Novel hemizygous single-nucleotide duplication in RPGR in a patient with retinal dystrophy and sensorineural hearing loss
Texome Project Members listed in Table S2.
Abstract
Background
The RPGR gene has been associated with X-linked cone-rod dystrophy. This report describes a variant in RPGR detected with exome sequencing (ES). Genes like RPGR have not always been included in panel-based testing and thus genome-wide tests such as ES may be required for accurate diagnosis.
Methods
The Texome Project is studying the impact of ES in medically underserved patients who are in need of genomic testing to guide diagnosis and medical management. The hypothesis is that ES could uncover diagnoses not made by standard medical care.
Results
A 58-year-old male presented with retinitis pigmentosa, sensorineural hearing loss, and a family history of retinal diseases. A previous targeted gene panel for retinal disorders had not identified a molecular cause. ES through the Texome Project identified a novel, hemizygous variant in RPGR (NM_000328.3: c.1302dup, p.L435Sfs*18) that explained the ocular phenotype.
Conclusions
Continued genetics evaluation can help to end diagnostic odysseys of patients. Careful consideration of genes represented when utilizing gene panels is crucial to ensure an accurate diagnosis. Medically underserved populations are less likely to receive comprehensive genetic testing in their diagnostic workup. Our report is an example of the medical impact of genomic medicine implementation.
1 INTRODUCTION
Recent studies have shown that an increasing proportion of blindness among younger and working-age adults is caused by genetic disorders of the eye including inherited retinal dystrophy, which has an estimated prevalence between 0.06% and 0.2% (Hanany et al., 2020; Heath Jeffery et al., 2021). Inherited retinal dystrophy comprises a group of genetically heterogeneous disorders and to date, pathogenic variants in over 700 genes have been discovered as causal for these disorders. Retinitis pigmentosa GTPase regulator (RPGR), located on Xp11.4, is associated with a wide phenotypic spectrum of retinal diseases, including retinitis pigmentosa 3(MIM300029), X-linked retinitis pigmentosa and sinorespiratory infections with or without deafness (MIM300455), X-linked cone-rod dystrophy (MIM304020), and X-linked atrophic macular degeneration (MIM300834) (Ayyagari et al., 2002; Demirci et al., 2002; Roepman et al., 1996; Zito et al., 2003). Pathogenic variants in RPGR have been reported to cause nearly two-thirds of all X-linked retinitis pigmentosa (Khanna et al., 2005; Mihailovic et al., 2022; Vervoort et al., 2000). Individuals with RPGR-related X-linked retinitis pigmentosa (XLRP) have an aggressive form of the disease, characterized by night blindness in the first or second decades of life, followed by progressive loss of visual acuity and legal blindness by the third or fourth decades of life (Di Iorio et al., 2020; Georgiou et al., 2021; Nguyen et al., 2020; Sandberg et al., 2007). Individuals with RPGR-related retinal diseases can also manifest extraocular phenotypes, including hearing loss, chronic otitis media, sinusitis, bronchitis, and bronchiectasis (Georgiou et al., 2021; Zito et al., 2003). Due to X-linked inheritance, most affected individuals are hemizygous males. However, heterozygous female carriers can also have ocular findings which can range from minimal visual impairment to severe retinitis pigmentosa (RP), sometimes with extraocular findings, particularly sinusitis, otitis media, and hearing loss (Georgiou et al., 2021; Tuupanen et al., 2022; Zito et al., 2003).
As there is significant clinical variability and phenotypic overlap between inherited retinal disorders, an accurate diagnosis is often dependent on molecular genetic testing; gene panel-based testing has become a standard approach in many clinics. This creates a disparity for medically underserved and under-resourced groups of the population who do not have access to genomic medicine. Such disparity occurs due to many factors, including decreased ability to navigate the healthcare system due to impaired vision, financial difficulty resulting from RP, and lack of access to genetic services. Additionally, long wait times to obtain a genetics referral and delays associated with getting appropriate comprehensive testing are barriers to having timely access to therapeutic approaches such as cell therapy and gene therapy (Cross et al., 2022).
Access to comprehensive genetic testing such as exome sequencing (ES) and whole genome sequencing (WGS) is crucial to find answers for patients and families with undiagnosed inherited retinal disorders. For many individuals such genome-wide genetic testing has been inaccessible because of a lack of resources in their communities, lack of insurance coverage, and inability to travel to medical providers with genetics expertise (Chou et al., 2021; National Academies of Sciences, 2018; Reuter et al., 2019; Schoch et al., 2021). XLRP confers a significant emotional, social, physical, and economic burden on patients because of its progressive nature that interferes with one's ability to independently perform activities of daily living (Chaumet-Riffaud et al., 2017). Several studies have shown that RP severely impacts quality of life as the condition presents barriers to traditional employment, hobbies, and family life. Furthermore, caregivers are also impacted by secondary exclusion from activities in which their loved one(s) cannot participate and by barriers to employment because of their caregiving role (Chivers et al., 2021; Watanabe et al., 2023). However, with support from appropriate social, healthcare, and educational services, some of these burdens can be decreased (Bertelsen et al., 2015). The Texome Project is a genomic implementation program focused on providing ES from a clinical genetics diagnostic laboratory to investigate the molecular cause(s) of undiagnosed diseases in populations that have not historically had access to genomic medicine due to a variety of barriers. The Texome Project includes the opportunity for reanalysis of genetic data and model organism study of candidate variants if ES is nondiagnostic. Participants are also engaged in a 2-year longitudinal follow-up of the perceptions, utility, and barriers they associate with genomic medicine. Here, we report on a molecular genetic diagnosis with ES in an individual with RP, in whom a molecular cause had not been identified after standard, panel-based testing.
2 MATERIALS AND METHODS
2.1 Ethical compliance
Informed consent was obtained from the participant under the Baylor College of Medicine Institutional Review Board Protocol H-49277 prior to participation in the Texome Project, a National Human Genome Research Institute (NHGRI) and private philanthropic-funded study that explores the diagnostic value of WES on the care of medically underserved children and adults with genetic diseases. The study enrolls individuals with phenotype compatible with a Mendelian genetic disease and individuals with complex clinical presentations where one diagnosis is not likely to explain the phenotype. Medical history was obtained by a review of the medical records.
ES was performed on DNA isolated from a buccal swab at Baylor Genetics Laboratory, a tertiary that is certified by the College of American Pathologists and is compliant with the Clinical Laboratory Improvement Amendments. The isolated DNA was indexed into separate libraries using the KAPA Hyper Prep Kit. Illumina paired-end massively parallel sequencing with 100 bp insert size was performed. The sequencing had a mean read depth of 159× with 98.9% of exons having at least 20× coverage. Data were aligned to the human reference genome build GRCh37 (Hg19). Data analysis and filtration were performed using the Illumina Dragen BioIT platform, Illumina Dragen haplotype-based variant caller and Illumina Dragen genome-wide depth-based CNV caller. Variant interpretation was performed using EMEDGENE software (Illumina), and curation was performed in accordance with the ACMG guidelines (Richards et al., 2015).
3 RESULTS
The participant was accepted into the Texome Project based on objective abnormal findings that indicated a genetic etiology, unique clinical presentation, multiple organ system involvement, and family history of retinal disease.
3.1 Clinical presentation
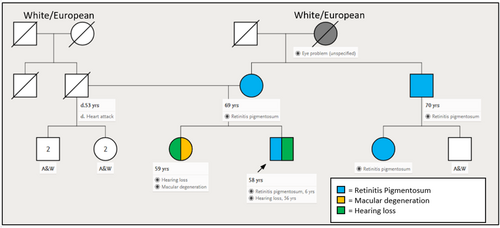
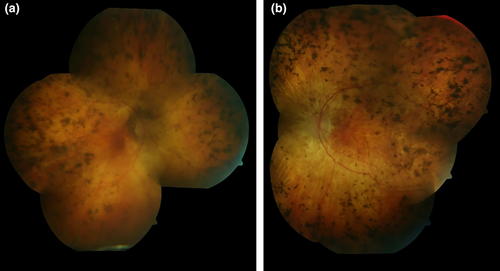
A 58-year-old male with cone-rod dystrophy and sensorineural hearing loss was referred to the Texome Project. This individual was diagnosed with retinitis pigmentosa at the age of 6 years as he had developed night blindness and had clinical findings suggestive of progressive bilateral retinal dystrophy. His vision had progressively deteriorated and at the time of evaluation at the genetics clinic, at the age of 57 years, he was legally blind. At 55 years of age, he also developed vertigo and hearing loss which had progressed rapidly over the preceeding two years. Additionally, throughout his life, he exhibited right-sided hemihypertrophy and macrocephaly. The family history was significant for a history of RP in the mother (age of onset 40 years), maternal uncle (age of onset unknown), and a female cousin through this maternal uncle (age of onset unknown). The sister of the proband had a diagnosis of “macular degeneration” (Figure 1). Physical examination at 57 years of age revealed mild hemihypertrophy with the right arm and forearms measuring 4 cm larger than the left. Mild hemihypertrophy was also noted in the right thigh and leg regions. An ophthalmology examination showed light perception in the right eye and no light perception in the left eye. The fundi exhibited vitreous syneresis, scattered pigmented cells, and features of classic end-stage retinitis pigmentosa (Figure 2).
Based on the phenotype, a commercially available inherited retinal disorders panel was ordered. This panel, which interrogated sequence variants and copy number variants in 293 genes known to cause retinal dystrophies (Table S1), did not identify a molecular diagnosis. We were unable to perform a genome-wide genetic test because of the complex logistical and financial challenges surrounding access to genomic medicine in the United States. Thus, the individual was enrolled in the Texome Project (Figure 3).
3.2 Genetic diagnosis
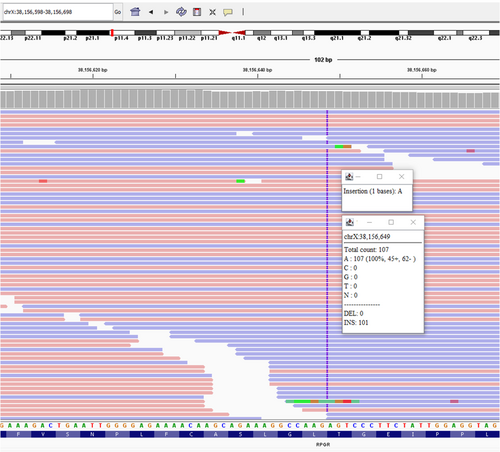
ES identified a single-nucleotide duplication in RPGR (c.1302dup, p.L435Sfs*18) (ClinVar ID:1707409). This variant causes a frameshift in exon 11, leading to a premature termination codon which is predicted to result in nonsense-mediated decay. This novel change is absent from GnomAD and has not been described previously in Clinvar. This result provides a molecular cause for the retinal dystrophy and may explain the hearing loss in our patient as no other genetic etiology was found. The exome sequencing did not identify any variants in known genes associated with hearing loss genes other than RPGR. Due to the later onset hearing loss for our patient, age may also be a factor in this case. However, we were not able to identify a molecular cause for the hemihypertrophy and macrocephaly observed in the proband. The RPGR gene was not included on the commercially available panel by the laboratory because of technical challenges associated with sequencing the repetitive glutamic acid and glycine-rich areas involved in the open reading frame of RPGRORF15 (Maggi et al., 2020). As a result, the RPGR-related RP diagnosis was missed with panel testing.
4 DISCUSSION
The RPGR protein plays an important role in the function of photoreceptors connecting cilia that are responsible for trafficking proteins to the outer segments of photoreceptors in the retina. Furthermore, RPGR has kinociliary functions in nonocular tissue, including the testis, kidneys, brain, lungs, and pancreas (Georgiou et al., 2021; Ghosh et al., 2010; Moreno-Leon et al., 2021; Zito et al., 2003).
There are two well-described isoforms of RPGR. The first isoform has 19 exons encoding 815 amino acids and is constitutively expressed, while the second shares the first 14 exons and then has an open reading frame at exon 15 (RPGRORF15) that includes part of intron 15 as the last exon. RPGRORF15 is expressed exclusively in the connecting cilium of photoreceptors of the retina (Di Iorio et al., 2020; Georgiou et al., 2021; Khanna et al., 2005; Moreno-Leon et al., 2021; Vervoort et al., 2000). The terminal exon 15 is a hotspot for mutations due to glutamate-glycine repeats and accounts for up to 80% of disease-causing variants (Di Iorio et al., 2020; Nguyen et al., 2020). The proband's variant falls in the second quartile of exon 11. While there are various reports on whether the phenotypes of individuals with mutations in exons 1–14 are more severe than those with mutations in ORF15, the extraocular findings such as in our patient have been described only in variants in exons 1–14 (Di Iorio et al., 2020; Sandberg et al., 2007; Talib et al., 2019; Zou et al., 2020).
When utilizing targeted panels in patients with rare diseases, cases with etiologies related to lesser-known genes or recently established gene-disease relationships can be missed since the process of adding genes to panels can be lengthy. In the case of RPGR, almost 10 years passed from the time a definitive disease relationship was outlined by Branham et al. to the time that RPGR was added to many retinal disease gene panels. Comprehensive genetic testing using ES or WGS can address this concern with complete coverage of the exome or genome, respectively, enabling evidence to be built from candidate genes. However, for individuals from underserved populations, access to panel testing may be significantly higher than access to ES or WGS because of current insurance payor coverage guidelines for public insurance. Particularly in scenarios where there is discordance from the expected inheritance pattern, as seen in female carriers of X-linked RPGR who can have similar presentations to affected hemizygous males, the diagnostic process can prove difficult because of variable expressivity in both males and females (Georgiou et al., 2021; Nanda et al., 2018; Tuupanen et al., 2022). Thus, the implementation of genetics services and comprehensive genetic testing in underserved populations, such as through the Texome Project, is key to transitioning to a more equitable era of genomic medicine that serves to guide care and management of disease in diverse groups.
Genomic medicine has been an instrumental tool in diagnosing individuals with rare diseases and limiting the diagnostic odyssey phenomenon (El Naofal et al., 2023; Wiener et al., 2023). Genetic testing has proved to be a cost-effective way to diagnose conditions and direct medical resources toward appropriate therapies, surveillance, or treatment, though many barriers and limitations remain implementation of such testing (Muth et al., 2019; O'Brien et al., 2021; Serrano et al., 2023). The COVID-19 pandemic both exposed these access disparities and eased some barriers by increasing the uptake of telemedicine in genetics clinics, willingness to share genetic information, and to undergoing genetic testing (Deruelle et al., 2023; Macalino et al., 2023). However, some challenges were exacerbated such as the exclusion of patients without reliable access to the internet or technological literacy to utilize telemedicine (Mann et al., 2021). Insurance payor limitations may also restrict the effectiveness of genetic services as they may stipulate narrow coverage guidelines (Amendola et al., 2019; Suther & Kiros, 2009). Additional strain can be observed in obtaining an appropriate genetics referral due to the lack of familiarity with genetic indications or limited accessibility of genetics providers outside of large medical centers. Intentional and structured collaboration between genetics and nongenetics providers can be effective at bridging this gap (Morrow et al., 2023; Truong et al., 2021). Lastly, genetics studies have had suboptimal inclusion of nonwhite populations which contributes to difficulty interpreting the genetic findings in diverse populations without the diverse background population data to serve as a control (Appelbaum et al., 2022; Fine et al., 2005; Gerido et al., 2023; Glenn et al., 2012).
5 CONCLUSION
To date, there are 593 pathogenic or likely pathogenic RPGR variants registered in ClinVar, and RPGR variants are noted to be causal in 15%–20% of all individuals with retinitis pigmentosa cases (Branham et al., 2012; Shu et al., 2005). Thus, RPGR should be considered a first-tier gene when screening males with retinal dystrophy consistent with X-linked inheritance. In the clinic, it may be difficult to discern a clear X-linked recessive pattern with a limited number of affected family members especially in the presence of affected females. Such pedigrees can also suggest an autosomal dominant inheritance with variable expressivity and incomplete penetrance. In this report, we highlight how a comprehensive retinal disorders gene panel from a major commercial genetics laboratory ordered in 2021 did not include RPGR. Access to more comprehensive genetic testing such as ES is important for the diagnosis of genetic disorders especially when considering diseases that have a short window in which curative genomic therapies can be implemented. Our report highlights the need for close consideration of gene inclusion on panels both for laboratories and for clinicians who order genetics testing.
AUTHOR CONTRIBUTIONS
R.J.G., B.V. wrote the first draft of the manuscript. L.S., S.N. recruited the human subject and provided clinical evaluation. L.V., N.O. performed and interpreted the exome sequencing. R.A.L. provided ophthalmic evaluation and photographs. M.F.W., S.N. oversaw the project. All authors edited and contributed to the final draft.
ACKNOWLEDGMENTS
First and foremost, we would like to recognize the research participant for his crucial role in our research of genetic disease. This work was supported by the Texome Project which is funded by the National Human Genome Research Institute (NHGRI) under R01HG011795. The Texome Project also receives philanthropic funding through the Jan and Dan Duncan Neurological Research Institute of Texas Children's Hospital. This work was supported in part by funding of The Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (P50HD103555) from the Eunice Kennedy Shriver NICHD. The contents of this publication do not necessarily reflect the views or policies of the NIH. The mention of trade names, commercial products, or organizations do not imply endorsement by the US Government. A full list of collaborators is provided in Supplemental Table S2.
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest to disclose.
ETHICS STATEMENT
Consent for participation in clinical research and publication was obtained from the individual presented in this case report. The Texome Project protocol has been reviewed and approved for use with human subjects by the Baylor College of Medicine Institutional Review Board (Protocol # H-49277).
Open Research
DATA AVAILABILITY STATEMENT
Variant level data from this report are available on Clinvar at reference number: SCV002574700.1.




