Tuning of Ultrasmall Gold Nanoparticles Surface Properties Affect Their Biological Fate
Abstract
Ultrasmall nanoparticles of 10 nm or less in size have been shown to have great potential in the biomedical field due to their high surface area and strong tissue penetration. Their easy functionalization and unique behavior at the nanoscale, such as the reduced corona formation and lower liver retention allow them to be a potential tool for precision targeting. In this study, PEGylated ultrasmall gold nanoparticles (GNPs) with a 4 nm core size are developed. They are functionalized with the cyclic RGD (cRGD) targeting peptide, which provides high binding affinity toward αVβ3 integrin receptor, often overexpressed in solid tumors. Further evidence is presented that cRGD functionalized GNPs partially escape lysosomes while penetrating deeper into the liver parenchyma. These particles provide a potential future strategy for specific αVβ3 integrin targeting.
1 Introduction
Gold nanoparticles (GNPs) have become extremely popular in nanomedicine, because of their great biocompatibility, surface plasmon resonance, and optical properties that make GNPs very attractive for several distinct biomedical applications.
Because of their well-established synthesis, which allows controlled changes in size and shape, multiple chemical routes are available for their surface functionalization with polymers, such as polyethylene glycol (PEG).[1-6]
PEG is a commonly used polymer, generally well tolerated by the organisms, that increases the nanomaterial circulation half-life and reduces interactions with proteins from the biological milieu,[6] decreasing the material's opsonization.[7-9] PEG used as a linker serves as a binding site for active biomolecules that can be functionalized to enhance the specificity of the interactions between the nanomaterial and its designed target. Despite the progress that has been achieved in the use of nanoparticles (NPs) for drug delivery, there are still unmet challenges delaying clinical translation for medical treatments. In biological matrices, biomolecules from the surrounding media strongly interact with the NP's surface, forming a biomolecular corona, often affecting the NP's physico-chemical properties,[10-12] thus hindering their ability to reach their intended target.[13-15] In addition, most NPs, following intracellular uptake, accumulate in lysosomes, limiting the therapeutic opportunities for drug delivery applications.
Recent studies have highlighted the potential of using small and ultrasmall GNPs (core < 10 nm) over larger NPs. Advantages such as efficient renal clearance, high tissue penetration, and a higher surface area for conjugation make these constructs attractive for drug delivery.[16-18]
Moreover, when compared to bigger NPs, ultrasmall constructs appear to differ in the endocytosis route. Although clathrin-mediated uptake has been identified as the most common pathway for NPs internalization,[19] small nanomaterials can enter cells through caveolin-dependent pathways as well.[20] These differences could lead to variations in intracellular trafficking that can be further exploited for therapeutic goals.[21, 22] Additionally, ultrasmall nanomaterials’ size is comparable to large protein aggregates and can lead to completely different interactions with blood proteins compared to larger ones. For instance, it has been reported that ultrasmall NPs may act as load rather than cargo due to being smaller than some proteins.[23, 24] While larger NPs translocate in the liver after short exposure times and are retained for a long period without degradation,[25] ultrasmall NPs have the potential to have a lower liver accumulation and therefore have a longer circulation half-life, which may promote their active targeting capability with potential application in biosensing and imaging to cancer therapies.[17] In addition, ultrasmall NPs also have high cargo-loading efficiency and can avoid the formation of a stable protein corona, unlike their larger counterparts.[26]
Integrins are a family of transmembrane receptors that are involved in both cell-cell and cell-extracellular matrix (ECM) protein interactions. In particular, integrin αVβ3 is overexpressed in many distinct pathological conditions, from solid tumors to stroke, and it has become a very attractive target receptor.[24-27] One of the main ligands used to target several integrins is the peptide sequence arginine-glycine-aspartate, commonly known as RGD. This motif can be found in several components of the ECM and contributes to the modulations of cellular behaviors, such as signaling, proliferation, migration, or differentiation. While being recognized by several integrins, the affinity of RGD toward different isoforms varies.[28-31] In the context of NPs surface functionalization, using a small ligand rather than a full protein offers several advantages. These include, in addition to simple synthesis and characterization, low steric hindrance, which allows the attachment of multiple copies of the ligand and the ideal ligand orientation for optimal interaction with the targeted molecule.[32-35]
In this study, we aimed to combine the enhanced properties of ultrasmall-sized constructs with the active targeting of the RGD peptide. For that purpose, we developed ultrasmall GNPs conjugated with cRGD peptide, which interacts more strongly with the integrin αVβ3 than its linear conformation.[36] Here, we show that the greater affinity of cRGD GNPs toward αVβ3 integrin depends on the peptide density. For this reason, after multiple characterization assays, the GNPs solution with the highest efficiency was selected. As a negative control, cyclic arginine-glycine-aspartate (cRAD) was also attached to the nanomaterial surface, where the replacement of glycine with alanine causes a substantial loss of affinity. Moreover, we developed a broad characterization study that successfully identified the ideal surface coating that would lead to optimal receptor recognition and cellular uptake, including in the presence of complex media. Our data show that cRGD GNPs were capable of partially avoiding lysosomal colocalization, and in vivo results point to different behavior between cRGD and cRAD constructs, highlighting the potential of ultrasmall GNPs as an attractive platform for targeted and specific delivery.
2 Results
2.1 NPs Synthesis, Characterization and Functionalization
In this study, we synthesized plasmonic ultrasmall GNPs with an overall diameter of ≈4 nm through a modified Brust Schiffrin synthesis, as described in previously published works.[37] Briefly, HAuCl4 gold salts were reduced in situ with sodium borohydride in the presence of a substoichiometric amount of a bifunctional (carboxylated and thiolated) PEG of 0.5 kDa, along with thiolated α-galactose. The latter was used as a spacer to control the PEG packing density. The size of the GNP core was controlled by tuning several parameters, such as pH, temperature, and reaction time, and by changing the ratio between the two thiolated ligands (Table S1, Supporting Information). Among the several candidates, we chose GNP-5 for further functionalization because of its optimal colloidal stability and a desirable core size of <5 nm.
Next, PEGylated GNPs were functionalized with the cRGD integrin-specific tripeptide modified with an amino-terminal linker to ensure correct peptide conjugation and orientation. The peptide conjugation was performed through EDC-NHS coupling of the PEG previously attached on the gold core. In the same way, the integrin non-specific cRAD peptide[38-40] was conjugated to the PEGylated GNPs. Following peptide functionalization, the fluorophore Sulfo-Cyanine5 was linked to the GNPs via the same EDC-NHS chemistry to allow the GNPs detection and tracking (Figure 1A).
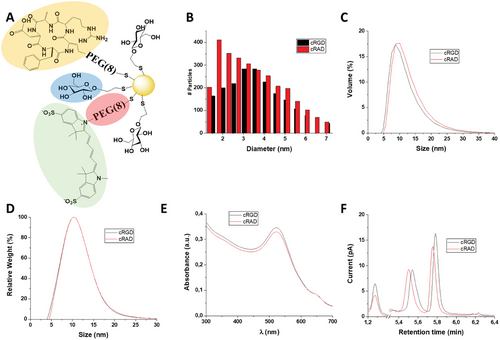
The multi-step functionalization process of the GNP core did not alter the shape and size of the particles, as confirmed by transmission electron microscopy (TEM) analysis (Figure 1B; Figure S1A and Table S1B, Supporting Information), Dynamic Light Scattering (DLS), and Differential Centrifugal Sedimentation (DCS). Collectively, the data show comparable size distributions to the non-functionalized particles (Figure 1C,D).
To further characterize the peptide conjugation on the GNPs’ surface, we employed Liquid Chromatography (LC) and Mass Spectrometry (MS). LC coupled with charged aerosol detector (LC-CAD) analysis indicated the presence of α-galactose along with the functionalized PEG with cRGD or cRAD, and the presence of unreacted PEG (Figure 1F).
Finally, the presence of the Sulfo-Cy5 conjugates was confirmed by UV–vis spectroscopy as the presence of a second band ≈ λ = 650 nm, while preserving the core's SPR peak at λ = 520 nm (Figure 1E).
The presence of the fluorophore was further confirmed by agarose gel electrophoresis. The GNP migrating band, detectable by the eye, overlapped with the fluorescent band from the fluorophore (Figure S2, Supporting Information). These characterization data are in agreement with a previous study, confirming the robustness of the synthesis and functionalization process.[35]
2.2 Measurement of Affinity Via Biolayer Interferometry
After confirming the successful peptide conjugation, we further variated their amount in the reaction to obtain particles with different peptide densities. To evaluate whether different levels of the peptide would affect the interaction with the αVβ3 integrin receptor, we used BioLayer Interferometry (BLI) as previously described.[37] Briefly, integrin αVβ3 was immobilized on high-precision streptavidin tips, followed by exposure to different concentrations of GNPs carrying increasing levels of peptide. For these experiments, several parameters, such as integrin loading or exposure time, were optimized to obtain more reliable GNPs/receptor binding affinity measurements by retrieving the values of the dissociation constant (KD).
Interestingly, the GNPs KD toward the receptor decreased in the range of 0.1–0.5 equivalents of cRGD per PEG. Above that range, no change in the binding affinity was observed, despite the continuous increase in the packing density of the peptide (Figure S3A, Supporting Information). On the other hand, cRAD GNPs showed a much weaker interaction with the receptor, as KD values were over an order of magnitude higher (Figure S3B, Supporting Information), in agreement with a previous study where a similar characterization approach was applied.[32] Based on these findings, 0.5 equivalents of each peptide were used for functionalization scaling-up and were used for the following experiments.
The dose-response over time for the cRGD GNP toward the immobilized integrin receptor is depicted in Figure 2A, and its corresponding fitting is described in Figure 2B. The calculated KD value of 236 pM is in agreement with other studies in the literature.[37]
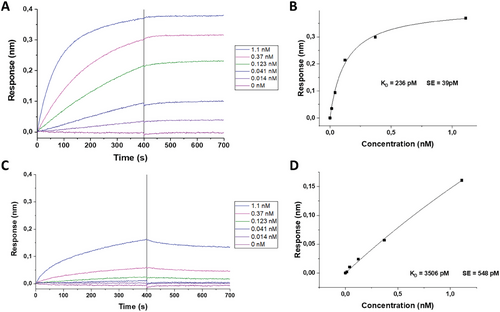
In contrast, cRAD GNPs’ dose-response interaction with the same integrin resulted in a higher KD value of 3506 pM, indicating a weaker affinity with the receptor (Figure 2C,D). Overall, these findings confirmed the proper orientation of the functionalized cRGD peptide and its potential for the GNP's specific targeting toward the αVβ3 integrin.
2.3 In Vitro Cell Viability of HUVECs and HepG2 Cells After GNPs Exposure
To evaluate the toxicity of each GNP group functionalized with the peptides, either cRGD or cRAD, in vitro, human umbilical vein endothelial cells (HUVECs) and the hepatocytic cell line (HepG2) were used. Both cell lines were exposed to the nanomaterials varying exposure time and GNPs concentration and on the resulting media we carried out LDH assay. In the two cases, cRGD and cRAD conjugated GNPs showed good biocompatibility regardless of the incubation time (Figure S4A,B, Supporting Information). A dose-response toxicity was not observed, as varying the concentration of GNPs did not impact cell viability in HUVECs (Figure S4C, Supporting Information). Likewise, cell viability was not affected by the presence of the fluorophore (Figure S4D, Supporting Information). Collectively, these results demonstrated that all the developed formulations were biocompatible, as the viability did not drop below the 85% threshold.[41-43]
2.4 In Vitro Uptake of GNPs in HUVECs and HepG2 Cells
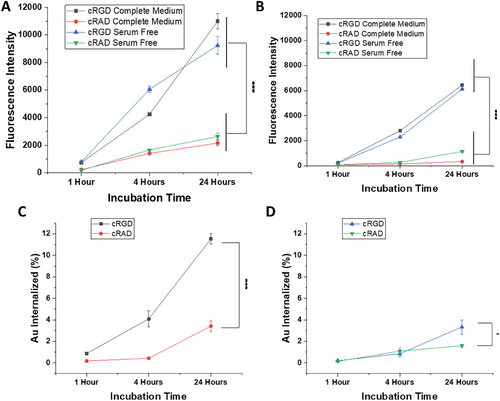
To investigate the effect of the peptides on GNPs uptake and the influence of the biomolecular corona, we performed cell uptake experiments using both HUVEC and HepG2 cell lines in serum-free (no biomolecular corona formation) and complete medium (biomolecular corona formation). The uptake was measured with flow cytometry following Cy5 signal intensity. When HUVECs were exposed to GNPs, a clear time-dependent effect was detected for both cRGD and cRAD conjugates indicating that a continuous cellular uptake occurred. A significant difference in uptake rate was observed between cRGD GNPs, which are being uptaken more than cRAD GNPs. These results suggest different cellular mechanisms of internalization. In addition, no difference was observed in cell uptake when the GNPs were exposed to serum-free or complete medium, indicating that the surrounding media or the potential corona formation did not affect the GNP-receptor binding affinity. Interestingly, for all conditions tested, a higher uptake in HUVECs was observed in comparison with HepG2 (Figure 3A.B). To confirm the flow cytometry data, inductively coupled plasma–mass spectrometry (ICP-MS) was used to directly determine the concentration of gold present in the cells. ICP-MS analysis confirmed the uptake pattern over time, as well as the same differences between cRGD and cRAD GNPs previously measured by flow cytometry (Figure 3C,D). Overall, these data showed the key role played by cRGD in cell internalization, as well as the lack of impact in uptake caused by the presence of serum in the medium, suggesting low influence of the protein corona in the nanoparticle cellular recognition.
2.5 Cellular Internalization and Lysosomal Colocalization
To elucidate the mechanism of intracellular trafficking that GNPs used to be internalized inside cells, we imaged HUVECs in vitro cultures in the presence of NPs and lysotracker, a specific lysosome dye, by confocal microscopy. We then, through computational analysis, defined the number of colocalizing peptide GNPs and quantified the ratios of colocalized and escaped GNPs. Representative images are shown in Figures 4A,B, and S6 (Supporting Information), where lysosomes are labelled in green, clusters of GNPs in red and colocalized objects in yellow after 1, 4 and 24 h of incubation time, respectively. Visually, it is possible to appreciate a difference after 1 h of exposure, as more cRAD GNPs were colocalized with lysosomes (Figure 4A), while both particles behave similarly after 24 h (Figure 4B). Image analysis was carried out to quantify the ratio between the colocalized GNPs signal over the total GNPs detected intracellularly. cRAD GNPs show a higher value, indicating a faster colocalization into the lysosome compared to cRGD GNPs (Figure 4C).
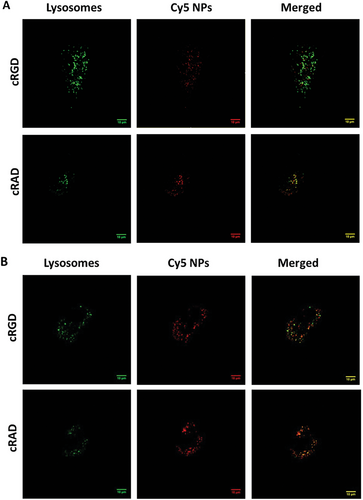
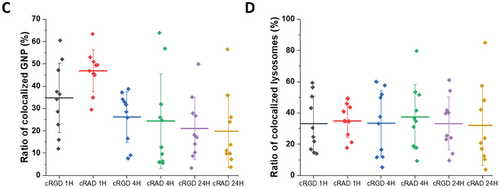
Interestingly, no changes in the percentage of lysosomes colocalized with GNPs were observed, regardless of GNPs or exposure time (Figure 4D).
2.6 In Vivo Biodistribution
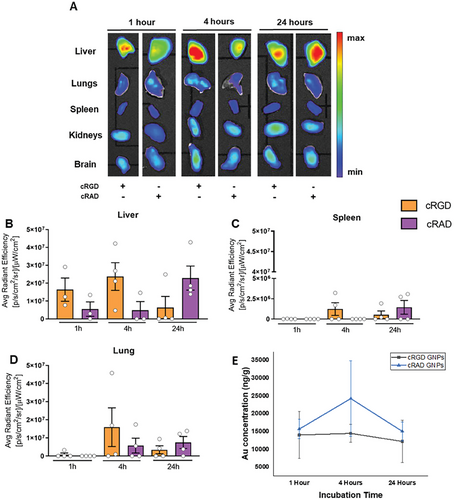
To study the biodistribution of cRGD GNPs, healthy immunocompetent mice were treated with a single dose of 2 × 1013 GNPs/mouse by intravenous injection at three different time points of 1, 4 and 24 h. Then, ex vivo analysis of the liver, spleen, kidneys, lungs, and brain from treated mice provided an overall idea of the accumulation and biodistribution of the GNPs in vivo. (Figure 5). As expected, the liver was the organ with the highest level of GNP signal, followed by the lungs. A rapid interaction between cRGD GNPs and hepatic cells caused immediate clearance of the GNPs from circulation. No signal was detected in the kidneys, spleen, or brain (Figure 5A). Based on the fluorescence signal, GNPs functionalized with cRGD have a faster penetration inside the liver, while cRAD GNPs accumulated progressively over time, showing a non-specific interaction with hepatic cells (Figure 5B). On the other hand, gold quantification in livers through ICP-MS (Figure 5E) showed very similar behavior of both GNPs. Moreover, differences between the tested time points were also not observed.
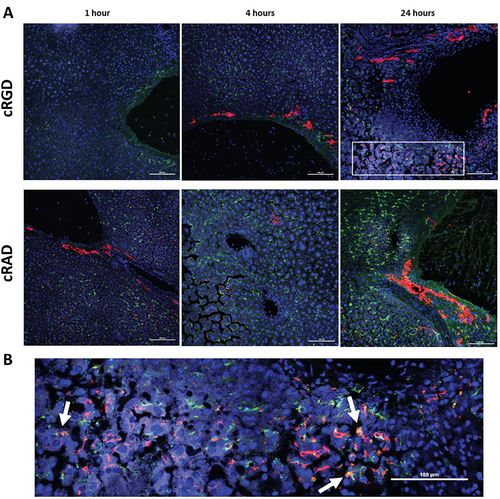
To determine the GNPs distribution in the hepatic tissue and possible interactions with the cells present in the liver, we performed immunostaining on liver sections after 1, 4, and 24 h of incubation (Figure 6A). Nuclei were stained blue, macrophages green, and GNPs, due to the functionalized fluorophore, were red. As was observed by ex vivo fluorescent analysis, cRGD GNPs were detected after 1 h of administration in the hepatic tissue, with most of the signal being in the peripheral area of the sinusoids, suggesting that GNPs were coming from the circulation into the liver. However, a substantial portion of GNPs were already inside the hepatic parenchyma, due to interaction with hepatic cells. No colocalization with macrophages (CD68 positive cells) was observed. After 4 h, it was possible to observe GNPs attached to the membrane of cells with big, round nuclei – hepatocytes – which appeared to be dying by necrosis. The presence of smaller nuclei in areas with a big accumulation of GNPs was detected due to the infiltration of leukocytes in the tissue parenchyma. After 24 h, a higher number of GNPs accumulated in the areas near the blood vessels, corresponding with their dilation and an overall increase in the number of macrophages, but without colocalization of the two. In mice treated with cRAD-bearing NPs, the signal from GNPs was almost absent, remaining confined near the vessels at early time points. Interestingly, after 24 h of incubation, colocalization of cRAD GNPs with macrophages was detected, further suggesting different behaviors between the GNPs functionalized with each peptide (Figure 6B). On the other hand, GNPs functionalized with the cRGD peptide were shown to selectively bind to the integrin αVβ3, expressed mostly in liver sinusoidal endothelial cells (LSECs). This result hints on the targeted GNPs entering the cytoplasm of the cells through a specific interaction with the transmembrane receptor.
Since GNPs in this size range might have a quenching effect on fluorophores, auto metallography (AMG) (histology with silver staining) was performed to confirm the presence of gold in the hepatic tissue seen with the immunohistochemistry experiments (Figure 7A,B). Both GNPs, bearing cRAD and cRGD, came from the circulation, displaying a homogeneous distribution within the hepatic sinusoids at early time points (1 h) with no difference between the two peptides. After 24 h, GNPs were mostly detected inside elongated cells from the liver parenchyma, possibly Kupffer cells or LSECs.
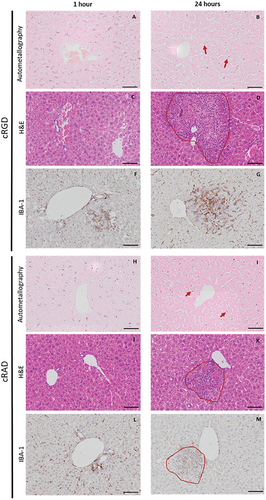
To ensure that no toxic effects in the liver were caused by the administration of both types of constructs, haematoxylin and eosin staining (H&E) and IBA-1 staining, a marker for macrophages, were performed at two different time points (Figure 7C–J). No substantial changes were observed after 1 h of administration for any of the peptides. However, after 24 h, microgranulomas were observed in the hepatic parenchyma near the surrounding areas of the vessels (Figure 7D–K). IBA-1 staining showed colocalized macrophages from the parenchyma and/or circulation in the inflamed tissue areas (Figure 7G–M). No macroscopic changes, weight loss, diarrhea, inability to walk, or other clinical signs were observed, demonstrating the transitory inflammatory response.
3 Discussion
In this study, we successfully synthesized a series of 4 nm ultrasmall GNPs and functionalized them with the αVβ3 integrin-targeting peptide cRGD. The particles were designed by finely tuning the peptide's packing density and availability using PEG as a spacer from the core and 𝛼-galactose as a spacer on the surface to allow efficient peptide functionalization, recognition, and binding. Moreover, GNPs were labelled with the fluorophore Cy5 for their detection. We showed that the synthesis of ultrasmall GNPs with a 4 nm core is extremely robust. In particular, small changes in critical process settings, such as decreasing the time or the temperature, lead to larger or lower core sizes, resulting in different ratios between linkers (PEG) and spacers (𝛼-galactose). Similarly, the functionalization process showed great reproducibility and tunability.
The particles’ synthesis and functionalization validation were carried out using an extensive array of physicochemical characterization techniques suitable for their size range. These techniques build a solid profiling and characterization platform to validate future complex ultrasmall GNP designs.
To prove the efficacy of the targeting capacity of the GNPs, we used an on-chip detection method, immobilizing the αVβ3 integrin and choosing the candidate with the higher affinity toward the receptor. The isolated and immobilized receptors interacted specifically with GNPs functionalized with the cRGD peptide in the range of 0.1–0.5 equivalents per PEG. A lower level of peptide functionalization resulted in a lower recognition efficiency of the nanocarrier, while higher peptide levels did not show any greater affinity of the nanoparticle toward the receptor. These findings indicate that overcrowding the surface with the peptide would not allocate any more benefits to the NPs’ targeting capacity. As expected, the specificity of the binding pocket of the αVβ3 integrin toward the tripeptide Arg-Gly-Asp resulted in an almost non-detectable binding of the cRAD.
In more complex environments, such as the bloodstream, the NP's potential in reaching and interacting with the target is mitigated by the biomolecular corona formation, a layer of biomolecules that absorbs on the NPs’ surface. Corona formation changes their biological identities decreasing the targeting efficiency and leading to subsequent sequestration of the nanoparticles by resident macrophages in the tissues and the liver.[43, 44]
To evaluate the NPs’ behavior in the biological milieu, in this study, we used as a cell model, HUVECs and HepG2, as they are known to express the αVβ3 integrin. In our study, we detected a significant difference in the uptake rate between cRGD and cRAD conjugated, indicating a difference in the uptake mechanisms. Additionally, the uptake rate for the cRGD was not affected by the presence or absence of the serum protein in the cell culture media indicating a less corona attenuation effect that is seen for larger nanomaterials.[45] After interacting with the receptor, most nanoparticles are internalized by the cell through a series of mechanisms, with clathrin-mediated endocytosis being the predominant pathway. This process typically directs its cargo toward lysosomes, where they are broken down and degraded. In this context, another advantage reported in the literature for nanomaterials of the ultrasmall size range (<10 nm) is that the uptake process can vary based on the specific formulation, possibly including some clathrin-independent that can escape the lysosomes.
To this extent, we performed a series of colocalization experiments to assess whether our GNPs bearing the cRGD peptide would overlap with the lysosomes or could be internalized through alternative routes. These showed no significant difference between the tested conditions of cRGD versus cRAD GNPs (Figure 4C,D), indicating, however, that not all the particles fully colocalized with the lysosomes. These results suggest the simultaneous presence of clathrin-mediated and alternative mechanisms used in the NPs internalization process.
Collectively, with the previously discussed in vitro results, we show that our cRGD ultrasmall GNPs effectively target parenchymal cells and are not affected in their targeting capability by the biomolecular corona formation and have the potential to partially avoid the lysosome trap and therefore they open new opportunities for new nanomaterials with active targeting properties and effective drug delivery.
While clathrin-mediated uptake, leading to lysosome formation, occurs in every mammal cell, alternative ways such as caveolin-dependent endocytosis are only specific to some cell types, such as endothelial cells.[46] The pathway through which a construct is internalized has a significant impact on its intracellular fate.[22] In particular, clathrin-dependent uptake leads to lysosomal translocation and degradation via the vesicles’ acidic environment and hydrolytic enzymes. On the other hand, NPs that are internalized through clathrin-independent routes can lead to a decrease in lysosomal entry, hence colocalization, thereby avoiding this final targeting barrier.[47] For example, Reilly et al. have shown that caveolin endocytosis can traffic to the endoplasmic reticulum and Golgi complex, partially through direct transport from the plasma membrane.[48]
Using targeting moieties for the right receptors in nanocarrier designs can unlock the delivery of therapies through specific internalization mechanisms and aim for the release in defined cell organelles or subcompartments. Integrins such as αVβ3 are knowingly associated with structures involved in caveolin-mediated endocytosis, particularly the protein caveolin-1.[49] Zhang et al. showed that the use of cRGD as a targeting moiety through liposomes bearing the tripeptide can increase internalization via caveolin-dependent mechanisms.[40] Similarly, Alam et al. showed, through the use of a different construct, siRNA assembled clusters with cRGD, the tripeptide's potential in exploiting this clathrin-independent uptake mechanism.[50] This information raises the question of what is the NPs fate after entering the cell.
Future studies could elucidate the endocytosis pathways driven by the cRGD peptide. While the most common strategies for these investigations require the use of pharmacological inhibitors, recent evidence demonstrated that off-site effects and the involvement of multiple internalization pathways could hide more complex mechanisms[51] Thus, further studies aiming to elucidate the path of GNPs, especially in this size range, would benefit of colocalization analysis of the particles with other cellular structures involved in endocytic processes.
One of the main issues regarding GNPs for in vivo use is the predictability of their behavior. To prove that our ultrasmall cRGD-bearing GNPs could specifically target parenchymal cells, we assessed the accumulation of GNPs in vivo in the liver, the spleen, and the lungs.
cRGD GNPs were found to be distributed in the organ parenchyma, unlike cRAD GNPs, which remained confined near the blood vessels even after long exposure. These findings are in accordance with what was observed in vitro, with higher uptake of cRGD GNPs uptake after short incubation times for both endothelial cells and hepatocytes. Moreover, the fact that cRGD GNPs’ signal in the parenchyma did not colocalize with CD68+ macrophages, while cRAD GNPs’ did, highlights the potential of these particles to overcome this barrier.
However, the high amount of peptide in the GNPs surface also caused an acute inflammatory response due to specific interactions with hepatic cells.
In conclusion, here we present a robust system to produce, characterize, and test, in vitro and in vivo, ultrasmall cRGD bearing GNPs that have great targeting efficiency and good abilities to overcome some of the main targeting barriers. Due to their characteristics, they not only emerge as an interesting tool for drug delivery but as a tool that could be exploited to study specific peptide targeting and cell uptake mechanisms.
4 Experimental Section
GNPs Synthesis and Functionalization
GNPs were synthesized following a previously described modified Brust–Schiffrin Synthesis method.[37] Briefly, HAuCl4 gold salts were reduced in situ with sodium borohydride in the presence of a substoichiometric amount of thiol ligands. For this purpose, a bifunctional PEG of 0.5 kDa with a thiolated end for GNPs surface binding and growth core capping, as well as a carboxylated end for further functionalization, along with a thiolated α-galactose, used as a capping agent but also as a spacer that could control the PEG packing density, were used. The impact of changes in key parameters (temperature, pH, and reaction time) were assessed.
Synthesized GNPs were then functionalized with moieties of interest following a previously reported method. Briefly, a mixture of EDC/NHS was added to GNPs, followed by the addition of either cRGD, cRAD, or Sulfo-Cyanine5 Amine.
Analytical Characterization
GNPs were characterized with the following techniques.
UV–Vis
The UV–vis spectra (200–700 nm) of the GNPs were obtained using Perkin–Elmer Lambda 35 UV–vis Spectrophotometer with 2 mL of 20 mg mL−1 Au GNPs in a quartz cuvette.
DLS
The hydrodynamic diameter of GNPs was determined by Dynamic Light Scattering using Malvern Zetasizer Nano-ZS. Measurements were performed in 1 mL of 200 µg mL−1 Au solutions in 1x PBS.
TEM
Samples were prepared from the aqueous particle solution under ambient conditions by desiccating a 0.35 mL droplet of a 150 mg mL−1 GNPs solution on a hydrophilized carbon film surface. Ultrathin film supports of type #01824 were used (Ted Pella Inc.). The hydrophilization was performed for 2 min with 25 mA strong glow discharge treatment in a K100X plasma chamber (Quorum Technologies Ltd). Image data was acquired in a transmission electron microscope of type JEM-2100F [Model EM-20014, UHR, 200 kV] (JEOL) equipped with a digital camera of type F-216 (TVIPS). Usually, wider field of view images at 150k magnification were assembled with the spotscan utility of the TVIPS EMMENU4 software from a beam shift-based 4 × 4 images matrix. Analysis was performed at CIC Biomagune (San Sebastian, Spain). Data processing was performed using ImageJ software.
DCS
Sizing analysis was performed using a CPS DC24000UHR disc centrifuge (CPS Instruments, Inc.). An 8−24% sucrose gradient was created in 11 mL water. A series of solutions of varying sucrose concentration were injected sequentially (from high to low concentrations) to generate the gradient. This was followed by a dodecane injection (500 µL), to reduce gradient evaporation. The gradient was allowed to stabilize and reach thermal equilibrium for ≈30 min prior to data acquisition. Polyvinylchloride (PVC) calibration standards (0.237 µm, 50 µL injection volume) were analyzed prior to each GNPs sample (100 µL, 100 mg mL−1 Au) to ensure that the instrument was operating optimally and with a high level of accuracy. Analysis was performed at 24 000 rpm with the light detector adjusted to a position suitable for the analysis of ultrasmall GNPs. Particle size was calculated based on an assumed GNPs density of 5.0 g cm−3.
1H NMR
10 mg Au of GNPs were freeze-dried, and the resulting powder incubated with 600 µL of KCN 0.3 m in KOH 0.1 m (solvent D2O). Particles were incubated at 60 °C for 4–6 h with strong agitation (950 rpm) to prevent pelleting using Thermo Scientific Digital Heating Shaking Drybath. The complete etching was achieved when the solution was transparent and without any pellet, and thus, visually checkable.
Experiments were performed at 298 K on a Bruker AVANCE III 500 spectrometer at CIC Biomagune (San Sebastian, Spain) (500 MHz, D2O). Data processing was performed using MestReNova software.
LC-CAD-MS
Au (350 µg) of GNPs were incubated with 15 µL of 0.3 m KCN and 0.01 m KOH and H2O up to 190 µL. Mixing was carried out by vortexing. Particles were incubated at 80 °C for 10 min with strong agitation (950 rpm) to prevent pelleting, using Thermo Scientific Digital Heating Shaking Drybath. The complete etching was achieved when the solution was transparent and without any pellet, and thus, visually checkable. To the etched solution, 10 µL TCEP 0.05 m from a commercial neutral 0.5 m solution (pH 7.0, aqueous solution; pH adjusted with ammonium hydroxide) were added. Thermo UltiMate 3000 Rapid Separation Liquid Chromatography system comprising a dual gradient standard pump, Corona Veo RS CAD detector (Chromeleon 7.0 software) in line with LCQ Fleet Ion trap Mass Spectrometer detector (Xcalibur 2.2 SP1 software), and Thermo Viper tubing (0.13 mm ID) were used. Separations were performed on an Acquity UPLC BEH C18 column, 130 Å, (100 × 2.1 mm i.d., 1.7 µm particle size) and an Acquity UPLC BEH C18 VanGuard precolumn, 130 Å, (5 × 2.1 mm i.d., 1.7 µm particle size) (Waters). Solvents used as mobile phase were as follows: A: 0.1% formic acid in H2O; B: 0.1% formic acid in acetonitrile. Elution conditions applied were as follows: 0-0.5 min, 5% B isocratic; 0.5-6 min, linear gradient 5–98% B; 6–7 min, 98% B isocratic; washing and reconditioning of the column.
The Flow rate was 0.350 mL min−1 and the injection volume 5 µL. The system operated at 35 °C. The Corona Veo RS Evaporation temperature was set at 35 °C; Power function: 1.0; Data collection Rate: 2 Hz; Signal Filter: 3.6 s. ESI-MS analysis was performed in the positive ion mode. Nitrogen was used as desolvation gas. The ESI parameters of the source were: capillary temperature 150 °C, the source heater temperature was held at 45 °C, a potential of 3.8 kV was used on the capillary for positive ion mode. MS spectra, within the m/z range 150–2000 amu, were obtained at 35 V cone voltage.
BLI Assays
Binding interactions were measured using Octet Red 96 (FortéBio) with High Precision Streptavidin (SAX) biosensors as per the manufacturer's guidelines. Briefly, biosensors were hydrated in binding buffer (1% (w/v) BSA, 0.5% (v/v) Tween 20, in Dulbecco's PBS) for 10 min, then washed twice in binding buffer (60 s, then 180 s), loaded with 22.5 nm biotinylated-αVβ3 integrin in binding buffer for 1500 s, quenched with 300 µm biocytin (Fisher) in binding buffer for 120 s, washed with binding buffer for 30 s and the baseline in binding buffer measured for 180 s. αVβ3-loaded biosensors were allowed to associate with GNPs in the binding buffer for 400 s and dissociate in the buffer used for baseline measurements for 300 s. All measurements were performed at 30 °C. Data were analyzed using Data Analysis software (FortéBio).
Cell Culture
HUVECs were purchased from PromoCell and cultured in ECGM supplemented with foetal calf serum, epidermal growth factor, basic fibroblast growth factor, heparin, hydrocortisone, and 1% v/v – Penicillin/Streptomycin. HepG2 was obtained from ATCC and cultured in DMEM supplemented with 10% v/v FBS and 1% v/v – Penicillin/Streptomycin. Both cells were cultured in a humidified incubator at 37 °C and 5% CO2. Medium was changed every 2 to 3 days. Experiments with HUVECs were performed using cells from passages–9. Regarding HepG2, passages 2–19 were used.
Cytotoxicity
HUVEC and HepG2 were seeded at 10,000 cells/well in 96-well plates and left to attach overnight. Cytotoxicity was assessed using CyQUANT LDH Cytotoxicity Assay Kit, as described by the manufacturer. The samples were analyzed using a CLARIOstar plate reader and software (BMG Labtech). The percentage of cell viability was calculated based on the ratio between the absorbance of each sample compared with the negative and positive controls. Experiments were performed in triplicates.
Uptake Studies
For HUVECs, wells were pre-coated with gelatin (1% v/v) for at least 30 min at 37 °C. Gelatin was removed, and the wells were washed twice with PBS. HUVECs were seeded at 200,000 cells/well and HepG2 at 100,000 cells/well in 12-well plates and left to attach overnight. Media was removed and the wells were washed twice with PBS. Cells were treated with the respective nanoparticle suspension (2.5, 5, or 10 µg mL−1) for either 1, 4, or 24 h in the corresponding culture media, in either the presence or absence of supplements.
Flow Cytometry
Following any uptake study, cells were detached using 0.025% trypsin and 0.01% EDTA in PBS and centrifuged at 1500 rpm for 5 min. The pellet was re-suspended in 200 µL of PBS supplemented with 5% FBS. This process was repeated for a total of three centrifugations and re-suspensions. Flow cytometry was carried out using Attune NxT Flow Cytometer (ThermoFisher). Data were analyzed with FlowJo software (v.10). Gating and analysis parameters were determined using untreated cells as controls. Experiments were performed in triplicates.
ICP-MS
To quantify the amount of gold, cells were collected as previously described. Samples were frozen at −80 °C until further use. Before analysis by ICP-MS, samples were digested by microwave irradiation. This was carried out using speedwave XPERT (Berghof). Briefly, 750 µL of HNO3 and 250 µL of HCl were added to 1.5 mL of samples, and then submitted to cycles of high temperature and pressure for 1 hour. Digested samples were then collected and, if necessary, diluted in diluted aqua regia, before injection in iCap-Q ICP-MS (ThermoFisher).
Livers were similarly digested, except an increased volume of acidic mixture (4.5 mL of HNO3 and 1.5 mL of HCl) was used instead.
Confocal Scanning Laser Microscopy Images Acquisition and Processing
Colocalization studies were carried out by confocal scanning laser microscopy. Cells were grown in ibiTreat µ-Slides (Ibidi) and exposed to GNPs as previously described. Following incubation, cells were washed with PBS, stained with LysoTracker Green DND-26 (Invitrogen) at 10 nM, at 37 °C, for 30 min and then fixed with PFA (4% in PBS) for 10 min, at room temperature, protected from light. Before and after every staining, cells were briefly washed with PBS. Confocal images were acquired with Stellaris 8 (Leica) equipped with different lasers and detectors that allowed the detection of the different fluorophores in the sample. Images were analyzed with ImageJ software. Briefly, an automatic threshold (Renyi Entropy) was applied to reduce the background signal, followed by the application of a region of interest (ROI), and the deletion of slices with a low signal-to-noise ratio. Colocalization was assessed on one slice per cell and analyzed a total of ten cells, calculating the ratio between the GNP fluorescence detected in the lysosome over the total GNP fluorescence detected in the cell (Ratio of colocalized GNPs). The ratio between the GNP fluorescence in lysosome over the total lysosome fluorescence (Ratio of Colocalized lysosomes) was also measured.
In Vivo
Eight-week-old female and male CD1 mice were housed in “specific pathogen-free” animal rooms at a constant temperature of 21 ± 1 °C, humidity 55 ± 10%, with a 12 h light/dark cycle, and ad libitum access to food and water. Mice were randomly divided into groups receiving cRGD and cRAD GNPs at three different time points (1, 4, and 24 h) (n = 4 for each experimental group). Briefly, all animals received by intravenous injection the same dose of GNPs preparation (1 × 1014 GNPs mL−1) diluted in 200 µL of injection-grade distilled water. At the selected time points 1, 4, and 24 h after GNPs injection, four mice for each group were sacrificed and organs were collected for histological analysis.
Ex Vivo Imaging Fluorescence
A total of 40 CD1 animals were used for the biodistribution study. Mice were euthanized after 1, 4, and 24 h of treatment with 4 nm ultrasmall GNPs functionalized with cRGD and cRAD, which were also functionalized with a Cy5 fluorophore. Liver, spleen, kidneys, lungs, and brain were collected without perfusing and scanned for ex vivo imaging. Fluorescence images were acquired with an IVIS Lumina III imaging system (PerkinElmer). The following acquisition parameters were used: excitation filter 620 nm, emission filter 670 nm, exposure time auto, binning factor medium, and f/Stop 2. Spectral un-mixing, image processing, and analysis were done using Living Image 4.3.1 software (PerkinElmer).
Histological Analysis
At the time of autopsy for each mouse, livers were sampled, fixed in 10% neutral buffered formalin (Bio-Optica, Italy) for at least 24 h at RT and then processed for paraffin embedding. Tissue micrometric sections (4 µm in thickness) were cut with Leica RM55 microtome (Leica Microsystem, Italy) and dried at 37 °C overnight. Hematoxylin and eosin (H&E) staining was performed in liver sections treated with GNPs and vehicle-treated mice by staining cells nuclei blue with Mayer's hematoxylin solution (Bio-Optica, Italy) for 2 min and 30 sec, and then washed with water. The counterstain was carried out in eosin Y solution (Bio-Optica, Italy) for 1 min and 20 sec to stain the cytoplasm in pink. The slides were then washed in tap water until discoloration occurred. IBA-1 staining was performed in liver sections treated with GNPs and vehicle-treated mice, to stain macrophages brown. Nuclei were stained blue with hematoxylin. HIER was performed with citrate buffer pH 6 for 30 min at 95 °C, inhibition of endogenous peroxidase with H2O2 3% for 10 min at RT, and incubation with blocking solution (PBS-NGS 10%-Tween 20 0.05%) for 30 min at RT. For subcellular localization, IBA-1 (1:200, Wako Chemicals, USA) was used to label the macrophage calcium-binding protein, followed by an amplification step with the labelling system ABC (Vectastain Elite) and the chromogenic reaction with DAB (Sigma-Aldrich). Cell nuclei were stained with Mayer's hematoxylin solution (Bio-Optica, Italy) for 30 s and then washed with water until discolored. To visualize the presence of gold agglomerates in the liver parenchyma, AMG staining was performed at RT for 45 min, as previously described.[25] All samples were dehydrated and fixed with a xylene-based mounting medium (DPX, Sigma). All images were acquired using Olympus BX61VS.
Immunohistochemical Analysis
Immunofluorescence in liver was performed in tissue slides of 10 µm cut by a cryostat. Tissue sections were fixed in 10% neutral buffered formalin (Bio-Optica, Italy) for 20 min and washed with phosphate-buffered saline (PBS) for 5 min. Finally, the sections were incubated for 1 h with a blocking solution (PBS-NGS 10%–Triton X-100 0.1%) and washed again with PBS. For subcellular localization, anti-CD68 (1:200, Serotec, Kidlington, UK) was used to label lysosomes and endosome membranes of macrophages and Hoechst-33258 (1 µg mL−1 in PBS, Thermo Fisher Scientific) to label nuclei.
Ethics Approval
The in vivo work is covered under the associated code 9F5F5.176, project number 49/2021-PR.
Acknowledgements
This project received funding from the European Commission under grant no. 814236 of the H2020 Marie-Curie European Training Network (MC-ETN) NanoCarb. Confocal microscopy images were acquired in the RCSI Super Resolution Imaging Consortium funded by Science Foundation Ireland (18/RI/5723). Training and consultation provided by Massimiliano Garre from the RCSI Imaging facility is highly appreciated. S.E.M. thanks the PID2020-114356RB-100 project from the Ministry of Science and Innovation of the Government of Spain.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
A.F. performed investigation, visualization and wrote the original draft. J.F.A. and M.B.V. performed in vivo studies and reviewed and edited the final manuscript. F.G., A.M., and L.R. performed in vivo studies. V.C. performed investigation and reviewed and edited the final manuscript. A.B. performed investigation, conceptualization. M.S. performed ICP MS measurements. M.D. performed flow cytometry analysis. S.E.M. performed supervision, funding acquisition, and reviewed the final manuscript. P.B. performed investigation, in vivo conceptualization, supervision, funding acquisition, and reviewed the final manuscript. M.P.M. helped in conceptualization, supervision, project administration, funding acquisition, and reviewed and edited the final manuscript.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.




