainless steel MALDI targets, and analyzed using either a 4700 or 4800 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems) in the linear mode. For mass spectrometry analysis of the intact OpuA complex, the purified protein was spotted using the ultra thin layer method to increase the signal to noise levels.23
Crystallization and structure determination
Crystals of SeMet-substituted OppA* were grown by vapor diffusion in hanging drops. The drops consisted of 1 μL protein (10 mg/mL OppA*, 9 mM Na-MES, pH 6.0, 9 mM NaCl, and 1 mM peptide (YGGFL)) and 1 μL reservoir solution (0.2M NaCl, 0.1M Na-Hepes, pH 7.0, 20% PEG 6000). Diffracting crystals were obtained after 24 h of incubation at room temperature. Crystals were soaked in mother liquor supplemented with 42% PEG 6000 for 30 s and then flash cooled in liquid nitrogen. Single-wavelength dispersion (SAD) data to 2.38 Å resolution were collected at the selenium K-absorption edge (wavelength: 0.979 Å) on beamline BM16 at the ESRF, Grenoble, using a single crystal (0.15 mm × 0.15 mm × 0.2 mm). Data processing and reduction were carried out using HKL2000.24 Data were submitted to Auto-Rickshaw, and the structure was solved using the SAS protocol of Auto-Rickshaw: the EMBL-Hamburg automated crystal structure determination platform.12 The input diffraction data were prepared and converted for use in Auto-Rickshaw using programs of the CCP4 suite.25 FA values were calculated using the program SHELXC. Based on an initial analysis of the data, the maximum resolution for substructure determination and initial phase calculation was set to 2.38 Å. Thirteen heavy atoms out of the maximum number of 14 were found using the program SHELXD.26 The correct hand for the substructure was determined using the programs ABS27 and SHELXE.26 The occupancy of all substructure atoms was refined and initial phases were calculated using the program MLPHARE.25 Density modification and phase extension were performed using the program DM.28 The model (39.8%) was built using the program ARP/warp.29 Auto-Rickshaw returned the partially built model, which was subsequently resubmitted to Auto-Rickshaw using the MRSAD protocol, combining SAD data with molecular replacement of the partially built model. The calculated electron density was of sufficient quality to let ARP/wARP build 89% of the residues, which Auto-Rickshaw returned. A few cycles of refinement using Refmac5,30 interspersed with manual model building using Coot,31 were necessary to complete the model. Water molecules were placed automatically in Fo−Fc Fourier difference maps at a 3σ cutoff level and validated to ensure correct coordination geometries using Coot. Relevant statistics of the data collection, phasing, and model refinement is given in Table I.
Accession Codes
The coordinates have been deposited in the Protein Data Bank with accession code 3FTO.
Acknowledgements
The authors thank Dr. Hjalmar Permentier and Wim Huibers for help with mass spectrometry analysis and the ESRF for providing excellent beam line facilities.




