Characterization of a novel COL10A1 variant associated with Schmid-type metaphyseal chondrodysplasia and a literature review
Huixiao Wu and, Shuping Wang contributed equally to this work.
Abstract
Background
Schmid-type metaphyseal chondrodysplasia (SMCD) is a rare autosomal dominant skeletal dysplasia caused by heterozygous mutations in COL10A1, the gene which encodes collagen type X alpha 1 chain. However, its genotype–phenotype relationship has not been fully determined.
Subjects and Methods
The proband is a 2-year-old boy, born of non-consanguineous Chinese parents. We conducted a systematic analysis of the clinical and radiological characteristics and a follow-up study of the proband. Whole-exome sequencing was applied for the genetic analysis, together with bioinformatic analysis of predicted consequences of the identified variant. A homotrimer model was built to visualize the affected region and predict possible outcomes of this variant. Furthermore, a literature review and genotype–phenotype analysis were performed by online searching all cases with SMCD.
Results
A novel heterozygous variant (NM_000493.4: c.1863_1866delAATG, NP_000484.2: p.(Met622 Thrfs*54)) was identified in COL10A1 gene in the affected child. And it was predicted to be pathogenic by in silico analysis. Protein modeling revealed that the variant was located in the NC1 domain, which was predicted to produce truncated collagen and impair the trimerization of collagen type X alpha 1 chain and combination with molecules in the matrix. Moreover, genotype–phenotype correlation analysis demonstrated that patients with truncating variants or variants in NC1 domain often presented earlier onset and severer symptoms compared with those with non-truncating or variants in non-NC1 domains.
Conclusion
The NC1 domain of COL10A1 was proved to be the hotspot region underlying SMCD, patients with variants in NC1 domain were more likely to present severer manifestations at an earlier age.
1 INTRODUCTION
Short stature (SS) in childhood is one of the most common chief complaints about a referral to pediatric endocrinologists, which is defined as actual height below 2 SDS of the average height for age, sex, and ethnic group established norm (Allen & Cuttler, 2013; Rogol & Hayden, 2014). There are a number of etiologies attributing to short stature which can be either congenital or acquired, and both can cause physical deformities and place a huge burden on patients and their families. Early diagnosis is quite necessary for these patients to accept early and effective intervention. Nevertheless, many disorders associated with short stature present similar phenotypes, making it more difficult to determine the cause.
Schmid-type metaphyseal chondrodysplasia (SMCD, OMIM 156500) is a rare autosomal dominant disease that is characterized by short stature, long bone deformities such as genu varum and genu valgum, and typical radiographic manifestations of metaphyseal dysplasia of the tubular bones especially the femur (e.g., splaying, flaring, widening, cupping; Richmond & Savarirayan, 2019). This skeletal dysplasia is attributed to the mutation of the COL10A1 gene (OMIM 120110) which encodes collagen type X alpha 1 chain and is featured by the assembly defect of type X collagen in the growth plate (Warman et al., 1993; Wilson et al., 2002). Human type X collagen is a homotrimer of three collagen type X alpha 1 chains, each crude chain composed of a signal peptide (amino acids 1–18) at the N-terminal followed by a non-collagenous 2 (NC2) region (amino acids 19–56), a triple helix-forming Gly-X-Y repeats region (amino acids 57–519) and a non-collagenous 1 (NC1) region (amino acids 520–680) at the C-terminal responsible for initiating trimerization of type X collagen and forming specific supramolecular structures within the cartilage matrix, which plays an important role in fetal chondrogenesis and endochondral ossification (Bogin et al., 2002). Till now, only 59 heterozygous COL10A1 variants have been reported to cause SMCD. Most SMCD-related variants identified recently cluster in the C-terminal NC1 trimerization domain (Bateman et al., 2005). Since the genotype–phenotype correlation in SMCD is still not clear, it will be helpful to better understand the genotype–phenotype relationship if more cases are found and studied.
Interestingly, here we found a SMCD patient with a new heterozygous frameshift variant of the COL10A1 gene which was predicted to be pathogenic possibly through NMD. Moreover, we summarized all previously reported variants as well as their phenotypic information, and further analyzed the genotype–phenotype correlation. Our results enrich the gene mutation spectrum to help identify the COL10A1 gene function and reveal the pathogenesis underlying SMCD.
2 PATIENTS AND METHODS
2.1 Ethical compliance
This study has been approved by the Ethics Committee of Shandong Provincial Hospital affiliated to Shandong University. The study protocol was in line with the Declaration of Helsinki (as revised in Brazil 2013). Informed consent was obtained from all individual participants included in the study and written informed consent was received from participants prior to inclusion in the study.
2.2 Patient
A detailed history about the onset and progression of growth failure and lower limb deformities as well as family history were obtained. Physical examination, laboratory detection, and X-rays were performed to confirm the diagnosis. Peripheral blood samples were obtained from the patient and his family members for genetic testing.
2.3 DNA extraction and whole-exome sequencing
Genomic DNA was isolated from peripheral blood leukocytes using the QIAamp DNA Mini Kit (Qiagen, Germany) following the manufacturer's instructions. Whole-exome sequencing (WES) was performed on DNA from peripheral blood. After genomic DNA fragmentation, paired-end adaptor ligation, amplification, and purification, the human exons were captured by using the SeqCap EZ Med Exome Enrichment Kit (Roche NimbleGen). The DNA library was generated by postcapture amplification and purification and then sequenced on the Illumina HiSeq sequencing platform. Sequence data alignment to the human genome reference (hg19) and variant-calling were performed with NextGene V2.3.4 software to obtain the coverage and mean read depth of the target regions. The average coverage of the exome was >100×, which permitted the examination of the target region with enough depth to exactly match >99% of the target exome. To ensure the accuracy of data analysis, mutations with low coverage in the target area will be filtered out.
Additionally, annotation information, including the conservation of nucleotide bases and amino acids, predictions of biological functions, the frequency in normal populations (Genome Aggregation Database [GenomAD], Trans-Omics for Precision Medicine [TOPMED], the Exome Aggregation Consortium [ExAC]), and data from the Human Gene Mutation Database (HGMD), Clinvar and Online Mendelian Inheritance in Man (OMIM) databases, was performed by NextGene V2.3.4 and our in-house scripts. A variant was recognized as a mutant when it was not found in dbSNP (http://www.ncbi.nlm.nih.gov/snp/), in the exome variant server (http://evs.gs.washington.edu/EVS/), in the Ensembl database and in 500 Chinese controls, or alternatively, the allele frequency was found to be less than 0.001 in the database. Pathogenic variants were determined according to the Standards and Guidelines for the Interpretation of Sequence Variants published by the American College of Medical Genetics and Genomics (ACMG) in 2015 with the Human Genome Variation Society (HGVS) nomenclature (den Dunnen et al., 2016; Richards et al., 2015).
When the detected pathogenic or suspected pathogenic variants exists, the laboratory verified it by Sanger sequencing and ensured that the coverage of the gene coding sequence reached 100%. Using Primer3 version 1.1.4 (http://www.sourceforge.net) and GeneDistiller 2014 (http://www.genedistiller.org/), tagged sequencing primers of COL10A1 were designed. Polymerase chain reaction (PCR) was performed in a 50 μL system including 4 μL genomic DNA, 1 μl forward and reverse primers, 5 μL 10 × PCR buffer, 4 μL dNTPs, and 0.3 μL Taq Hot Start (Takara Bio). The PCR conditions were as follows: an initial denaturation step (95°C for 5 min), followed by 40 cycles of denaturation (95°C for 30 s), annealing (65°C for 30 s), and elongation (72°C for 30 s). Amplicons were sequenced using an ABI 3730 system (Applied Biosystems), and sequence analysis was performed using the autoassembler software Chromas 2.6.6 (Technelysium Pty Ltd. Available at www.technelysium.com.au/chromas.html.) and visual inspection.
2.4 Bioinformatic analysis
The bioinformatic analysis of COL10A1 (Gene ID:1300, NCBI Reference sequence: NG_008032.1) variant was performed by two software tools, MutationTaster (http://www.mutationtaster.org/), PROVEAN (http://provean.jcvi.org) to predict disease-causing effects of the variant. Furthermore, SWISS-MODEL software was used to build a model of the homotrimer of type X collagen trimerization domain and DeepView software was used for visualizing the spatial structure and altered residues of the protein model.
2.5 Follow-up study
After his first clinical evaluation, the patient was followed-up every 3 months in our hospital to closely keep track of his growth and development, therapeutic regimens, and potential complications as well as biochemical tests and radiographs performed if necessary. If there was an emergency (such as acute joint pain), the patient would be taken to the local hospital for examination and treatment.
2.6 Statistical analysis
Statistical analysis was performed by SPSS 19.0 software package (SPSS Inc.). The Kolmogorov–Smirnov test was used to determine the distribution of continuous variables. Continuous variables with normal distribution were given as mean±SD and compared by independent samples Student's t-test, while those with non-normal distribution were given as median (25th, 75th percentiles) and compared by Mann–Whitney U-test. p < .05 was considered statistically significant.
3 RESULTS
3.1 Clinical features
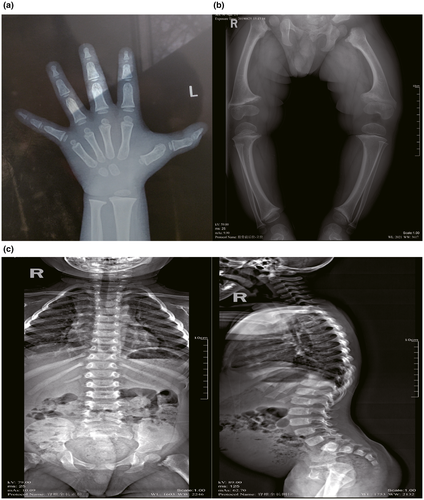
The patient, a 2-year-old Chinese boy, was first admitted to our hospital for genu varum and waddling gait. After a 40-week gestation without any abnormality, he was born with a body length of 51 cm (+0.3 SD), and a body weight of 3.8 kg (+1.2 SD). When he could first walk at the age of 1-year-old, he was found genu varum and waddling gait. He was only 81 cm (−2.1 SD) tall and weighed 13 kg (+0.3 SD) when he was 2 years old. The serum biochemical and hormone assays at the first clinical examination showed normal serum calcium, phosphate, vitamin D, parathyroid hormone, and a slightly increased alkaline phosphatase with normal bone mineral density and bone age (Table 1). The measurements of different body segments were listed in Table 2. Radiographs showed mild dorsal scoliosis, genu varum, and metaphyseal widening and irregularity (Figure 1). There were no extra-skeletal manifestations. The cytogenetic analysis confirmed a 46,XY karyotype. His parents were not consanguineous and there was no special medical or family history. His older brother was both physically and mentally healthy.
| Parameters | Results | Reference range |
|---|---|---|
| Serum calcium (mmol/L) | 2.44 | 2.2–2.7 |
| Serum phosphate (mmol/L) | 1.49 | 0.85–1.51 |
| Vitamin D (ng/mL) | 51.04 | 15–100 |
| Parathyroid hormone (pg/mL) | 16.7 | 15–65 |
| Alkaline phosphatase (U/L) | 333 | 45–125 |
| Creatinine (μmol/L) | 25 | 40–135 |
| Body segments | Results | SD |
|---|---|---|
| Height (cm) | 81.3 | −2.1 |
| Weight (kg) | 13 | +0.3 |
| Head circumference (cm) | 47 | −1.0 |
| Upper segment (cm) | 47 | / |
| Lower segment (cm) | 36 | / |
Note
- Age of the patient is 2 years old.
3.2 Follow-up
During the follow-up, the patient has taken long-acting recombinant growth hormone at a dose of 0.17 IU/kg/day once a week together with daily calcium and vitamin D supplements since July 2019. The height of the patient was increased obviously after treatment without side effects. At the last follow-up, his height was up to 86 cm at the age of 2 years and 6 months old (−1.9 SD) with a weight of 16.1 kg (+1.5 SD). Besides, the results of biochemical and hormonal test were normal and the radiograph showed similar results compared to that of the first examination mentioned before. There was no evidence for treatment with recombinant growth hormone systematically evaluated in children with SMCD yet. Therefore, growth hormone seems to be effective, at least in our present study, to treat short stature in SMCD children.
3.3 Genetic analysis of COL10A1 gene
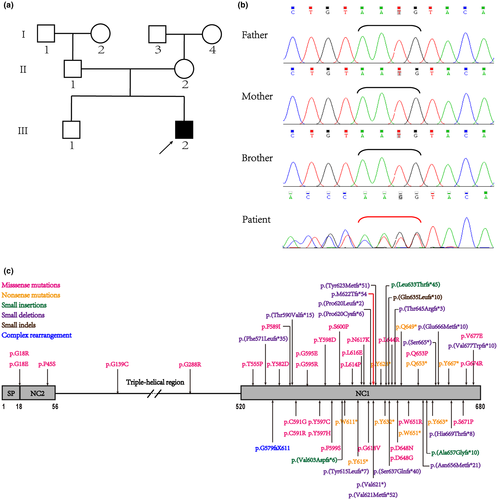
The patient's clinical features, as well as biochemical and hormone profiles, were typical of SMCD. The patient was subject to whole-exome sequencing and the detected variant was further confirmed via Sanger sequencing in all available family members. We found the patient carried a novel heterozygous variant c.1863_1866delAATG, p.(Met622 Thrfs*54) while it was absent in his family members (Figure 2a,b), indicating that this variant was probably a de novo variant (PS2). And this variant has not been reported before in the HGMD, TOPMED, ExAC, and 1000Genome databases (PM2). The novel c.1863_1866delAATG, p.(Met622 Thrfs*54) frameshift variant was located in the NC1 domain (Figure 2c). It caused a shift in the normal open reading frame of mRNA codons and created a premature stop codon at amino acid 675, producing a slightly truncated protein, which was 6 amino acids shorter than the wild type (PVS1). Moreover, this frameshift variant was strongly predicted to be pathogenic and deleterious using two online bioinformatic software—MutationTaster and PROVEAN with a PROVEAN score of −93.9 far below the cutoff (cutoff = −2.5). This might cause NMD according to MutationTaster. All of the above suggested that the novel COL10A1 variant of the patient was pathogenic (PVS1+PS2+PM2) according to the criteria for classifying pathogenic variants established by American College of Medical Genetics and Genomics (ACMG; Richards et al., 2015).
3.4 Protein structural model
The model of type X collagen NC1 homotrimer with three identical chains was built by SWISSMODEL automatically based on the template and visualized by DeepView. The NC1 trimers were formed by very tight interaction of each subunit, creating a central solvent-filled channel lined predominantly by strand F, which becomes more hydrophilic at the apex of the trimer where a cluster of four Ca2+ is presently contributing to the stability of type X collagen NC1 trimer, and an interaction surface composed of three strips of exposed aromatic residues, which are likely to be involved in the higher-order assembly of type X collagen in the extracellular matrix. The NC1 region consists of a 10-stranded β-sandwich with a jellyroll topology similar to the globular domain (gC1q) of the complement protein C1q. Ten β-strands are respectively labeled A, A’, B, B’,C, D, E, F, G, and H, among of which strands A, H, C, and F are mostly buried inside, whereas strands A’, B, B’, G, D, and E form the solvent-accessible surface of the NC1 trimer. It is of note that the mutated region started from amino acid 622 to the end of COL10A1 (Figure 3a), including both solvent-filled channels and the surface of the homotrimer (Figure 3b,c). The slightly truncated mutant protein lacking normal E-H strands (Figure 3a) will cause misfolding and trimer assembly failure of COL10A1 subunits, so we naturally predict this variant to cause SMCD by haploinsufficiency mechanism.
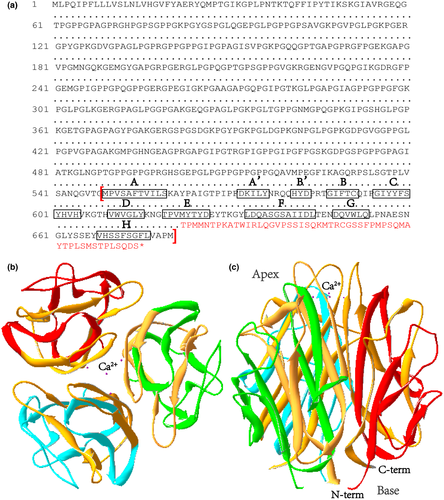
Therefore, all of the information indicates that the p.(Met622 Thrfs*54) variant is predicted to impair the hydrophobicity of the subunit core residues to prevent protein folding and the collagen X supra-structure in a haploinsufficiency manner via changing the structure of both central solvent-filled channel and patches of aromatic residues on the surface of NC1 trimer.
3.5 Literature review on COL10A1 variants related to SMCD
Here, we listed and summarized all the published literature about COL10A1 variants related to SMCD including our newly discovered variant (Table S1). As only the proband's information was given detailedly in most literature, mutation frequencies and the sample size in the genotype–phenotype correlation were both calculated by pedigrees, rather than individuals. All variants could be grouped based on different criteria (Table 3). There were 41 point variants and 19 frameshift variants, these respectively including 29 missense, 10 nonsense, 2 single-base deletions as well as 3 large insertions, 14 large deletions, and 2 complex rearrangements. 91.7% of the variants were located in NC1 domain of collagen type X alpha 1 chain with only 5 variants in non-NC1 domains. Since blood samples of the proband's parents were only available in 49 of 60 pedigrees, among which inherited and de novo variants took a nearly identical share. The vast majority of patients had short stature, bowed legs, waddling gait and metaphyseal irregularities and genu varum was the most prevalent lower limb deformity.
| Variant type | Numbers | Percentage (%) |
|---|---|---|
| Point variants | ||
| Missense variants | 29 | 48.4 |
| Nonsensevariants | 10 | 16.7 |
| Single-base deletions | 2 | 3.3 |
| Frameshift variants | ||
| Large insertions | 3 | 5 |
| Large deletions | 14 | 23.3 |
| Complex variants | 2 | 3.3 |
| NC1 domain variants | 55 | 91.7 |
| Non-NC1 domain variants | 5 | 8.3 |
| Inherited variants | 26 | 43.3 |
| De novo variants | 23 | 38.3 |
| Variants of unclear origin | 11 | 18.4 |
Then we performed a simple genotype–phenotype correlation analysis. Based on different variant types, we further stratified patients into subgroups. It was shown in Table 4 that there existed significant phenotypic differences between different groups of patients. On the one hand, patients with truncating variants presented lower limb deformity at a significantly earlier age (p = .020) compared to those with non-truncating variants. In addition, patients carrying truncating variants demonstrated slightly earlier onset (p = .149) and shorter stature (p = .082) in comparison with those carrying non-truncating variants. On the other hand, patients with variants in NC1 domain showed significantly earlier onset (p = .008) and shorter stature (p = .022) compared with those with variants in non-NC1 domains. However, due to the unavailability of patient information in some pedigrees, the sample size included in the analysis was a bit small.
| Truncating variants | Non-truncating variants | p value | NC1 domain variant | Non-NC1 domain variant | p value | |
|---|---|---|---|---|---|---|
| Age when signs and symptoms were first noticed |
19.5 (12, 24) (N = 16) |
38.56 ± 25.76 (N = 16) |
.149 |
21(12, 36) (N = 28) |
60 (60, 78) (N = 4) |
.008* |
| Age when lower limb deformity first appeared |
12 (12,24) (N = 14) |
35.92 ± 25.99 (N = 13) |
.020* |
17(12,32.5) (N = 24) |
60 ± 0 (N = 3) |
.014* |
| Height SDS |
−3.38 ± 1.13 (N = 15) |
−2.72 ± 0.92 (N = 16) |
.082 |
−3.18 ± 1.01 (N = 28) |
−1.73 ± 0.60 (N = 3) |
.022* |
Note
- Ages are displayed in months.
- * Significant difference exists between subgroups (p < .05).
4 DISCUSSION
As it is widely known that SMCD is a rare skeletal disorder caused by COL10A1 gene mutation and inherited in an autosomal dominant manner (Bateman et al., 2005), our study reported a SMCD patient who carried a novel frameshift variant c.1863_1866delAATG,p.(Met622 Thrfs*54) with the heterozygous substitution of a large segment in the NC1 domain of collagen type X alpha 1 chain, leading to early onset at birth in this male child patient. The SMCD diagnosis was established in the proband with characteristic clinical, laboratory, and radiographic features together with a genetic test. In addition, the novel COL10A1 variant of the patient was pathogenic according to the criteria for classifying pathogenic variants established by ACMG and it was predicted to induce NMD which is the pathogenic mechanism of nonsense variants in SMCD development.
To our knowledge, our study is the first to review all published papers on COL10A1 variants associated with SMCD and analyze the genotype–phenotype correlation, thus providing a comprehensive characterization of patients with SMCD. Our results indicated that significant phenotypic differences existed between different subgroups of patients. Patients with truncating or NC1 domain variants tended to show earlier onset and severer symptoms compared with those with non-truncating or non-NC1 domains variants. Nevertheless, as a result of the small sample size in some subgroups, more cases need to be found and studied in further studies to have a deeper understanding of SMCD.
Up to date, all reported dominantly inherited variants in SMCD patients are located in the NC1 region of COL10A1 except five variants, two in the signal peptide (p.Gly18Arg, p.Gly18Glu; Ikegawa et al., 1997), one in the NC2 domain (p.P45S; ul Ain et al., 2018) and two in the triple helical domain (p.G139C, p.Gly288Arg) of COL10A1 (Park et al., 2015; Zhang et al., 2019; Figure 2c), which are associated with later onset and milder symptoms of MCDS (Marks et al., 1999; Park et al., 2015). Almost half variants detected in the probands were inherited from parents while the other half were de novo variants. Frameshift variants were much less frequent than point variants, among which missense variants took a dominant share. Most patients with COL10A1 variants previously reported presented similar phenotypes seen in our case, such as short stature, genu varum, waddling gait, and metaphyseal irregularities. But the severity and some uncommon manifestations like lumbar lordosis and arthralgia demonstrated great variability between individuals and families.
Since clinical phenotypes vary among SMCD patients with different COL10A1 variants, there maybe great complexity in the pathogenic mechanism (Bateman et al., 2003). And the pathogenic mechanism through which the frameshift variant p.(Met622 Thrfs*54) leads to lower limb deformity in our patient remains to be elucidated. NC1 homotrimer formation creates a central solvent-filled channel and an external surface of each NC1 trimer containing three hydrophobic patches which participate in initiating the supramolecular assembly between collagen type X alpha 1 chains or between collagen type X alpha 1 chains and other molecules for the higher-order structure of the collagen X network in bone matrix (Bateman et al., 2004). Critical hydrophobic interface residues near the base include Ala-553, Val-556, Ile-557, Leu-575, Ile-641, Phe-675, Val-677, and Ala-678 which accounted for the close packing between the NC1 subunits to generate a tight and stable collagen X protein structure (McLaughlin et al., 1999). And the most prominent feature of the trimer external surface is the presence of three strips of aromatic residues extending across each subunit interface (Figure 3b,c), with each strip containing eight partially exposed residues which are Trp-611 (9%-63% of the side chain accessible to a 1.4 A˚ probe), Tyr-615, Tyr-623, Tyr-625, and Trp-651 from one subunit as well as Tyr-562, Tyr-663, and Tyr-667 from the adjacent subunit (Shapiro & Scherer, 1998). So the novel frameshift variant c.1863_1866delAATG, p.(Met622 Thrfs*54) which altered most of the amino acid sequence after residue 621 (residue 622–680 altered) covering the entire central channel and partially exposed surface (Figure 3b,c), and introduced a premature stop codon in the NC1 domain to produce a shortened collagen type X alpha 1 chain (Figure 3a), was likely to tremendously damage collagen X structure and function by inhibiting collagen X homotrimer assembly preceding adverse clinical symptoms like lower limb deformity in patients with SMCD.
Haploinsufficiency has been recently recognized as one of the most probable cause of SMCD (Bateman et al., 2003; Chen et al., 2020). NC1 missense variants lead to misfolding, improper trimerization, intracellular retention of mutated collagen, and eventual collagen X haploinsufficiency (Mullan et al., 2017; Wilson et al., 2005). In addition, the process of nonsense-mediated decay (NMD) in which mutant mRNAs are determined to be completely degraded in chondrocytes gets involved in the molecular mechanism of SMCD caused by nonsense variants (Ho et al., 2007; Tan et al., 2008). Besides haploinsufficiency, the dominant-negative impairment of protein folding has been proposed to be involved in the pathogenesis of SMCD but this still needs more evidence to verify (Bateman et al., 2003; Gregory et al., 2000; Makitie et al., 2005). Compared to missense variants, patients with non-sense or frameshift variants generally show earlier onset and more severe manifestations, as present in the patient of our study (Higuchi et al., 2016; Makitie et al., 2005; Woelfle et al., 2011).
The lack of in vitro functional study was an obvious limitation in our study and we will investigate the mutant type X collagen biosynthesis and assembly in our further study. Not all COL10A1 variants have been studied in detail and there was no protein analysis on mutant collagen X in cartilage tissue or chondrocytes from the patients, therefore studying the function of the variants on collagen X secretion and assembly had to depend on in vitro experiments at present.
In summary, our study is the first to identify and report a novel COL10A1 heterozygous frameshift variant c.1863_1866delAATG (p.Met622 Thrfs*54) in a Chinese pedigree with SMCD which expands the mutation spectrum of COL10A1 related to SMCD and promotes the understanding of genotype–phenotype correlations. Moreover, predicted pathophysiological consequences secondary to the variant help explain the underlying mechanisms of SMCD in this patient. Further functional studies remain to be completed to elucidate the pathogenic mechanism of SMCD.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation (No. 81974124) and National Key Research and Development Project (No. 2016YFC0901503), together with special funds for Taishan Scholar Project (No. tsqn20161071), Shandong Provincial Key Research and Development Project (No. 2017GSF18118). The authors are sincerely grateful to all of the participants in this study. And the first author would like to heartfelt thank her parents for their maximum support in all aspects and also give special thanks to JJ Lin (a famous Singapore pop singer) for the spiritual support and great power got from his songs.
CONFLICTS OF INTEREST
All authors have no conflicts of interest.
AUTHOR CONTRIBUTIONS
C.X. and Y.W. designed and supervised the study as well as revised the manuscript. H.W. performed data analysis and literature review and wrote the manuscript. S.W. assisted data analysis. G.L. and J.Z. made the diagnosis. Y.Y. and X.J. collected clinical data. N.W. collected peripheral blood sample and contributed to genetic analysis. J.Z., X.S., and L.F. assisted the acquisition of data. All authors read and approved the final manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




