EXIST: EXamining rIsk of excesS adiposiTy—Machine learning to predict obesity-related complications
Abstract
Background
Obesity is associated with an increased risk of multiple conditions, ranging from heart disease to cancer. However, there are few predictive models for these outcomes that have been developed specifically for people with overweight/obesity.
Objective
To develop predictive models for obesity-related complications in patients with overweight and obesity.
Methods
Electronic health record data of adults with body mass index 25–80 kg/m2 treated in primary care practices between 2000 and 2019 were utilized to develop and evaluate predictive models for nine long-term clinical outcomes using a) Lasso-Cox models and b) a machine-learning method random survival forests (RSF). Models were trained on a training dataset and evaluated on a test dataset over 100 replicates. Parsimonious models of <10 variables were also developed using Lasso-Cox.
Results
Over a median follow-up of 5.6 years, study outcome incidence in the cohort of 433,272 patients ranged from 1.8% for knee replacement to 11.7% for atherosclerotic cardiovascular disease. Harrell C-index averaged over replicates ranged from 0.702 for liver outcomes to 0.896 for death for RSF, and from 0.694 for liver outcomes to 0.891 for death for Lasso-Cox. The Harrell C-index for parsimonious models ranged from 0.675 for liver outcomes to 0.850 for knee replacement.
Conclusions
Predictive modeling can identify patients at high risk of obesity-related complications. Interpretable Cox models achieve results close to those of machine learning methods and could be helpful for population health management and clinical treatment decisions.
Graphical Abstract
Overweight and obesity are common conditions associated with multiple adverse outcomes. This study found that it is possible to predict the development of many adverse outcomes associated with overweight and obesity. Traditional statistical models achieved similar accuracy to machine learning-based models while facilitating interpretability.
1 INTRODUCTION
There is a broad spectrum of conditions whose incidence is increased in people with obesity, including coronary artery disease, stroke, diabetes mellitus, chronic kidney disease, liver disease, and certain types of cancer. Comprehensive lifestyle intervention, including caloric restriction, increased physical activity, and behavioral modification counseling, is the cornerstone of obesity treatment. Studies show that these interventions can reduce the risk of diabetes.1 Pharmacological options are also available, including pancreatic lipase inhibitors, GLP-1 receptor agonists, sympathomimetics, and combination drugs. Finally, bariatric surgical procedures are an effective, albeit invasive, means of reducing excess weight and have been shown to reduce the incidence of a broad range of obesity-related complications.2, 3 On the other hand, many of these interventions are costly,4-7 and may also carry a risk of adverse events.8-11 Therefore, being able to identify patients who are likely to reap the greatest benefits from the intervention could help direct healthcare resources. This could be accomplished using predictive models that can assess the risk of obesity-related complications. While several predictive models for individual obesity-related complications exist,12, 13 for many of these complications, there are no predictive models that incorporate measures of the patient's weight. Additionally, no published risk prediction models for obesity-related complications have been specifically developed in people with overweight or obesity.
Increasing availability of electronic health records (EHR) data provides an opportunity to assess adverse clinical outcomes and their risk factors across large populations over extended periods of time.14, 15 A number of methods have been used to develop these models, including both traditional statistical (e.g., regression) and machine learning techniques, each of which has its own strengths and weaknesses.16, 17 Therefore, this study was conducted to leverage a large EHR dataset to develop risk prediction models for long-term obesity-related complications using both machine learning and traditional statistical approaches.
2 MATERIALS AND METHODS
2.1 Study design
Electronic health records data of a cohort of patients with overweight and obesity were used to develop and evaluate predictive models for nine long-term clinical outcomes using a) Lasso-Cox proportional hazards models and b) random survival forests (RSF).
2.2 Study cohort
Study participants included adults (age ≥18 years) with body mass index (BMI) between 25 and 80 kg/m2 who were being treated in primary care practices affiliated with Mass General Brigham, a large integrated healthcare delivery network in Massachusetts founded by Brigham and Women's Hospital and Massachusetts General Hospital, between 01/01/2000 and 12/31/2019. Patients were excluded from the study if they: a) were older than 80 years old, b) had missing demographic information, or c) were diagnosed with a disease outcome of interest at baseline. The last exclusion criterion was applied on a per-analysis basis (e.g., patients with history of cancer were only excluded from the analysis where incidence of cancer served as the outcome).
2.3 Study measurements
A patient was entered into the study (Index Date) on the first date when they met all of the following criteria: 1) had had at least one primary care encounter (to ensure availability of baseline clinical characteristics, as patients who only receive specialist care at the study institution are less likely to have their medical history comprehensively documented in the EHR); 2) was ≥18 years old; 3) had first of two consecutive BMI measurements ≥25 kg/m2; and 4) on or after Study Start date (01/01/2000). Patients exited the study (Study Exit Date) on the first of the following dates: a) reaching an outcome endpoint (this criterion was specific to the outcome being analyzed); b) death (even when it is not a component of the outcome being analyzed); c) 24 months after the last primary care note (i.e., lost to follow-up) or d) Study End (12/31/2019). Patients were enrolled into the study until 12/31/2018 (to allow at least 12 months of follow-up to assess study outcomes).
Outcomes were obtained from EHR data (both in- and outpatient). Dates of death were obtained from the Social Security Death Master File. The following endpoints were assessed: a) atherosclerotic cardiovascular disease (ASCVD); b) heart failure (HF); c) diabetes mellitus type 2 (T2DM); d) non-alcoholic steatohepatitis (NASH)/ non-alcoholic fatty liver disease (NAFLD); e) sleep apnea; f) cancer (excluding non-melanoma skin cancer); g) degenerative joint disease (DJD); h) knee replacement and i) all-cause mortality. The length of time (days) from the Index Date to each of the clinical events listed above served as study outcomes.
A literature search was performed to identify the candidate predictor variables for study outcomes (Table 1). These predictor variables included demographic characteristics, vital signs and laboratory measurements, past medical history, family history, and history of substance abuse. Predictor variables were also assessed based on EHR data. All variables were assessed at baseline, defined as the most recent record/measurement prior to study entry. For quantitative variables, if no measurements prior to study entry were available, the earliest measurement within 1 month after study entry was used. International classification of diseases codes used to identify candidate predictor variables and study outcomes in the EHR data are provided in Supplemental Table 1.
| Variable | Outcomes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ASCVD | Heart failure | T2DM | NASH/NAFLD | Sleep apnea | Cancer | DJD | Knee replacement | Death | |
| Age | X | X | X | X | X | X | X | X | X |
| Sex | X | X | X | X | X | X | X | X | X |
| Marital status | X | X | X | X | X | X | X | X | X |
| Commercial insurance | X | X | X | X | X | X | X | X | |
| BMI | X | X | X | X | X | X | X | X | X |
| SBP | X | X | X | X | X | X | X | ||
| DBP | X | X | X | X | X | X | X | X | |
| eGFR | X | X | X | X | X | X | |||
| HbA1c | X | X | X | X | X | X | X | X | X |
| LDL-C | X | X | X | X | X | X | X | ||
| Proteinuria | X | X | X | X | X | X | X | X | |
| ASCVD | X | X | X | X | X | X | X | X | |
| Heart failure | X | X | X | X | X | ||||
| T2DM | X | X | X | X | X | X | X | X | |
| NASH/NAFLD | X | X | X | X | X | X | |||
| Sleep apnea | X | X | X | X | |||||
| Cancer | X | X | X | X | X | ||||
| DJD | X | X | X | X | X | X | X | ||
| Knee replacement | X | X | X | X | X | X | X | ||
| Hepatitis B | X | X | X | X | X | ||||
| Hepatitis C | X | X | X | X | X | X | X | X | X |
| HIV | X | X | X | X | X | X | X | X | |
| Chronic inflammation | X | X | X | X | X | X | X | X | |
| H. pylori | X | X | X | ||||||
| PCOS | X | X | X | X | |||||
| GDM | X | X | |||||||
| COPD | X | X | X | X | X | X | X | ||
| Dementia | X | X | X | X | X | X | |||
| Sepsis | X | X | X | X | X | X | X | ||
| Knee injury | X | X | X | X | X | X | X | ||
| Joint infection | X | X | X | X | X | X | X | X | |
| Valvular heart disease | X | X | X | X | X | X | |||
| Hypertension | X | X | X | X | X | X | X | X | X |
| Hypercholesterolemia | X | X | X | X | X | X | X | ||
| Stimulant abuse | X | X | X | X | X | X | |||
| Alcohol abuse | X | X | X | X | X | X | X | X | |
| Smoking | X | X | X | X | X | X | X | X | X |
| Family history of ASCVD | X | X | X | X | X | X | X | X | X |
| Family history of DM | X | X | X | X | X | X | X | X | |
| Family history of cancer | X | X | X | X | X | X | X | X | X |
- Note: Cells corresponding to identical predictor and outcome variables are grayed out.
- Abbreviations: ASCVD, atherosclerotic cardiovascular disease; BMI, body mass index; COPD, chronic obstructive pulmonary disease; DBP, diastolic blood pressure; DJD, degenerative joint disease; eGFR, estimated glomerular filtration rate; GDM, gestational diabetes mellitus; HIV, human immunodeficiency virus; LDL-C, low density lipoprotein cholesterol; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; PCOS, polycystic ovarian syndrome; SBP, systolic blood pressure; T2DM, type 2 diabetes mellitus.
Missing information for predictor variables was handled using imputation. Imputation was first performed for HbA1c (which was missing for 290,131/67.0% of observations). HbA1c measurements were imputed using random draws from normal distributions based on published data for similar populations, separately for patients with (mean 8.3% and SD 1.7%) versus without (mean 5.6% and SD 0.7%) diabetes.18, 19 Imputed HbA1c data were left truncated at 4.0%, which is generally considered to be the lower limit of normal.
Random forest imputation was not used for HbA1c because of the high missingness (67%) of this variable (primarily because it is not typically measured in patients who are not suspected to have diabetes). Previously published reports showed that at this level of missingness random forest imputation performs no better than “strawman imputation” (i.e., imputation of the mean or the median of the non-missing population).20 Under these circumstances, the approach of using data (mean +SD) from the published literature for (separately) patients with versus without diabetes was preferable.
All other missing data were imputed using a random forest algorithm applied to all covariates (using already imputed HbA1c values). MissForest random forest imputation algorithm21 was used to impute missing variables on the entire dataset (prior to it being split into training, testing and validation datasets). The MissForest algorithm regresses each variable against all other variables and then predicts missing data for the dependent variable using fitted forest.20, 21 The following variables had missing values that were imputed using this approach: a) systolic blood pressure, b) diastolic blood pressure, c) estimated glomerular filtration rate, and d) low-density lipoprotein cholesterol.
2.4 Predictive models
To identify the initial set of variables for the risk prediction model, bivariate analyses of the relationships between each of the candidate variables and each of the outcomes being analyzed were conducted. Candidate variables that were associated with one of the risk prediction model outcomes with a p-value <0.15 were selected for further development of the risk prediction model for that outcome. This approach was taken in part because the ultimate goal of the study was to develop compact predictive models that could be used in patient-facing applications requiring manual data entry and in provider-facing applications drawing data from EHRs, which in some cases may only have complete information on a limited number of variables. Therefore, we prioritized candidate variables likely to have the strongest predictive relationship with obesity-related complications.
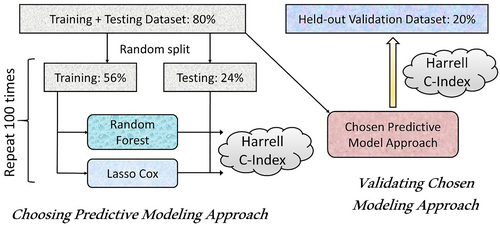
The entire dataset was randomly split into 80% training and testing versus 20% that were held out for validation after model development was complete. The training and testing 80% were further split into 70% training and 30% testing (56% and 24% of the entire dataset, respectively) datasets. The overall approach to model training and selection is illustrated in Figure 1.
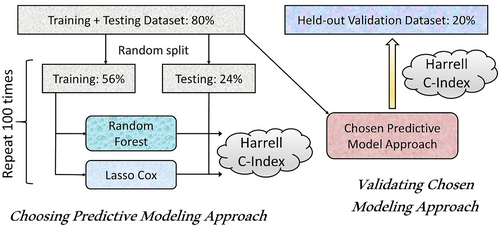
Selection and validation of predictive models.
The training dataset was utilized to build models using Lasso-Cox regression and RSF22 (RSF) methods for variable selection and prediction modeling. Lasso-Cox model regularization selects variables by shrinking the coefficients of less important variables to zero.23 The optimal penalty parameter lambda for the variable selection was estimated using cross-validation. Variable selection for the RSF model was conducted using the minimal depth approach.24 For every variable used in the growing of the tree, the minimum depth of the variable is the minimum depth of tree nodes split on this variable. Variables used for splitting closer to the root of the tree are applied to larger subsets than those used later in the growing process and are thus expected to have greater predictive power. Variable selection took place in the process of model building. After constructing an RSF model, the variable selection was conducted based on the minimal depth threshold of mean + one standard deviation of minimal depth, and a new RSF model was generated using the selected variables. Both the Lasso Cox regression and RSF models were developed using the 56% of patients included in the training dataset.
The relationship between the variables and the outcome of interest was assessed using a) Lasso-Cox proportional hazards model and b) a machine learning–based method, RSF model. A RSF is an ensemble classification method that determines a consensus prediction by averaging the results of many individual decision trees.22 Lasso-Cox and RSF predictive models were evaluated for each study outcome using multiple replicates of the variable selection and model fitting process (Figure 1). Each replicate included as the first step a random split of the 80% training + test dataset into 56% training dataset and 24% test dataset. Subsequently, both Lasso-Cox and RSF models were developed (including variable selection and model fitting) on the 56% training dataset. These models were then evaluated by calculating Harrell's C-index,25 on the corresponding 24% test dataset. Replicates were stopped once the average prediction error (calculated as 1—Harrell's C-index) stabilized; 100 replicates were used.
The performance of the Lasso-Cox and RSF predictive models was compared using the mean Harrell concordance index (C-index)25 for each outcome across all replicates. This selection process was used to identify the optimal variable selection strategy + modeling approach (Cox regression with Lasso vs. random survival forests) based on both the value of the mean Harrell concordance index across all replicates for the particular outcome as well as on computational resource utilization and interpretability (and ultimately face validity) of the predictive modeling approach. Once the best modeling approach was identified, the final set of variables was selected and coefficients for the model that included these variables were re-estimated using the combined 80% training + testing datasets. Specifically, Lasso-Cox model was computed on the 80% training + testing dataset with cross-validation to select the final set of variables. These variables were then used in the final Cox model that included the evaluation of competing risk of death (except the model for the mortality outcome) as previously described26 and was fitted on the 80% training + testing dataset. Тhe final Harrell C-index was assessed for this final model on the 20% held-out validation dataset.
After the models were developed and evaluated as described above, risk models that included a more limited number of variables appropriate for implementation of provider- and/or patient-facing risk calculators were subsequently selected and evaluated. These (parsimonious) models were developed by using the lasso procedure to identify the order in which variables are added to the model in an incremental fashion to maximize the model fit for any given number of variables (using penalized partial likelihood function) while taking into account the variables' clinical relevance, similar to the previously described approach.27, 28 Parsimonious predictive models were developed based on the combined training + testing dataset and evaluated on the held-out validation dataset. The development of the models started with the calculation of the Harrell C-index for each model that had ≤10 variables using the combined training + testing dataset. Body mass index was forced into the parsimonious model if not included using the lasso variable selection procedure. The discriminative ability of models with different number of variables was manually reviewed and a representative model was selected for each study outcome that included the lowest number of variables when the inclusion of additional variables did not lead to significant increases in the model's discriminative ability. A qualitative approach that took into account the trade-off between the increase in the model's discriminative ability (as represented by Harrell's C-index) versus the required increase in the number of variables that would make the predictive model's practical implementation more challenging as well as clinically perceived relevance of the variable being added was used to identify the final set of variables for each parsimonious predictive model. Each parsimonious predictive model developed in this way was then evaluated on the held-out validation dataset.
Analyses were conducted using R (R Foundation for Statistical Computing, Vienna, Austria). The RSF models and random forest imputation were implemented using the randomForestSRC package (R version 3.6.3 and randomForestSRC version 2.9.3 were used).29 Lasso-Cox modeling was implemented using the R package glmnet (R version 4.0.3 and glmnet package version 4.0.2 were used).30
3 RESULTS
3.1 Study cohort and selection of predictor variables
A total of 448,639 adults with BMI between 25 and 80 kg/m2 who were being treated in primary care practices affiliated with Mass General Brigham were identified. After excluding patients who a) were older than 80 or b) did not have information available for demographic characteristics, 433,272 patients were included in the study (Table 2). The median age of the study patients (Table 3) was 48 years and their median BMI 28.9 kg/m2; they were followed for a median of 2030 days (5.6 years). Over this period of time, the incidence of study outcomes ranged from 1.8% for knee replacement to 11.7% for ASCVD (Table 4). Based on the analysis of bivariate relationships between candidate predictor variables and study outcomes, all candidate predictor variables (Table 1) were selected for further analysis.
| Exclusion criterion | N after criterion applied | N excluded | % Excluded |
|---|---|---|---|
| Patients who met inclusion criteria | 448,639 | ||
| Patients >80 years old | 433,283 | 15,356 | 3.4% |
| Patients with missing gender | 433,272 | 11 | 0.003% |
| Variable | Mean (SD) or N (%) | Missing, N (%) |
|---|---|---|
| Study population | 433,272 | |
| Age, years | 47.9 (15.7) | 0 |
| Female | 226,172 (52.2) | 0 |
| Race/Ethnicity | ||
| Asian | 13,715 (3.2) | |
| Black | 34,813 (8.0) | |
| Hispanic | 28,136 (6.5) | |
| White | 324,836 (75.0) | |
| Other (includes unknown) | 31,772 (7.3) | |
| Partnereda | 240,465 (55.5) | 0 |
| Smoking | 179,587 (41.5) | 0 |
| Alcohol abuse | 10,417 (2.4) | 0 |
| Stimulant abuse | 2047 (0.47) | 0 |
| Commercial insurance | 271,755 (62.7) | 0 |
| ASCVD | 33,912 (7.8) | 0 |
| Heart failure | 11,668 (2.7) | 0 |
| Diabetes mellitus | 33,561 (7.8) | 0 |
| NASH/NAFLD | 4274 (1.0) | 0 |
| Sleep apnea | 14,484 (3.3) | 0 |
| Cancer | 39,111 (9.0) | 0 |
| DJD | 29,410 (6.8) | 0 |
| Knee replacement | 3937 (0.9) | 0 |
| Proteinuria | 27,489 (6.3) | 0 |
| Hepatitis B | 2037 (0.47) | 0 |
| Hepatitis C | 4359 (1.0) | 0 |
| HIV | 1305 (0.30) | 0 |
| Chronic inflammationb | 11,431 (2.6) | 0 |
| Helicobacter pylori infection | 2487 (0.57) | 0 |
| PCOS | 3132 (0.72) | 0 |
| COPD | 4589 (1.1) | 0 |
| Dementia | 1033 (0.24) | 0 |
| Sepsis | 3964 (0.91) | 0 |
| Gestational diabetes | 1125 (0.26) | 0 |
| Knee injury | 20,537 (4.7) | 0 |
| Joint infection | 2936 (0.68) | 0 |
| Valvular heart disease | 17,575 (4.1) | 0 |
| Hypertension | 120,583 (27.8) | 0 |
| Hypercholesterolemia | 57,175 (13.2) | 0 |
| Family history of ASCVD | 114,409 (26.4) | 0 |
| Family history of diabetes | 133,844 (30.9) | 0 |
| Family history of cancer | 185,788 (42.9) | 0 |
| BMI, kg/m2 | 30.5 (5.4) | 0 |
| 25.0–29.9 kg/m2 | 254,153 (58.7%) | |
| 30.0–34.9 k/m2 | 109,090 (25.2%) | |
| 35.0–39.9 kg/m2 | 43,099 (9.9%) | |
| ≥40.0 kg/m2 | 26,930 (6.2%) | |
| SBP, mm Hg | 125(16) | 52,531 (12.1) |
| DBP, mm Hg | 77(10) | 52,531 (12.1) |
| HbA1c, % | 6.0 (1.4) | 290,131 (67.0) |
| LDL-C, mg/dL | 111(34) | 114,248 (26.4) |
| eGFR, mL/min/1.73 m2 | 89(21) | 78,825 (18.2) |
| Length of follow-up, days | 2669 (1937) | 0 |
- a Partnered: married or living with a partner. Excludes single, separated, divorced and widowed.
- b Chronic inflammation incudes a) inflammatory bowel disease; b) rheumatoid arthritis; c) systemic lupus erythematosus; and d) multiple sclerosis.
- Abbreviations: ASCVD, atherosclerotic cardiovascular disease; BMI, body mass index; COPD, chronic obstructive pulmonary disease; DBP, diastolic blood pressure; DJD, degenerative joint disease; eGFR, estimated glomerular filtration rate; HIV, human immunodeficiency virus; LDL-C, low density lipoprotein cholesterol; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; PCOS, polycystic ovarian syndrome; SBP, systolic blood pressure.
| Outcome | Population | Patients who reached the outcome | Fraction who reached the outcome | Patient-years of follow-up | Annual incidence rate |
|---|---|---|---|---|---|
| ASCVD | 399,360 | 46,779 | 11.7% | 2,704,042 | 1.7% |
| Heart failure | 421,604 | 21,205 | 5.0% | 3,010,103 | 0.70% |
| Diabetes | 399,711 | 38,989 | 9.8% | 2,698,197 | 1.5% |
| NASH/NAFLD | 418,581 | 19,217 | 4.6% | 2,969,560 | 0.65% |
| Sleep apnea | 418,788 | 38,374 | 9.2% | 2,901,024 | 1.3% |
| Cancer | 394,161 | 34,679 | 8.8% | 2,756,490 | 1.3% |
| DJD | 403,862 | 50,952 | 12.6% | 2,587,897 | 2.0% |
| Knee replacement | 429,335 | 7872 | 1.8% | 3,099,043 | 0.25% |
| All-cause mortality | 433,272 | 21,014 | 4.9% | 3,176,315 | 0.66% |
- Abbreviations: ASCVD, atherosclerotic cardiovascular disease; DJD, degenerative joint disease; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis.
3.2 Machine learning predictive models
Training of 100 replicates of RSF predictive models for a single outcome took approximately 24 h on 80 central processing unit (CPU) cores of a computational cluster, totaling c. 1920 h (80 days) of computing time. For most outcomes studied, in each 100 replicates, the RSF model selected all available variables for the predictive model. The only exceptions were the predictive models of HF, knee replacement and all-cause mortality, for which the RSF model did not select gestational diabetes mellitus for 37%, 28% and 31% of replicates, respectively. The mean Harrell C-index for RSF predictive models over 100 replicates on the test dataset ranged from 0.702 for NASH/NAFLD to 0.896 for all-cause mortality (Table 5).
| Outcome | On testing (24%) dataseta | On validation (held-out 20%) dataset | ||
|---|---|---|---|---|
| Lasso-cox | RSF | Lasso-cox: Full | Lasso-cox: Parsimonious | |
| ASCVD | 0.805 | 0.812 | 0.801 | 0.788 |
| Heart failure | 0.862 | 0.871 | 0.856 | 0.840 |
| Diabetes | 0.809 | 0.823 | 0.803 | 0.782 |
| NASH/NAFLD | 0.702 | 0.702 | 0.694 | 0.671 |
| Sleep apnea | 0.730 | 0.731 | 0.725 | 0.721 |
| Cancer | 0.727 | 0.734 | 0.724 | 0.720 |
| DJD | 0.749 | 0.757 | 0.741 | 0.736 |
| Knee replacement | 0.865 | 0.862 | 0.847 | 0.843 |
| All-cause mortality | 0.888 | 0.896 | 0.891 | 0.850 |
- a Mean over 100 replicates.
- Abbreviations: ASCVD, atherosclerotic cardiovascular disease; DJD, degenerative joint disease; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; RSF, random survival forest.
3.3 Traditional statistical predictive models
Training of 100 replicates of Lasso-Cox models for a single outcome took approximately 160 s on a single CPU. The mean Harrell C-index for Lasso-Cox predictive models on the test dataset over 100 replicates ranged from 0.694 for NASH/NAFLD to 0.891 for all-cause mortality (Table 5). Across all study outcomes, the average mean Harrell C-index for Lasso-Cox predictive models was lower than the Harrell C-index for the corresponding RSF predictive models by 0.012, ranging from 0.005 for all-cause mortality to 0.020 for T2DM.
Based on its predictive accuracy (slightly lower but comparable to RSF), significantly lower computational requirements and greater interpretability, Lasso-Cox modeling approach was selected for the final predictive model validation. On the training + testing (80% of the overall dataset) dataset, the final Lasso-Cox models were selected from 26 (knee replacement) to 35 (ASCVD) out of 40 available predictor variables (Table 1). Body mass index, age, sex, marital status, HbA1c, hepatitis C, hypertension, smoking status, family history of ASCVD and family history of cancer were included in models for all study outcomes based on Lasso variable selection. On the held-out validation dataset, the Harrell C-index for these Lasso-Cox predictive models, trained on the training + testing dataset, ranged from 0.675 for NASH/NAFLD to 0.849 for all-cause mortality. Body mass index was significantly (p < 0.0001) associated with all study outcomes. Hazard ratios for the relationship between BMI and study outcomes (using one standard deviation of BMI in each respective study population) varied from 1.060 for cancer to 1.602 for sleep apnea (Table 6).
| Outcome | 1 SD of BMI, kg/m2 | Hazard ratio (95% CI) |
|---|---|---|
| Sleep apnea | 5.189 | 1.602 (1.587–1.618) |
| Diabetes | 5.169 | 1.502 (1.486–1.517) |
| NASH/NAFLD | 5.346 | 1.433 (1.416–1.450) |
| Knee replacement | 5.348 | 1.395 (1.370–1.421) |
| Heart failure | 5.311 | 1.343 (1.318–1.369) |
| DJD | 5.286 | 1.235 (1.224–1.246) |
| ASCVD | 5.339 | 1.148 (1.137–1.160) |
| All-cause mortality | 5.364 | 1.142 (1.125–1.159) |
| Cancer | 5.380 | 1.060 (1.048–1.073) |
- Abbreviations: ASCVD, atherosclerotic cardiovascular disease; DJD, degenerative joint disease; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis.
3.4 Parsimonious risk predictive models
Parsimonious Lasso-Cox models included four (cancer and knee replacement) to seven (ASCVD, HF, NASH/NAFLD, DJD and all-cause mortality) variables (Table 7). Body mass index was selected by the lasso procedure as one of the first 10 variables for all models except all-cause mortality and was forced into the all-cause mortality model. Among the rest of the predictor variables, age was included most frequently (eight out of nine predictive models) followed by hypertension (five models). Assessment of model accuracy on the validation (held-out) dataset demonstrated a Harrell C-index ranging from 0.675 for NASH/NAFLD to 0.850 for knee replacement (Table 5).
| Variable | ASCVD | Heart failure | T2DM | Sleep apnea | NASH/NAFLD | Cancer | DJD | Knee replacement | Death |
|---|---|---|---|---|---|---|---|---|---|
| BMI | X | X | X | X | X | X | X | X | X |
| Age | X | X | X | X | X | X | X | X | |
| Sex | X | X | X | X | X | ||||
| Smoking status | X | X | X | X | |||||
| Family history of cancer | X | X | X | ||||||
| Family history of diabetes | X | X | X | ||||||
| Family history of ASCVD | X | ||||||||
| ASCVD | X | X | |||||||
| Heart failure | X | ||||||||
| T2DM | X | X | X | ||||||
| Cancer | X | ||||||||
| DJD | X | ||||||||
| Hypertension | X | X | X | X | X | ||||
| Valvular heart disease | X | ||||||||
| Knee injury | X | X | |||||||
| Hepatitis B | X | ||||||||
| Chronic inflammation | X | ||||||||
| eGFR | X | X | X | ||||||
| HbA1c | X | X |
- Abbreviations: ASCVD, atherosclerotic cardiovascular disease; BMI, body mass index; DJD, degenerative joint disease; eGFR, estimated glomerular filtration rate; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis.
4 DISCUSSION
This large observational study of over 400,000 overweight and obese patients successfully developed predictive models for a broad range of obesity-related complications. Most predictive models achieved good to strong discriminative ability, confirmed on the held-out validation dataset that was not used in model development. Model accuracy was largely retained in the corresponding parsimonious predictive models that only used four to seven predictor variables, facilitating incorporation into medical decision-making.
Models developed using the machine learning technique RSF attained the highest discriminative ability. However, predictive models that utilized Cox proportional hazards regression were only slightly less accurate, with the majority of models having a Harrell C-index above 0.8 (considered strong discriminative ability) and only one slightly below 0.7 (considered good discriminative ability). One possible explanation for this small difference is the nature of the data, where most variables (e.g., diagnoses) were binary. One important advantage of machine learning methods such as RSF is their superior ability to model non-linear relationships. However, data with a predominance of binary variables will not have extensive non-linear relationships, thus decreasing the possible gains from machine learning analytical techniques. Another possible explanation is that the number of variables in the models was relatively small, minimizing the opportunity to leverage interactions between multiple variables—another potential strength of machine learning techniques.
The accuracy of predictive models ranged (in their most accurate, RSF implementation) from 0.702 for NASH/NAFLD to 0.896 for mortality. Lower discriminative ability for conditions like NASH/NAFLD and sleep apnea could be explained, in part, by their likely underdiagnosis.31, 32 On the other hand, the incidence of death was ascertained from a combination of local hospital data and the Social Security Death Master File, leading to a fairly comprehensive identification of deceased individuals, which could potentially explain the particularly high performance of the predictive models in this area. The accuracy of the predictive models developed in this study was comparable to other previously published predictive models for the same outcomes.13, 33, 34
Even though multiple variables were selected into the predictive models by the lasso procedure and many of these had highly significant relationships with the corresponding outcome, parsimonious risk models that only included four to seven predictor variables had only slightly lower discriminative ability. One potential explanation could be the relatively low prevalence of many of the patient characteristics that were highly predictive of complications (e.g., human immunodeficiency virus infection) and relatively low strength of the effect of others (e.g., elevated blood pressure or hypercholesterolemia) that were not ultimately included in the parsimonious models. On the other hand, relatively common risk factors such as age, smoking and family history of related conditions were included in many of the parsimonious risk models.
Body mass index was significantly associated with all nine outcomes studied. This finding underscores the importance of excess weight as a major contributor to human health and quality of life. It is thought that overweight and obesity play a significant role in the lag of life expectancy in the U.S. compared to many other developed countries.35 Nevertheless, this major health problem is often not addressed by treating clinicians36 and potential treatments are not being discussed with patients,37 reflecting possibly a mix of social stigma and therapeutic inertia.
The development of compact and highly accurate predictive models for obesity-related complications could have important clinical applications. These models could be used by clinicians at the point of care to identify patients most likely to benefit from anti-obesity interventions; by patients and their family members to decide which treatment options, including bariatric surgery and anti-obesity medications, to pursue (patients with overweight/obesity initiate many discussions of anti-obesity treatments38, 39); and in population management to make decisions on allocation of healthcare resources.40
The present study had a number of strengths. It was based on a large population of over 400,000 patients, the majority of whom were followed for over five years. It drew on a rich dataset of 40 variables selected based on published literature and compared both traditional regression-based and machine learning methodologies. Predictive models that were generated in the course of the study were rigorously validated on a held-out dataset that was not used in the development of the models. Finally, the study also developed parsimonious predictive models that utilized a limited number of variables in order to facilitate practical implementation while retaining most of the accuracy.
The findings of this study should be interpreted in light of several limitations. The study was conducted in Eastern Massachusetts (a state with lower-than-average prevalence of overweight and obesity) and therefore, the findings may not be fully generalizable to the rest of the U.S. The analysis used data from EHR which might have been incomplete or sometimes inaccurate with respect to either outcomes or patient characteristics, affecting the accuracy of the predictive models. In particular, HbA1c measurements were missing for 67% of the study patients, and using imputation for this variable may have impacted the model accuracy. Finally, the racial/ethnic diversity of study patients was not representative of the U.S. population (75% white vs. 65% white for the U.S. population).
The findings of this study could serve as the foundation for the next steps towards the identification of patients who could derive the greatest benefits from weight loss interventions, including anti-obesity medications. These could include the development of risk calculators for both healthcare providers and patients, medical decision aides as well as cost-effectiveness assessments. The development of these tools could be further facilitated by increasing availability of EHR information that also served as the source of data for the predictive models developed in this study.
5 CONCLUSIONS
Predictive modeling can identify patients with overweight and obesity at a high risk of obesity-related complications. These Cox models, including those with parsimonious lists of predictors, achieve high accuracy and can be used to identify patient characteristics indicative of higher risk, potentially helpful for population health management and clinical treatment decisions.
AUTHOR CONTRIBUTIONS
All authors participated in the design of the study. AT and FM collected the data. FM and MS analyzed the data. All authors participated in data interpretation. AT drafted the manuscript. All authors critically reviewed the manuscript and have given the final approval for the manuscript to be published. AT obtained funding and supervised the study.
ACKNOWLEDGMENTS
The study was funded by Eli Lilly. The study funder was involved in the study design, interpretation of data and writing the manuscript. We would like to thank Dr. Guohai Zhou for verification of the statistical analysis.
CONFLICT OF INTEREST STATEMENT
Turchin reports equity in Brio Systems, consulting for Novo Nordisk and Proteomics International, and research support from Astra-Zeneca, Eli Lilly and Company and Novo Nordisk. Lipkovich, Shinde, Ahmad and Kan are employees and stockholders of Eli Lilly and Company. None of the other authors report any conflicts of interest.