Novel frameshift mutation in PURA gene causes severe encephalopathy of unclear cause
Abstract
Background
The etiology of many genetic diseases is challenging. This is especially true for developmental disorders of the central nervous system, since several genes can be involved. Many of such pathologies are considered rare diseases, since they affect less than 1 in 2000 people. Due to their low frequency, they present several difficulties for patients, from the delay in the diagnosis to the lack of treatments. Next-generation sequencing techniques have improved the search for diagnosis in several pathologies. Many studies have shown that the use of whole-exome/genome sequencing in rare Mendelian diseases has a diagnostic yield between 30% and 50% depending on the disease.
Methods
Here, we present the case of an undiagnosed 6-year-old boy with severe encephalopathy of unclear cause, whose etiological diagnosis was achieved by whole-genome sequencing.
Results
We found a novel variant that has not been previously reported in patients nor it has been described in GnomAD. Segregation analysis supports a de novo mutation, since it is not present in healthy parents. The change is predicted to be harmful to protein function, since it falls in the first quarter of the protein producing an altered reading frame and generating a premature stop codon. Additionally, the variant is classified as pathogenic according to ACMG criteria (PVS1, PM2, and PP3). Furthermore, there are several reported frameshift mutations in nearby codons as well as nonsense mutations that are predicted as pathogenic in other studies.
Conclusion
We found a novel de novo frameshift mutation in the PURA gene (MIM number 600473), c.151_161del, with sufficient evidence of its pathogenicity.
1 INTRODUCTION
Specific diagnosis of many genetic diseases is challenging. One area where greater difficulties are met is that of developmental disorders of the central nervous system, since many genes can be involved (van Loo & Martens, 2007) with many more to be discovered probably. In more broad terms, rare diseases (RD) are pathologies that affect less than 1 in 2000 people (Commission & – European Commission, 2020). Due to their low frequency, they present diverse difficulties, from the delay in the diagnosis to the lack of specific treatments. Most of them have a substantial impact on quality and life expectancy, affecting children and young people.
Since the diagnosis itself is the first obstacle that RD patients face, from which most medical decisions will rely on, it becomes of crucial importance to have a precise and accurate diagnosis. Molecular genomics approaches have helped in this sense. Next-generation sequencing (NGS) techniques have highly improved diagnosis of RD. Many studies have shown that the use of whole-exome sequencing (WES) in rare Mendelian diseases has a diagnostic yield between 30% and 50% depending on the disease (Clark et al., 2018; Yang et al., 2014). The use of whole-genome sequencing (WGS) in a cohort of pediatric patients has found causative genetic variants in 34% of the cases (Stavropoulos et al. 2016). These results encouraged our group to use these tools (both WES/WGS) to help with the diagnosis of pediatric patients with RD.
Here, we present the case of a 6-year-old boy presenting with severe encephalopathy with unclear cause, which started in the first months of life, whose etiological diagnosis was made by WGS.
2 METHODS
2.1 Ethical compliance
This project (URUGENOMES Ref IP011-17/CEI/LC/MB) was approved by the ethics committee of the Institut Pasteur de Montevideo.
A written informed consent was elaborated for this study and was signed by the patients’ parents.
2.2 NGS sequencing and bioinformatics analysis
Genomic DNA was extracted from 100 μl of whole blood using QIAamp® DNA Blood Mini kit (Qiagen, Germany) according to manufacturer instructions.
We did a whole-genome sequencing of the patient with 30X in a HiSeq X Ten Illumina sequencer. Quality of reads was analyzed using FastQC (Andrews, 2010), and they were mapped onto the human genome (GRCh37) using BWA (Li & Durbin, 2009). Variant calling was performed using GATK (best practices) (Mckenna et al., 2010). Annotation of found variants was done with ANNOVAR (Wang et al., 2010). Series of filters were applied in order to detect relevant mutations. Candidate mutation (frameshift deletion) was further evaluated with the SIFT Indel tool [9], in order to address pathogenicity.
This relevant mutation found in the patient was evaluated via Sanger sequencing.
Additionally, the mitochondrial genome was analyzed using MToolBox (Calabrese et al., 2012).
2.3 Variants filtering scheme
In order to filter and prioritize the variants found, we used the following rationale:
- Homozygous or hemizygous mutations with a frequency lower than 1% in coding/splicing region;
- Heterozygous mutations with at least two variants in the same gene with frequency lower than 1% (compound heterozygous) in coding/splicing region;
- Heterozygous mutations with frequency less than 0.5% in coding/splicing region;
- Mitochondrial mutations with high heteroplasmy (>10%) and in coding regions or tRNA and rRNA genes (and not part of the definition of the haplogroup), not in D-Loop region;
- Noncoding variants, either with “uncertain significance” (VUS) or “pathogenic/Likely pathogenic” or “conflicting interpretations of pathogenicity” classifications, as determined by ClinVar (Landrum et al., 2018).
3 RESULTS
3.1 Case report
A 6-year-old boy was referred to the medical genetics’ unit for evaluation for neurodevelopmental delay and epilepsy. He is the son of a non-consanguineous healthy couple who has three healthy children from other marriages, two from the mother's and one from the father's side. The child is the younger one and the product of a third uncomplicated pregnancy. The mother did not take alcohol or illegal drugs and was not exposed to teratogens during pregnancy. Fetal movements were normal as well as fetal ultrasounds. A C-section delivery was performed at week 40 due to cephalopelvic disproportion. The weight at birth was 4150 grams, length 50.5 cm, Apgar score: 3/8. He presented involuntary rhythmic movements of the diaphragm, difficulties in sucking, weak cry, and jaundice that required phototherapy. From the examination of this period we can highlight: myopathic facies, no spontaneous eye opening with intermittent eye fixation, severe hypotonia with quadriparesis, and increased tendon reflexes.
Excessive somnolence and lethargy were present in the neonatal period as well as in the first year of life. This feature showed slow improvement since the age of 2 years.
Generalized neurodevelopmental delay was evident afterwards. Regarding motor skills, he reached cephalic support at the age of one year, and he was able to sit unsupported at the age of two. He is still not able to walk. No verbal language was developed so far.
At 2 years old, he began to suffer from seizures which are multifocal and generalized. Some of these crises are nocturnal. Currently, he is under treatment with valproic acid and levetiracetam with amelioration of the number of crises but not achieving a complete remission. Sleep apneas and dystonic movements appeared in the last year. Apart from “hypotonic face” no other dysmorphic signs were evident. Growth was normal in all somatic and cranial parameters.
The exams performed showed additional findings as follows:
Abnormal (prechiasmatic) visual evoked potentials. Auditory evoked potentials were normal.
Electroencephalograms were normal at the beginning. The last one (induced sleep) showed bifocal epileptic activity: intense activity consisting of spike-and-slow-wave complexes in temporal and parietal regions of the left hemisphere, and moderate activity in temporal regions of the right hemisphere. Background rhythms were normal.
Polysomnography showed severe sleep apnea. Gastroesophageal reflux is present (coincident with apneas) as measured by esophageal pH.
Magnetic resonance imaging was performed twice (age 2 and 4), being normal in both cases. Previous genetic studies were performed with normal results: karyotype (46, XY), CGH array, and methylation studies for PWS/ANG region (MS-MLPA). Additionally, amino acid dosification in plasma and organic acids in urine were measured with normal results.
3.2 GS results
From a whole blood sample from the patient whole-genome sequencing (nuclear and mitochondrial DNA) was performed with a sequencing depth of 30x. We obtained 709.502.196 reads that passed QC-controls (according to samtools flagstat) and ~94% (669945761) were mapped onto the reference genome (GRCh37). Variant calling analysis detected 4925231 variants that were further annotated and prioritized (see 2.3). For the mitochondrial genome a high sequencing depth (2999x) and 100% coverage were obtained. A total of 273 variants were detected in the mtDNA.
3.3 Frameshift variant in the PURA gene is a candidate pathogenic mutation
Results of our filtering rationale (see methods 4.2), are found in Table 1. An autosomal recessive inheritance for the patient's phenotype was plausible, since both parents were unaffected. For this reason, on the one hand, coding variants in homozygosity with low frequency were considered. Out of the 24 (21 non-synonymous variants) none of them were related to a gene with a concordant phenotype. Additionally, compound heterozygous variants were considered. Out of the 75 genes, neither of them was previously associated with disease, or the phenotype associated was not concordant to the patient's. Another possibility was a de novo mutation, appearing for the first time in a heterozygous state in the patient. For this reason, we investigated all heterozygous mutations in coding regions with very low frequency (<0.5%). Here, we found a frameshift deletion of 11 nucleotides (CCAGGGGGGCT, c.151_161del) (Figure 1a) in the only exon of the PURA gene (chr5:139493917–139493927, GRCh37, PURA:NM_005859:exon1:c.151_161del:p.P51fs) not previously reported in any of the population genomic projects (such as 1000 Genomes (The 1000 Genomes Project Consortium, 2015)/GnomAD (Karczewski et al., 2020) /ExAC (Lek et al. 2016)). The deletion occurs between the 51th and the 54th amino acid (the first 16% of the protein), it causes a stop codon at codon 147 after the deletion. Therefore, not only the reading frame is altered for 147 amino acids, but also it results in a 130 amino acid shorter protein. Additionally, the variant was evaluated with SIFT Indel 2 (Pauline, 2003) to predict its pathogenicity from an in silico point of view. It was predicted as “damaging” with a confident score of 0.858.
| 1% hom | 1% comp. Het | 0.5% het | chrM | |
|---|---|---|---|---|
| # variants | 24 | 477 | 427 | 29 |
| # genes | 14 | 75 | 196 | 12 |
| Missense | 19 | 273 | 373 | 9 |
| Nonsense | 1 | 10 | 14 | 0 |
| Stoploss | 0 | 1 | 1 | 0 |
| fs indel | 1 | 15 | 17 | 0 |
| Non-fs indel | 0 | 2 | 0 | 0 |
| Silent | 3 | 162 | 0 | 18 |
| Splicing | 0 | 3 | 5 | 0 |
| Others | . | Unknown 10 | Unknown 17 | 2 tRNA |
- Columns show each of the four filtering categories: homozygous variants with a population frequency of less than 1%; heterozygous variants with a population frequency of less than 1%, with at least two variants in the same gene; heterozygous variants with a population frequency of less than 0.5%. Rows have the total number of variants found and the number of genes involved, and below the different types of variants found in each category. Unknown category corresponds to various ambiguities in the gene structure definition in the database file. fs, frameshift; non-fs, non-frameshift. # symbol represents number.
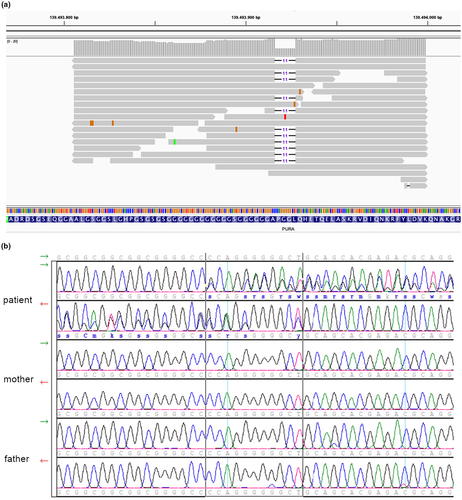
3.4 De novo status and confirmation of the variant
Even though the quality of the sequencing in the variant position was good (total coverage of the region is 19x: 11 reads with the deletion and 8 reads without), the deletion was validated and confirmed in the patient via Sanger sequencing. Additionally, both biological parents were analyzed for the presence of the variant with Sanger sequencing. The variant was not found in any of them in white blood cells (Figure 1b).
4 DISCUSSION
The PURA gene is a DNA/RNA-binding protein which functions as a transcription regulator also involved in mRNA localization (Weber, 2016). A number of pathologies, ranging from encephalopathies and mental retardation to leukemia and ALS, have been associated with heterozygous mutations in the PURA gene (Daniel, 2018; Johnson & Gordon, 2013). Its expression is ubiquitous with no clear tissue preference (human protein atlas, www.proteinatlas.org). The gene product is a protein with three domain repeats: repeats I and II are binding domains and repeat III is a dimerization domain (Johnson & Gordon., 2013; Weber, 2016). The crystal structure of the first two domains involved in nucleic acid binding from D. melanogaster complexed with ssDNA, has provided definitive evidence into the molecular interactions of the PURA protein (Weber, 2016), proving its purine base preference allows it for binding both DNA and RNA.
Mutations in the PURA gene were reported as a cause of dominant form of severe delayed psychomotor development and seizures a few years ago (Hunt et al., 2014; Lalani et al.,2014; Tanaka et al., 2015) and the PURA protein was shown to be a key player in brain development. PURA gene knockdown (−/−) mice show severe neurological and brain developmental anomalies and die within 4 weeks after birth (Hokkanen, 2012; Khalili et al., 2003).
Several lines of evidence point to the variant we found as a causative mutation. The change is predicted to be harmful to protein function, since it falls in the first quarter of the protein (AA 51 to 54 of 323 AA) producing an altered reading frame (for 147 AA following the deletion) and generating a premature stop codon (product is 130AA shorter), which leads to two possibly overlapping scenarios. On the one hand, the production of prematurely truncated mRNAs to be processed by the nonsense-mediated mRNA decay pathway (Brogna, & Wen, 2009). On the other hand, the production of a different protein, which we posit would be 195 amino acids: PURA gene's first 50 residues followed by a different sequence starting position 51, and therefore, affecting all of the three functional domains of the protein. This predicted protein has no detectable eukaryotic homologs, as checked by an HHpred (Söding, 2005) search over the Protein Data Bank or two iterations of jackhammer (Potter et al., 2018) over the Reference Proteome database. The production of an abnormal protein may result in multiple cellular problems, including accumulation of misfolded proteins. In this sense, the most probable transcript, if translated, is predicted to have high disorder propensity. Any of those mechanisms probably causes a total loss of function (LoF) of the product encoded by this allele. LoF has been demonstrated to be the main pathogenic mechanism in diseases associated with this gene (Bonaglia et al., 2015 and see below). Furthermore, microdeletions involving the PURA gene are also associated with a form of dominant epileptic encephalopathy (Hosoki et al., 2012; Shimojima et al., 2011). In most of the cases, these are de novo mutations with apparently complete penetrance.
Additionally, the variant fulfills following ACMG criteria for pathogenesis: (a) PVS1 (pathogenic very strong), since the deletion is a null variant in a gene for which LoF is a known mechanism of disease. According to the Varsome database (Kopanos et al., 2018) PURA gene has 70 pathogenic LoF variants and a significant LOF Z-Score of 2.77, associated with Mental retardation, autosomal dominant. (b) PS2 (pathogenic strong), since it is a de novo mutation (both maternally and paternally confirmed) in a patient with the disease and no family history. (c) PM2 (pathogenic moderate), since the variant has not been previously reported in patients nor it has been described in GnomAD and/or other large-scale population studies involving healthy individuals (the region has a good coverage in GnomAD (>25x) according to Varsome analysis). iv) PP3 (supporting evidence of pathogenicity), since an in silico score (GERP) predicted the variant as pathogenic (and no score predicted it as benign). Hence, all together the variant classifies as pathogenic according to ACMG.
There are several reported frameshift mutations in nearby codons (40, 46, 54, 59, and 83) as well as nonsense mutations (e.g., p. Gln55Ter, see https://www.ncbi.nlm.nih.gov/clinvar/variation/426145/) in that region of the protein, all of them reported as pathogenic. Other nonsense variants downstream in the protein (p. Gln163*, p. Gln186*, and p. Tyr261*) have also been reported as pathogenic (see: https://www.ncbi.nlm.nih.gov/clinvar/variation/582890/).
Finally, segregation analysis in the family (parents) supports a de novo mutation hypothesis which is consistent with both parents and half-siblings being healthy. In addition, no other variants in the PURA gene of less than 1% population frequency have been detected in this patient.
There are several patients reported with gene disrupting mutations which likely cause haploinsufficiency (nonsense and frameshift indels). In a larger series of patients reported so far (Reijnders et al., 2018 and Lee et al., 2018), no significant differences in clinical severity between mutation classes was found. As expected, in our patient, whose mutation causes a complete loss of function of the affected allele, whether caused by protein primary structure disruption or/and truncation or nonsense-mediated mRNA decay, the clinical presentation is severe with serious defect in motor skills, absence of language, respiratory problems, and intense epileptic activity.
Therefore, we think that the variant we found is a strong candidate pathogenic mutation probably explaining the patients’ phenotype marked by severe central neurological dysfunction.
ACKNOWLEDGMENTS
Funded by URUGENOMES Project IP011-17/CEI/LC/MB.
CONFLICT OF INTERESTS
Authors declare no conflict of interests.
Open Research
DATA AVAILABILITY STATEMENT
WGS is available upon request.




