Development and Validation of Mass Spectrometry-Based Targeted Analysis for Amyloid Proteins
Abstract
Purpose
Amyloidosis is caused by the extracellular deposition of insoluble fibrils and results in multiple organ dysfunctions. Accurate typing of amyloid is mandatory to decide the optimal treatment for amyloidosis.
Experimental design
A mass spectrometry-based test combined with laser microdissection (LMD/MS) has been widely used even in the tricky cases that cannot be discriminated by immunohistochemical staining. However, shotgun proteomics analysis obligatorily requires not only expensive instruments but also highly-experienced specialists to interpret the analysis. Thus, a method of LMD combined with multiple reaction monitoring mass spectrometry (MRM-MS) that can detect three most common types of amyloidosis is developed.
Results
The diagnostic method is tested in 16 Congo red-positive tissues and 10 Congo red-negative control tissues. The results showed better performance in amyloidosis typing than other analytical techniques.
Conclusion and clinical relevance
This novel diagnostic approach can achieve successful amyloidosis typing with high specificity and sensitivity and be implemented more easily in general clinical laboratories.
Amyloidosis is caused by the misfolding, aggregation, and accumulation of abnormal amyloid fibrils in tissues.1 To date, 31 extracellular fibril proteins have been identified.2 The three major types of systemic amyloidosis are as the following; light chain (AL), amyloid A (AA), and transthyretin (ATTR) amyloidosis, etc., and these types comprise more than 90% of amyloidosis.2, 3 Those deposited proteins are immunoglobulin light chains including the lambda (IGL) and kappa (IGK) in AL type, serum amyloid A (SAA) in AA type, and transthyretin (TTR) in ATTR type.
Accurate typing of amyloidosis is indispensable, because the treatment strategies are drastically different according to the types of amyloidosis. To differentiate the specific type of amyloidosis, a special staining of biopsy specimens with an immunohistochemical (IHC) stain is frequently used. However, the use of IHC for this purpose has several limitations: (1) the inevitable epitope loss during the process of protein cross-linking after formalin fixation and (2) the unmet specificity and sensitivity of the conventional antibody-based determination.4, 5 Furthermore, accurate visual discrimination of intensity among different IHC stains can be challenging. Thus, a novel method that combines laser microdissection (LMD) and MS has been introduced as an advanced method overcoming the limitations of antibody-based platforms and is currently considered as the gold-standard technique for identifying the causative protein of amyloidosis. In fact, these techniques have been successfully applied in typing amyloidosis and show better performance than IHC staining.6-8 Nevertheless, this method has some barriers to being adopted in all general clinical laboratories, because its interpretation is difficult and requires a well-trained mass spectrometrist. Standardization for clinical implementation is also difficult.8
Multiple reaction monitoring (MRM) has been widely used for the determination of a small molecule, and has recently been increasingly accepted for protein quantification due to the easy availability of development and standardization to allow clinical utilization. Therefore, we established an MRM assay combined with laser microdissection for the amyloidosis subtyping in a clinically applicable format. MRM-MS evaluated four amyloid proteins, IGL, IGK, SAA, and TTR, which are involved in the development of the three most common types of amyloidosis. The proteotryptic peptides with high ion efficiency in MS and MS/MS scans were selected. These MS/MS scans were performed for the acquisition of the best Q1/Q3 pairs (MRM transition) with high specificity for each amyloid protein (Supporting Information Figure S1). Four peptides were finally selected for amyloidosis typing, and at least three transitions from a proteotryptic peptide were chosen and optimized. Of these, the quantitative ion pair was used to quantify those proteins (Supporting Information Table S1).
The peak areas corresponding to each peptide selected were calculated by MultiQuant software (SCIEX, Framingham, MA, USA) with the following parameters: smoothing width, 2 points; minimum peak width, 3 points; noise percentage, 40%; baseline subtraction of window, 2 min; and peak splitting factor, 2 points. The peak area ratios were calculated to represent their concentrations in amyloid deposits as follows: the peak area of each peptide transition was divided by the peak area of the corresponding transition from stable isotope-labeled internal standard (SIS) peptide. The concentration of each of these proteins was calculated as the product of its peak area ratio and the concentration of SIS.
To validate the MRM assay developed, we used 16 amyloid and ten nonamyloid affected tissues from the kidney, heart, and liver, which were pathologically confirmed in their biopsy specimens and approved by the Institutional Review Board of Samsung Medical Center (Seoul, Korea)(IRB number 2013-04-087). We compared those results from MRM-MS with those of shotgun proteomics data and IHC.
Briefly, 10 μm thick paraffin-embedded tissue sections were made on the window slide for LMD (Jungwoo F&B, Incheon, Korea), and stained with hematoxylin and eosin. Pink amorphous materials in the interstitial space, which were birefringent under polarized microscope on the serial section stained by Congo-red, were micro-dissected with an LMD system (ION LMD-II, Jungwoo F&B). The dissected area of each case was 0.25∼1 × 105 μm2, and then applied to MRM-MS or shotgun proteomics analyses (Figure 1).
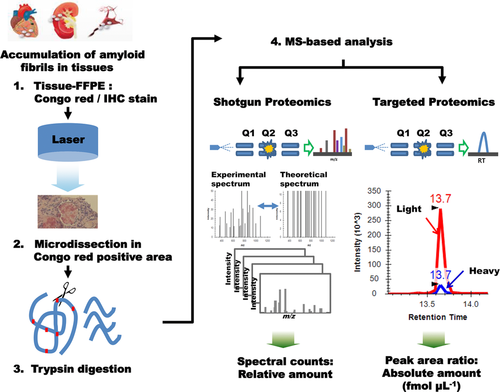
After that, each tryptic digestion sample was quantitatively analyzed after being spiked with SIS peptide mixture (100 fmol μL–1) (Supporting Information Table S1). The tryptic peptides were loaded onto nanocHiPLC columns (75 μm × 15 cm ChromXP C18-CL 3 μm 300 Å; SCIEX) and separated by the EksigentNanoLC-1D plus system combined with the cHiPLC-Nanoflex system. MRM scans were performed using a QTRAP 5500 (SCIEX) in the positive mode.
For shotgun proteomics analysis, samples were loaded at a 3 μL min–1 flow rate and eluted at 250 nL min–1 using a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer interfaced with an EASYnano LC system (Thermo Fisher Scientific, Waltham, MA, USA) equipped with an EASY-Spray column (50 cm × 75 μm internal diameter, PepMap RSLC C18, 2 μm). A full-scan Orbitrap mass analyzer run (400–2000 m/z, 70 000 resolution) and ten data-dependent MS/MS scans (27% normalized collision energy) were performed. All MS/MS data were searched against the Uniprot human database using SEQUEST(Thermo Fisher Scientific, San Jose, CA, USA; version 1.3.0.339). Scaffold (version Scaffold 3.6.4, Proteome Software Inc., Portland, OR, United States) was used to validate the MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the PeptideProphet algorithm.9 Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the ProteinProphet algorithm.
We established MRM assays for amyloidosis typing (e.g., distinguishing IGK, IGL, SAA, and TTR). To evaluate the assay performance, a set of linearity standards was tested in triplicate for each amyloidogenic protein. The assay performance was successfully validated and found to be linear in the concentration range of 1∼1000 fmol μL–1 (R2 = 0.999), 1∼1000 fmol μL–1 (R2 = 0.998), 0.5∼400 fmol μL–1 (R2 = 0.985), and 0.5∼400 fmol μL–1 (R2 = 0.999) for IGK, IGL, SAA, and TTR, respectively (Supporting Information Figure S2). The LOD (defined by a S/N greater than 10) were estimated to be 1, 1, 0.5, and 1 fmol μL–1 for IGK, IGL, SAA, and TTR, respectively. We also analyzed ten control tissues of nonamyloid patients to prevent unwanted interference by the components of blood, and the relatively low amyloidogenic protein levels in the control tissues ranged from 0.1 to 1 fmol μL–1. A cutoff value of 2 fmol μL–1 was adopted to identify each amyloidogenic protein.
We validated a total of 16 amyloidosis cases with Congo-red positive specimen using MRM-MS; nine patients had their subtypes confirmed by IHC methods, and for the remaining seven patients whose subtypes were not confirmed by IHC. Eleven cases were confirmed based on shotgun proteomics analysis (Table 1). When the IHC results showed strong positive staining for kappa or lambda in the AL type (suggesting monoclonal gammopathy), TTR in the TTR type and AA in the AA type, this was considered successful IHC typing. On the other hand, when no amyloidogenic proteins were specifically detected or two or more proteins were detected, the IHC findings were nondiagnostic. In shotgun proteomics analysis, amyloidosis types were identified based on the largest number of spectra matching to a protein.
| Case | Tissue | CR | IHC | Subtype by IHC | Shotgun (Spectral Counts) | Subtype by Shotgun | MRM-MS (fmol uL–1) | Subtype by MRM | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AL-IGK | AL-IGL | SAA | TTR | AL-IGK | AL-IGL | SAA | TTR | AL-IGK | AL-IGL | SAA | TTR | ||||||
| 1 | Kidney | + | − | − | + | − | AA | − | − | 11 | − | AA | 6 | − | 756 | − | AA |
| 2 | Kidney | + | NA | NA | NA | NA | NA | 7 | − | 45 | − | AA | − | − | 1194 | − | AA |
| 3 | Heart | + | − | − | + | − | AA | NA | NA | NA | NA | NA | − | − | 32 | − | AA |
| 4 | Heart | + | + | − | − | − | AL | 5 | − | − | − | AL | 1841 | − | − | − | AL |
| 5 | Liver | + | + | + | − | − | Unknown | 26 | − | − | − | AL | 43 | − | − | − | AL |
| 6 | Heart | + | + | − | − | − | AL | NA | NA | NA | NA | NA | 34 | − | − | − | AL |
| 7 | Liver | + | + | − | − | − | AL | NA | NA | NA | NA | NA | 183 | 2 | − | − | AL |
| 8 | Kidney | + | − | − | NA | NA | Unknown | 12 | 12 | − | − | AL | 2 | 13 | − | − | AL |
| 9 | Heart | + | − | + | − | − | AL | − | − | − | − | Unknown | − | 8 | − | − | AL |
| 10 | Heart | + | − | + | − | − | AL | NA | NA | NA | NA | NA | − | 7 | − | − | AL |
| 11 | Heart | + | − | − | − | + | ATTR | − | − | − | 7 | ATTR | − | − | − | 2 | ATTR |
| 12 | Colon | + | − | − | − | + | ATTR | NA | NA | NA | NA | NA | − | − | − | 46 | ATTR |
| 13 | Kidney | + | − | − | NA | NA | Unknown | 2 | − | − | − | AL | 3 | − | − | − | AL |
| 14 | Heart | + | + | + | − | − | Unknown | 24 | 40 | − | − | AL | 26 | 4 | − | − | AL |
| 15 | Heart | + | − | − | − | − | Unknown | 13 | 24 | − | − | AL | 500 | 183 | − | − | AL |
| 16 | Heart | + | − | − | − | − | Unknown | 17 | 13 | − | − | AL | 439 | 87 | − | − | AL |
- NOTE. A total of 16 patients with amyloidosis (ten patients with definite IHC results and six patients without) were examined by shotgun and MRM analysis. Negative indicates negative results for each protein in each analysis.
- Abbreviations: NA, not available; CR, congo red stain; AL, light chain amyloidosis; AA, amyloid A amyloidosis; ATTR, transthyretin amyloidosis.
MRM-MS identified all amyloidogenic protein in 16 cases, including seven cases not conclusive by IHC (cases 2, 5, 8, 13–16) and one case not detected by shotgun proteomics (case 9). Cases 5 and 13 demonstrated nonspecific intensities on both kappa and lambda staining, which indicate a failure to demonstrate light chain monoclonality on amyloidosis infiltration, but the MRM-MS successfully displayed the same results as shotgun proteomics. In case 9, shotgun MS failed to detect any amyloidogenic proteins, while the MRM-MS method successfully identified IGL, which indicates the excellent sensitivity of MRM-MS.
Key issues for a diagnostic test in routine clinical laboratory environments are its applicability, standardization and analytical sensitivity in a small tissue sample. MRM-MS has advantage over shotgun analysis in that it is easy to be standardized, can be constructed by less-experienced operator with less expensive instrument, and more sensitive than shotgun proteomics analysis. Our MRM-MS based amyloidosis typing showed much better performance than those of IHC or shotgun proteomics analysis.
Overall, we successfully established a novel assay to determine the most common types of amyloidosis with an easy and applicable technology.
Abbreviations
-
- AA
-
- amyloid A
-
- AL
-
- light chain amyloidosis
-
- ATTR
-
- transthyretin amyloidosis
-
- FFPE
-
- formalin-fixed and paraffin-embedded
-
- IGK
-
- immunoglobulin kappa chains
-
- IGL
-
- immunoglobulin lambda chains
-
- IHC
-
- immunohistochemical
-
- LMD
-
- laser microdissection
-
- MRM
-
- multiple reaction monitoring
-
- SAA
-
- serum amyloid A
-
- SIS
-
- isotope-labeled internal standard
-
- TTR
-
- transthyretin
Acknowledgments
J.P. and G.Y.L. contributed equally to this work. This work was supported by the Research Program funded by the Korea Centers for Disease Control and Prevention (2013-E63011-11) and a grant of Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HI13C2098, A120175).




