RETRACTED: Evaluation of a six-dye multiplex composed of 27 markers for forensic analysis and databasing
Shuangshuang Wang and Feng Song contributed equally to this work and should be considered the co-first author.
Funding information
This study was supported by the National Natural Science Foundation of China (NSFC, No. 81701866 and 81772030), the Postdoctoral Fund of Sichuan University (No. 2017SCU12049), and the Outstanding Youth Fund of Sichuan University (No. 2016SCU04A07)
Abstract
Background
Short tandem repeat (STR) markers play a significant role in genetic applications and have proved to be effective for the personal identification in forensic medicine. In this study, a six-dye multiplex composed of 23 autosomal STR loci (TH01, D3S1358, Penta D, D6S1043, D21S11, TPOX, D1S1656, D12S391, Penta E, D10S1248, D22S1045, D19S433, D8S1179, D2S1338, D2S441, D18S51, vWA, FGA, D16S539, CSF1PO, D13S317, D5S818, D7S820), one Y chromosome STR (DYS391), two internal quality control markers (Quality Sensor QS1 and QS2), and Amelogenin was evaluated.
Methods
Evaluation studies, including PCR-based studies, sensitivity studies, species specificity studies, stability studies, DNA mixture studies, concordance studies, and precision evaluations were performed according to the guidelines of “Validation Guidelines for Forensic DNA Analysis Methods (2016)” by the Scientific Working Group on DNA Analysis Methods (SWGDAM). In addition, the forensic characteristics of 357 unrelated male samples from Han and Hui populations in China were investigated using 27 markers.
Results
Full STR profiles were obtained from different reaction volumes (5 ~ 25 μl), cycle numbers (28 ~ 34 cycles) and annealing temperatures (58 ~ 62°C). All STR profiles were obtained at humic acid concentration of up to 200 ng/μl and hematin concentration of up to 500 μM. No peaks were observed in most common animal samples except two innovative internal PCR controls (Quality Sensor QS1 and QS2). The six-dye multiplex showed a notably high value for the combined probability of exclusion (CPE), exhibiting values of with 0.99999999977688 in the Han population and 0.999999999583875 in the Hui population. The values of combined probability of discrimination (CPD) were 0.999999999999999999999999999997453 in the Han population and 0. 999999999999999999999999999994398 in the Hui population. In addition, concordance studies showed that there was no difference with the AGCU Express Marker 22 Kit.
Conclusion
The results indicated that the Investigator® 26plex QS Kit is a robust, reliable, and suitable tool for forensic analysis and databasing.
1 BACKGROUND
Short tandem repeat (STR) was first applied in a forensic case in 1991. In subsequent years, STR has gradually become widely used in personal identification and paternity testing (Li et al., 2017). The autosomal STR profiles generated from biological material could be compared with the profiles of known suspects whose profiles provide a strong evidence under statistical support for personal identification (Jobling & Gill, 2004). Information on Y-STRs and X-STRs is also useful in forensic applications, especially Y-STRs. Y-STR is highly important for addressing DNA mixtures in rape cases. Genetic profile databases are constantly established and expanded with the widely used STR method (Kowalczyk, Zawadzka, Szewczuk, Gryzinska, & Jakubczak, 2018). STR genotyping has become one of the most scientifically rigorous and practical techniques in forensic science due to its widely recognized results (Zhou et al., 2014) and large STR database. Therefore, an increasing number of commercial kits have been developed for forensic application, such as the AGCU EX16 Kit, the AGCU EX22 Kit (AGCU ScienTech, Jiangsu, China), PowerPlex® Fusion System (Promega, WI, USA), and the GlobalFiler® kit (Applied Biosystems, CA, USA).
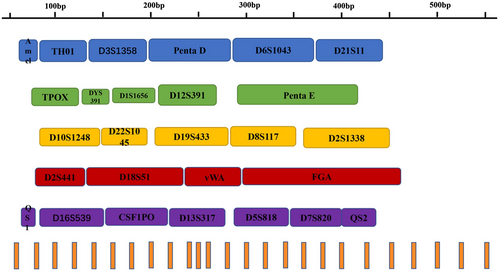
The Investigator® 26plex QS Kit was used for multiplex PCR in personal identification and paternity testing. This system simultaneously amplifies 23 autosomal STR loci (TH01, D3S1358, Penta D, D6S1043, D21S11, TPOX, D1S1656, D12S391, Penta E, D10S1248, D22S1045, D19S433, D8S1179, D2S1338, D2S441, D18S51, vWA, FGA, D16S539, CSF1PO, D13S317, D5S818, and D7S820), one Y chromosome STR (DYS391), two internal quality control markers (Quality Sensor QS1 and QS2) along with Amelogenin. Two innovative internal quality controls were added to this kit to monitor the efficiency of the PCR and the presence of PCR inhibitors. This kit was a six-dye multiplex system, which was compatible with the Applied Biosystems 3500 and 3730 Genetic Analyzers (Applied Biosystems, CA, USA). The arrangement of loci in each dye channel is shown in Figure 1. The Investigator® 26plex QS Kit was specifically designed for rapid and reliable generation of DNA profiles from blood, buccal swabs, and forensic stains. A previous study showed that the kit had high sensitivity, and had high robustness toward inhibitors (Vraneš et al., 2019). However, to determine the applicability of this kit in forensic practice, more factors should be considered. In this study, the Investigator® 26plex QS Kit were evaluated according to the guidelines of “Validation Guidelines for Forensic DNA Analysis Methods (2016)” by the Scientific Working Group on DNA Analysis Methods (SWGDAM) (https://www.swgdam.org/publications). Therefore, a series of validation tests were conducted, including PCR-based studies, sensitivity studies, species specificity studies, stability studies, DNA mixtures studies, precision evaluations, concordance studies, and population studies on 357 unrelated samples.
2 MATERIALS AND METHODS
2.1 DNA samples and ethics statement
With informed consent, 357 samples were collected upon the approval of the Ethics Committee at Sichuan University (K2019018), which included 137 saliva samples and 220 blood samples. These samples contained two Chinese populations, Han population (n = 220) and Hui population (n = 137) respectively. About 97 samples of Hui population were previously used in other study (Xie et al., 2020), and another 40 of which were newly collected. All genomic DNA was extracted by salting out method (Miller, Dykes, & Polesky, 1988). DNA was quantitated with a NanoDrop 1000 following the manufacturer's protocol. The typical control DNA included 9947A and 9948 (Applied Biosystems, CA, USA).
2.2 PCR amplification
Unless otherwise stated, PCR amplification was performed strictly according to the instructions of the Investigator® 26plex QS Kit Handbooks. PCR amplification was performed with a 25 μl volume system, including 7.5 μl Fast Reaction Mix 3.0, 2.5 μl primer mix, 1 ng template DNA, and nuclease-free water. PCR amplification was performed on ProFlex PCR System (Thermo Fisher Scientific, MA, USA) with the condition as follows: preincubation at 98°C for 8 min; followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 60°C for 55 s, and extension at 72°C for 5 s; then, 68°C for 2 min, 60°C for 2 min; finally, hold at 10°C for further analysis.
2.3 Capillary electrophoresis and data analysis
The Investigator® 26plex QS Kit was a six fluorescent dye system (6-FAM, BTG, BTY, BTR2, BTP, and BTO). To allow evaluation of each fluorescence dye contained in the kit, spectral resolution was established using the Matrix Standard BT6. Amplified PCR products were subsequently detected on Applied Biosystems 3500 Genetic Analyzers (Thermo Fisher Scientific, MA, USA) with a 36-cm capillary array and POP-4 polymer (Life Technologies, MA, USA). Then, 0.5 μl the amplified PCR products or 0.8 μl allelic ladders were added to a 9 μl mixture of Hi-Di formamide (Applied Biosystems, CA, USA) and DNA Size Standard 550 (BTO) (Hi-Di formamide: DNA Size Standard 550 = 100:1). Samples were injected at 1.2 kV for 10 s and electrophoresed at 15 kV for 1310 s at a run temperature of 60°C. The raw data were analyzed using GeneMapper ID Software v3.2.1 (Applied Biosystems, CA, USA) with the 26plex QS Panels, Bin sets and stutter files. All peaks were set with a threshold of 50 relative fluorescence units (RFU).
2.4 PCR-based study
To ensure the accuracy and stable amplification of the STR genotyping results obtained from forensic cases, it was necessary to value the performance of the key PCR parameters and establish the parameter range of amplification conditions. Several amplified parameters were in this study, including PCR reaction volume (25 μl, 10 μl, 5 μl), cycle number of PCR analyses(26c, 28c, 30c, 32c, 34c), denaturing temperature (54°C, 56°C, 58°C, 60°C, 62°C, 64°C, 66°C), concentration of the Fast Reaction Mix 3.0 (3.5 μl, 5.5 μl, 7.5 μl, 9.5 μl, 11.5 μl, 13.5 μl), concentration of the primer mix (0.625 μl, 1.25 μl, 2.5 μl, 5 μl, 7.5 μl, 10 μl). Each parameter was tested in triplicate. 9948 DNA was used as template DNA. 0.5 ng DNA was amplified in a 25 μL reaction volume, 0.2 ng DNA in a 10 μl reaction volume and 0.1 ng DNA in a 5 μl reaction volume. Other tested studies were performed in a 25 μl reaction volume with 0.5 ng 9948 DNA. The tested parameters were changed, in turn, while others were in keeping with the recommended conditions.
2.5 Sensitivity study
To evaluate the sensitivity of the Investigator® 26plex QS Kit, a serial concentration of control DNA 9948 (0.03125 ng, 0.0625 ng, 0.1 ng, 0.125 ng, 0.25 ng, 0.5 ng, 1 ng) was amplified and detected in triplicate. Amplification processes were carried out following the manufacturer's recommended conditions.
2.6 Species specificity study
To estimate the species specificity of the Investigator® 26plex QS Kit, DNA samples of common animal species (cat, cattle, cavy, chicken, duck, goat, mouse, pig, and rabbit) were prepared. The genomic DNA was extracted by the salting-out method (Miller et al., 1988), and quantitated with a NanoDrop 1000. Ten nanograms of genomic DNA was used for the species specificity study of the Investigator® 26plex QS Kit.
2.7 Stability study
The samples obtained from forensic casework usually contained PCR inhibitors, such as hematin and humic acid. Two PCR inhibition models were established to test the stability of the Investigator® 26plex QS Kit: hematin (Shanghai Aladdin Biochemical Technology, Shanghai, China) and humic acid (Sigma, St. Louis, MO USA). A total of 0.5 ng of control 9948 DNA was coamplified with various concentrations of hematin and humic acid according to the instructions of the Investigator® 26plex QS Kit Handbooks in triplicate. The concentrations of humic acid were 50 ng/μl, 100 ng/μl, 200 ng/μl, and those of hematin were 125 μM, 250 μM, 500 μM, and 1000 μM.
2.8 Mixtures study
To assess the Investigator® 26plex QS Kit's identification capability with DNA mixture samples. A mixed sample of 9947A and 9948 was tested at various ratios (1:1, 2:1, 4:1, 9:1, 19:1) with 1 ng of total DNA template in triplicate.
2.9 Precision
The precision study was tested with 22 allelic ladder samples, which were detected on an Applied Biosystems 3500 Genetic Analyzer. The mean and standard deviation of every allele were calculated.
2.10 Population and concordance study
Population studies were performed to study the genetic characteristics of the Investigator® 26plex QS Kit. A total of 357 unrelated male samples from the Chinese Han population (n = 220) and the Hui population (n = 137) was used in the population study. The parameters of autosome STR were assessed by Modified-Power states software, including matching probability (MP), probability of discrimination (PD), probability of exclusion (PE), homozygotes, heterozygotes, polymorphism information content (PIC), and probability values of Hardy–Weinberg equilibrium (P). The genetic diversity of Y-STR was calculated according to the formula  .
.
Concordance studies were carried out to estimate the stability and performance of this kit. This analysis was conducted by comparison with the AGCU Express Marker 22 Kit, which is a commonly used kit, and the capability of genotyping was widely recognized. The genotypes of 211 donors of the abovementioned 357 individuals were obtained from the Investigator® 26plex QS Kit and the AGCU Express Marker 22 Kit.
3 RESULTS AND DISCUSSION
3.1 PCR-based studies
3.1.1 PCR reaction volume
The recommended PCR reaction volume of the Investigator® 26plex QS Kit is 25 μl; however, many labs usually reduce the PCR reaction volume for cost saving. In this study, control DNA 9948 was employed with three different volumes (25 μl, 10 μl, and 5 μl) in which each PCR component ratio remained the same as the manufacturer's recommended protocol. Complete genotyping was obtained in all reaction volume. These results showed a high degree of consistency in the three reaction volumes. Meanwhile, no allelic dropout events or artifact peaks were observed, and the peak morphology was consistent. This result suggested that correct genotyping could be obtained when the PCR volume was reduced to 5 μl. In some forensic cases with trace or low levels of template DNA, the PCR reaction volume was often reduced to improve the reliability and sensitivity of STR profiling (Gibson-Daw, Crenshaw, & McCord, 2018; McNevin, Edson, Robertson, & Austin, 2015). A lower PCR reaction volume would sometimes be chosen to deal with low copy DNA (Zhang et al., 2015). The Investigator® 26plex QS Kit was able to meet requirements.
3.1.2 Cycle number of PCR
As the cycle number of the PCR ranged from 26 to 34 cycles, the peak heights increased gradually. When the cycle number turned to 26, allelic dropout events were observed, and the heights of detected peaks were between 650 RFU and 50 RFU. Full STR profiles were obtained from different cycle numbers (28, 30, 32, 34). However, stutter peaks were observed at 34 cycles. The cycle number can be decreased when the concertation of DNA was too high. In the same measure, to enhance the amplification signals of low-copy-number DNA, high cycle number could be applied. The results indicated that a change in cycle number can affect the amount of PCR products. The kit was adapted to changes in cycles number from 28 to 34 cycles; however, PCR amplification was better performed in 30 cycles for optimal STR profile peaks.
3.1.3 Denaturing temperature
Annealing temperature is a highly important parameter in thermal cycling. This temperature can affect the specificity, sensitivity, and robustness of amplification (Kraemer et al., 2017). All STR profiles were observed with no allele dropping out when the annealing temperature ranged from 58°C to 62°C. Alleles of D6S1043, DYS391, D22S1045, and D8S1179 loci dropped out, and an unexpected peak, allele “4” at CSF1PO loci, appeared when the annealing temperature was set at 54°C. The allele of FGA loci dropped out when the annealing temperature turned to 56°C. The allele of D22S1045 loci dropped out at an annealing temperature of 64°C. When the annealing temperature was 66°C, allelic dropouts were observed, including DYS391, PentaE, D22S1045, and the vWA locus, and there was an additional peak: allele “11” at TPOX loci. At annealing temperatures of 58°C and 62°C, the obtained STR profile peaks were balanced, and they worked almost equally well as the recommended annealing temperature of 60°C. However, facing relatively large changes in annealing temperature (52°C, 54°C, 64°C, 66°C), genotyping results were affected. Therefore, the change in annealing temperature should not exceed the range of 58–62°C. These results indicated that the Investigator® 26plex QS Kit was capable of withstanding slight change in annealing temperatures, which was able to reduce the possibility of unsuccessful amplification caused by inaccurate annealing temperatures.
3.1.4 Concentration of PCR component
The concentration of primers plays an important role in the amplification system, which can affect the specificity and sensitivity of STR analysis. Complete and correct genotyping was obtained from different concentrations of primer mix (1.25 μl, 2.5 μl, and 5 μl). About 28% of loci were dropped out when 0.625 μl primer mix was input, including Amel, TH01, DYS391, D1S1656, D12S391, D10S1248, and CSF1PO. When the amount of primer mix increased to 7.5 μl, 20% of loci (D21S11, PentaE, D2S1338, FGA, and D7S820) were dropped out. This phenomenon became considerably worse at the amount of 10 μl primer mix with only 36% of loci (Amel, TH01, D3S1358, TPOX, DYS391, D1S1656, D10S1248, D2S441, and D16S539) were detected and genotyped. The dropped-out loci at the amounts of 7.5 μl and 10 μl were all relatively large fragments, which may be observed because when the primer was high, short fragments amplified faster with more reaction mix consumption which caused inhibition of large fragment amplification. Sensitive and balanced amplification results were obtained from 1.25 μl of primer mix and 5 μl of primer mix as the recommended 2.5 μl of primer mix. Full and correct genotyping was obtained from different concentrations of the Fast Reaction Mix 3.0 (5.5 μl, 7.5 μl, 9.5 μl, 11.5 μl). Only 2 loci (TPOX, D16S539) were detected when the amount of Fast Reaction Mixes 3.0 reached 3.5 μl. The peak heights decreased as the Fast Reaction Mix 3.0 component increased to 9.5 μl and 11.5 μl. When this volume increased to 13.5 μl, allelic dropping out was observed. When the Fast Reaction Mixes 3.0 were 5.5 μl–9.5 μl, relatively good genotyping results could be obtained as the recommended volume of 7.5 μl. These results indicated that the amount of a PCR component can affect amplification and genotyping, however, the amount of a PCR component changes within a certain range, which may not have a strong impact on genotyping.
3.2 Sensitivity
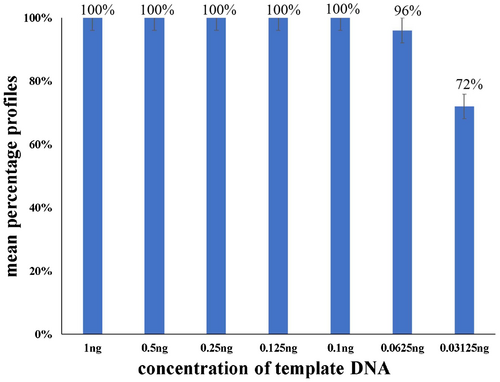
In forensic practice, samples collected from scenes are often diverse, and some of the samples may have lower DNA levels than the recommended quantity. The results showed that all STR profiles (25 loci) were obtained with a concentration of 9948 DNA equal to or above 0.1 ng using 50 RFU peak height analysis threshold. As the template DNA decreased gradually from 1 ng to 0.125 ng, the peak heights decreased gradually. The average peak heights of 0.1 ng template DNA were higher than those of 0.125 ng template DNA. This finding demonstrated that the average peak height nonlinearly decreased with decreasing template DNA input, which would influence mixture interpretation. When template DNA decreased to 0.0625 ng, 96% of the loci were detected and genotyped, excluding D21S11. At the amount of 0.03125 ng template DNA input, an average of 72% of the loci were detected and genotyped; seven loci (Amelogenin, DYS391, D10S1248, D19S433, D8S1179, D18S51, and D5S818) were dropped out (Figure 2). A sensitivity study can guide the upper and lower limits of the multiplex-PCR system. From the sensitivity testing results, the limit of template DNA for the Investigator® 26plex QS Kit was 0.1 ng.
3.3 Species specificity
DNA from several common animals was used to test its cross-reactivity with the peak height analysis threshold setting at 50 RFU. No peaks were detected in cat, cattle, cavy, duck, or rabbit except for two innovative internal PCR controls (Quality Sensor QS1 and QS2). Significative peaks above 50 RFU were detected in chickens, goats, mice, and pigs. For chickens, an allele “39.3” at D21S11 and “OL” peaks (TPOX, CSF1PO and D5S818 with peak height>50 RFU) were detected. In addition, one purple peak with a size of 403.27 bp was detected outside the range of normal amplification products. For goats, two “OL” peaks at D16S539 were detected. For mice, “OL” peaks at TPOX, D16S539, CSF1PO, and D5S818 were detected. For pigs, an “OL” peak was detected at CSF1PO. The results indicated that we should be cautious for samples possibly mixed with DNA from chickens, goats, mice, and pigs.
3.4 Stability
The samples obtained from forensic practice usually contained PCR inhibitors, and the most commonly observed inhibitors were humic acid from soil and hematin from blood (Bender, Farfan, & Schneider, 2004; Geng & Mathies, 2015). Humic acid affected amplification by binding to DNA while hematin inhibited Taq polymerase function to affect amplification (Opel, Chung, & McCord, 2010). The results of the stability study showed that all STR profiles were obtained at concentrations of humic acid up to 200 ng/μl and hematin up to 500 μM. Allelic dropout was observed at hematin concentrations up to 1000 μM. The peak heights gradually decreased with increasing inhibitor concentration. Therefore, the Investigator® 26plex QS Kit can handle samples with a certain degree of inhibition.
3.5 Mixtures
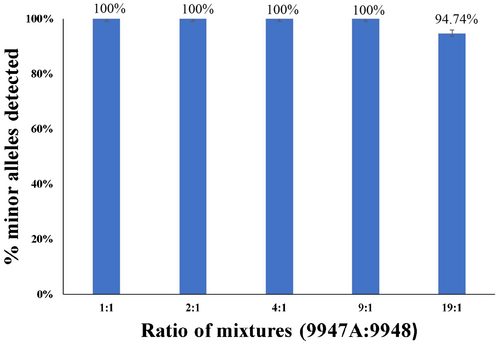
The samples obtained from forensic casework are usually mixture samples, which contain more than one individual's DNA. Especially in the case of sexual crimes (Purps, Geppert, Nagy, & Roewer, 2015), most samples were mixtures of female and male DNA, and usually, the amount of female DNA was considerably greater than that of male DNA. A total of 1 ng template DNA mixed with 9947A and 9948 DNA at various ratios (1:1, 2:1, 4:1, 9:1, 19:1) was tested. The peak heights of 9948 DNA decreased gradually with increasing ratio. To detect the minor component, nonoverlapping alleles of both DNAs were used. Therefore, the detection percentage of minor alleles was calculated from 19 loci. All minor allele profiles were observed at ratios of 1:1, 2:1, 4:1, and 9:1 (Figure 3). At a ratio of 9:1, minor alleles could be completely genotyped at the 50 RFU peak height analysis threshold, with peak heights between 800 RFU and 100 RFU, well below the peaks of 9947A DNA. It should be noted that those low peaks may be confused with the stutter peak. A drop-out of allele “Y” was observed at the ratio of 19:1, with 94.74% minor alleles being detected, and the detected peak heights were also between 800 RFU and 100 RFU, which were clearly lower than that of 9947A DNA. These results indicated that the Investigator® 26plex QS Kit can deal with mixture samples at a ratio of 9:1. In practical work, the multi-peak STR profiles with mixed low peaks need to be considered and are distinguished from stutter peaks combined with other case information.
3.6 Precision
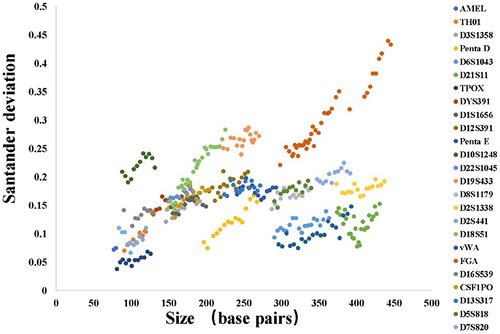
Sizing precision is an important indicator for reliable and accurate genotyping. Twenty-two allelic ladder samples were detected on a 3500 Genetic Analyzer. The standard deviation and average size of each allele were calculated and determined to be from 0.0377 at locus TPOX to 0.4392 at locus FGA (Figure 4), which demonstrated that this kit had sufficient sizing precision.
3.7 Population and concordance studies
A total of 357 unrelated male samples was included in the population study. The combined probability of exclusion (CPE) was 0.99999999977688 in the Han population and 0.999999999583875 in the Hui population. The combined probability of discrimination (CPD) were 0.999999999999999999999999999997453 in the Han population and 0. 999999999999999999999999999994398 in the Hui population. These results indicated that the system performance of the Investigator® 26plex QS Kit was notably high, and worked well in different populations.
The genotyping results from 211 unrelated male individuals detected by Investigator® 26plex QS Kit and AGCU Express Marker 22 Kit analyzed. There was no difference between the two kits. This result suggested that reliable results could be obtained with the Investigator® 26plex QS Kit.
4 CONCLUSION
The Investigator® 26plex QS Kit was developed for personal identification and paternity testing in human. The performance of this kit was tested in this study with the following testing: PCR-based studies, sensitivity studies, species specificity studies, stability studies, DNA mixtures studies, precision evaluations, concordance studies with AGCU Express Marker 22 Kit and population studies on 357 unrelated male samples. The results obtained from the validation study showed that the Investigator® 26plex QS Kit was a reliable, accurate, and sensitive system. In addition, the system had a high combined probability of discrimination and combined probability of exclusion value in the Chinese Han and Hui populations. Therefore, the six-dye multiplex assay was a useful tool for forensic analysis and databasing.
ACKNOWLEDGMENT
The authors are grateful to all the subjects for participating in this study. This study was supported by the National Natural Science Foundation of China (NSFC, No. 81701866 and 81772030), the Postdoctoral Fund of Sichuan University (No. 2017SCU12049), and the Outstanding Youth Fund of Sichuan University (No. 2016SCU04A07).
CONFLICT OF INTEREST
No potential conflict of interest was reported by the authors.
AUTHOR CONTRIBUTIONS
Haibo Luo and Feng Song conceived the idea for the study. Shuangshuang Wang and Zhanglong Huang performed or supervised laboratory work. Mingkun Xie, Ke Zhang, and Bowen Xie analyzed the data. Shuangshuang Wang and Feng Song wrote and edited the manuscript. All authors reviewed the final manuscript.