Inherited and de novo biallelic pathogenic variants in COL11A1 result in type 2 Stickler syndrome with severe hearing loss
Funding information
The Vitreoretinal Research Group are supported by the Retinal Research Fund at the University of Cambridge.
Abstract
Background
Type 2 Stickler syndrome is usually a dominant disorder resulting from pathogenic variants in COL11A1 encoding the alpha 1 chain of type XI collagen. Typical molecular changes result in either substitution of an obligate glycine within the Gly-Xaa-Yaa amino acid sequence repeat region of the molecule, mRNA missplicing or deletions/duplications that typically leaves the message in-frame. Clinical features include myopia, retinal detachment, craniofacial, joint, and hearing problems. Fibrochondrogenesis is also a COL11A1 related disorder, but here disease-associated variants are recessive and may be either null alleles or substitutions of glycine, and the condition is usually lethal in infancy.
Methods
The patient was assessed in the NHS England Stickler syndrome diagnostic service. DNA from the patient and family were analyzed with Next Generation Sequencing on a panel of genes known to cause Stickler Syndrome. The effect of sequence variants was assessed using minigene analysis. Allele-specific RT-PCR was performed.
Results
This patient had clinical type 2 Stickler syndrome but with severe hearing loss and severe ocular features including retinal atrophy and retinal tears in childhood. We identified a de novo in frame deletion of COL11A1 (c.4109_4126del) consistent with dominantly inherited Stickler syndrome but also a second inherited variant (c.1245+2T>C), on the other allele, affecting normal splicing of COL11A1 exon 9.
Conclusion
Exon 9 of COL11A1 is alternatively expressed and disease causing changes affecting only this exon modify the phenotype resulting from biallelic COL11A1 disease-associated variants and, instead of fibrochondrogenesis, produce a form of Stickler syndrome with severe hearing loss. Disease phenotypes from de novo pathogenic variants can be modified by inherited recessive variants on the other allele. This highlights the need for functional and family analysis to confirm the mode of inheritance in COL11A1-related disorders, particularly for those variants that may alter normal pre-mRNA splicing.
1 INTRODUCTION
Type 2 Stickler syndrome is a type XI collagenopathy, with clinical features that include myopia, retinal detachment, hearing problems, hypermobility, and premature arthritis, and changes in craniofacial development. Variants in COL11A1 that cause this disorder usually have a dominant negative effect typically resulting from changes that either substitute an obligate glycine within the repeating collagen Gly-Xaa-Yaa amino acid sequence, or from deletions or duplications to the mRNA which leave the message in frame (Richards et al., 2010). The deletions can result from either gross changes to the gene structure, or more frequently alterations to splice sites that cause exon skipping (collagen genes typically have exons coding for a complete set of Gly-Xaa-Yaa repeats). Type 1 Stickler syndrome, with similar clinical features but even more common retinal detachment, is caused by pathogenic variants in COL2A1, which encodes type II collagen (Snead et al., 2011). Stickler syndrome can also be inherited in a recessive form with homozygous variants in genes for type 9 collagen (COL9A1, COL9A2, COL9A3), LOXL3, and LRP2 (Nixon et al., 2019). Unlike COL2A1 where haploinsufficiency is associated with disease (Barat-Houari et al., 2016), carriers of null variants in COL11A1 are clinically unaffected with Stickler syndrome (but may have some mild hearing loss). However, biallelic null variants in COL11A1 result in fibrochondrogenesis, a usually lethal disorder, which can also be caused by a combination of null and missense variants (Tompson et al., 2010; Whitley et al., 1984).
Three exons in COL11A1 are alternatively spliced, and loss-of-function variants affecting one of these, exon 9, have been demonstrated to be associated with a form of recessive type 2 Stickler syndrome with profound hearing loss (Richards et al., 2013). This is because this exon is not expressed in mature cartilage but is expressed in Meckel's cartilage which develops into structures of the middle ear (Davies, Oxford, & Hausagus, 1998). Hence the naturally occurring alternative splicing of exon 9 modifies the expected outcome of biallelic COL11A1 loss-of-function variants from fibrochondrogenesis to Stickler syndrome with profound hearing loss, when one of the variants is located in or affects splicing of exon 9. Here we report a patient with a de novo in frame deletion in COL11A1 and an inherited exon 9 donor splice site alteration. The de novo deletion alone would be expected to result in dominant type 2 Stickler syndrome, but as described in the previous recessive examples, missplicing of exon 9 leads to additional severe hearing loss. This case illustrates the importance of the ACMG guidelines for DNA variant analysis and family studies to help determine the mode of inheritance for changes to splice sites in COL11A1 (Richards et al., 2015).
2 MATERIALS AND METHODS
2.1 Ethical compliance
The work in this study is approved by the local research ethics committee.
2.2 Clinical assessment
A patient was referred by a clinical geneticist to the NHS England Stickler syndrome diagnostic service for assessment including of her eyes, hearing, joints, and orofacial features of Stickler syndrome. The family pedigree was ascertained, and available family members were also examined. Hearing was assessed by pure tone audiometry, both air and conduction, at frequencies 250, 500, 1,000, 2,000, 4,000, and 8,000 Hz. Plain radiographs of the hips, knees, and spine were available. Ocular examination was at the slit-lamp, and the patient was also examined under general anesthetic including fundoscopy to assess the retina and, retinoscopy to assess refractive error.
2.3 Genetic analysis
The patient's DNA was analyzed by targeted next-generation sequencing using the Illumina TruSight One sequencing system and an in house bioinformatics pipeline to call variants in a virtual panel of genes known to cause Stickler syndrome (COL2A1, COL11A1, COL11A2, COL9A1, COL92, and COL9A3). The exons are numbered 1–68 as described in the reference sequences (NM_001854.3, NM_080692.2, and NM_080693.3), where exons 6, 7, and 9 are alternatively spliced, rather than numbered as exons 1–67 as in Annunen et al. (1999). Variant nomenclature is based on transcript NM_001854.3 which contains exons 6 and 9 (also referred to as exon 6A and 8, respectively) but not exon 7 (also referred to as exon 6B which is present in transcript NM_080629.2.).
2.4 Minigene analysis
The effect of sequence variants on pre-mRNA processing of exon 9 was assessed functionally using minigene analysis as described previously (Richards et al., 2012, 2013). Briefly, DNA encompassing COL11A1 exon 9 and surrounding intronic regions was amplified from the patient's DNA and ligated in between COL2A1 exons 43–46 in the splicing reporter USR13. Normal and variant clones were transfected into MIO-M1 cells, expressed and the resulting RNA analyzed by RT-PCR, agarose gel electrophoresis, and sequencing.
2.5 Allele specific RT-PCR
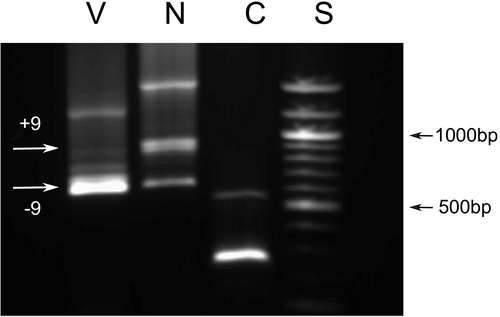
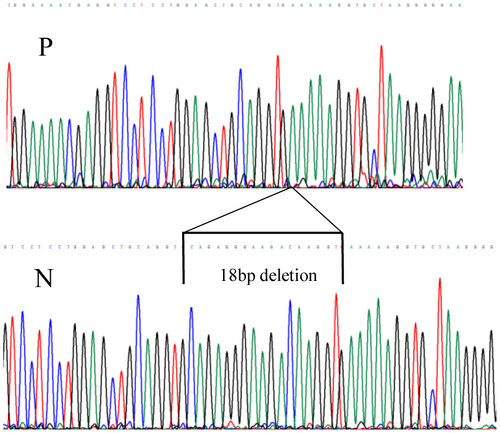
Cultured dermal fibroblasts obtained from the patient were used to prepare RNA and analyzed by reverse transcription PCR (RT-PCR) as previously described, where exon 9 was shown to be fully expressed in these cells (Martin et al., 1999; Richards et al., 2013), and can therefore be used as a reflection of expression of COL11A1 transcripts in other tissues/organs such as the ear. Two different amplifications were performed, first primers used for amplification were in exon 9 (5′GAAAACAAAGAAATAGACGGCAGG3′) and exon 59 (5′TTCACCCTTGGAGCCAGGGTCACC3′), so that exon 56 where the 18-bp deletion (see Results) was amplified within the amplicon. This ensured that only transcripts that contained exon 9 were amplified. Second, a primer covering the in frame deletion (see Results) in exon 56 (5′ACCTTTTTCACCTTGTCTTCCCTC3′) along with a primer in exon 6 (5′GACTATGAGTATGGGGAAGCAGCG3′), were used to amplify only cDNA which did not contain the 18-bp deletion in exon 56, but included the region encoded by exon 9. Amplified products were analyzed by agarose gel electrophoresis and Sanger sequencing.
3 RESULTS
3.1 Clinical assessment
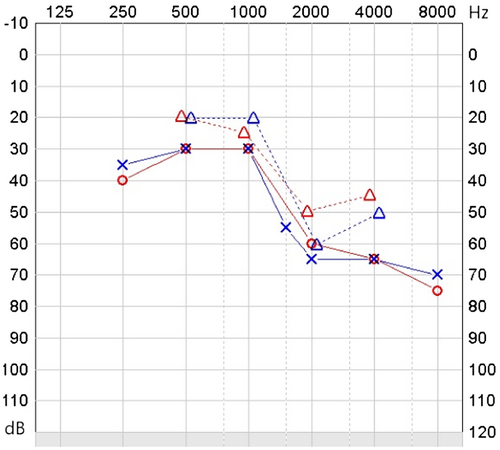
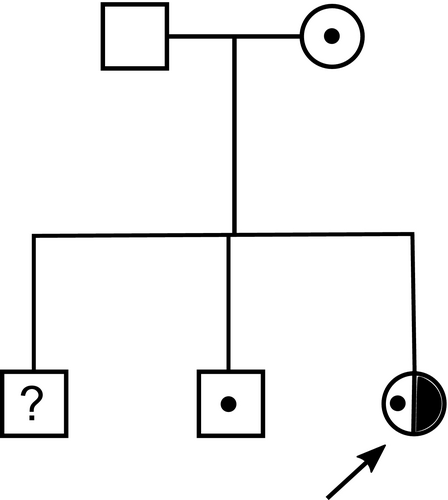
The patient had moved with her family to the UK from India aged 14. She was known to have poor hearing since birth, poor eyesight and global developmental delay, with limited communication using Signalong. She was given hearing aids and glasses for myopia (−5D) at age 15, and assessed in the Stickler diagnostic service aged 16. Ophthalmic features included visual acuity 6/48 in both eyes, bilateral sectoral lamellar and posterior subcapsular cataract, bilateral posterior vitreous detachment with pigment in the right vitreous, and widespread retinal atrophy. Examination under anesthetic was performed, as her learning difficulties made thorough assessment in clinic difficult. This demonstrated −10D myopia in the right eye, although retinoscopy was not possible in the left eye due to significant cataract. Horseshoe tears were identified in the left temporal retina just posterior to the ora serrata, and were treated with cryoretinopexy. Her audiogram showed severe (defined by the WHO as 61–80 dB of impairment) sensorineural hearing loss, particularly at high frequencies (Figure 1). Her joints were normal, with no abnormality on plain radiographs and no joint symptoms. Her palate was normal and there was no history of palate surgery. We were unable to obtain facial photographs. Her parents were nonconsanguineous and had no eye, hearing, joint, or palate abnormalities. Her older brother exhibited normal vitreous gel architecture and normal fundus examination. She has another brother who has not been assessed but is not known to be affected. The family pedigree is shown in Figure 2.
3.2 Genetic analysis
Next-generation sequencing detected two heterozygous variants in COL11A1 c.1245+2T>C and c.4109_4126del p.(Ala1370_Gly1375del). The c.1245+2T>C change affected the donor splice site of the alternatively spliced exon 9. The in frame deletion was situated in exon 56. Additional family analysis (Figure 2) showed that the c.1245+2T>C variant had been inherited from the patient's unaffected mother and was also carried by her unaffected brother. Neither parent possessed the c.4109_4126del p.(Ala1370_Gly1375del) variant, which was a de novo change in the patient with paternity having been confirmed molecularly (data not shown). The c.1245+2T>C variant was not present in the gnomAD database (https://gnomad.broadinstitute.org/) of sequence variants present in clinically normal individuals. Both variants were considered to be pathogenic, but it was not known if the c.1245+2T>C variant resulted in exon skipping or a frameshift due to a different missplicing event. These variants were submitted to the Leiden Open Variation Database (LOVD) and can be found at www.LOVD.nl (patient ID: 00238991). Minigene analysis of the c.1245+2T>C change to the exon 9 donor splice site determined that this change resulted in exon skipping, and not a missplicing event that caused a shift in the reading frame of the mRNA and subsequent nonsense mediated decay (Figure 3). Because the in frame deletion p.(Ala1370_Gly1375del) was de novo, and there were no heterozygous variants close to the deletion, it was not possible to use family studies to determine if the two changes were in cis or in trans. Instead we used allele specific RT-PCR using primers either in exon 9 or exon 56 for amplification. This demonstrated that the two changes were in trans (on different alleles), as amplified cDNA using a primer located in exon 9 enabled detection of the 18-bp deleted allele in exon 56 (Figure 4). As the donor splice site c.1245+2T>C variant causes skipping of exon 9, the cDNA would not be able to bind the exon 9 primer used for this amplification and transcripts from that allele would fail to amplify. Hence, since the assay was able to detect the 18-bp deletion, it must be on the opposite allele to the c.1245+2T>C variant. Similarly using a primer that included the region encoded by the de novo deletion in exon 56, cDNA amplified from the patient lacked exon 9, whereas cDNA amplified from normal individuals contained the 255-bp exon 9 (data not shown). This also confirmed the result obtained from the minigene analysis.
4 DISCUSSION
The ACMG guidelines for interpretation of DNA sequence variants (Richards et al., 2015) suggests using the criterion PVS1 (pathogenic very strong) for variants that alter acceptor and donor splice sites at the ±1 and 2 positions. This has been further refined by Tayoun et al. (2018) to take account of whether the reading frame is preserved or if the message is predicted to undergo nonsense mediated decay and the functional relevance of the region encoded by the exon. The case described here illustrates why this is particularly relevant to the type XI collagenopathies.
As disease-associated variants in COL11A1 can be both dominant and recessive careful consideration should be undertaken, particularly when analyzing variants that may lead to abnormal splicing of the pre-mRNA and especially when the variant affects an exon that is alternatively spliced. Many disease-associated variants in COL11A1 affect either donor or acceptor splice sites. In dominant disorders, the most common effect of these is exon skipping which leaves the message in-frame and leads to a dominant negative effect (Annunen et al., 1999; Booth et al., 2019; Griffith et al., 1998; Martin et al., 1999; Richards et al., 2013), where the shortened pro-alpha 1(XI) chain can coassemble with normal pro-alpha chains, resulting in abnormal procollagen molecules. However, missplicing events that lead to a frameshift and inclusion of a premature termination codon may not result in a discernible phenotype in a heterozygote as haploinsufficiency of COL11A1 is recessive and not associated with an obvious clinical phenotype (Akawi, Al-Gazali, & Ali, 2012; Richards et al., 2013; Tompson et al., 2010). Examination of the gnomAD DNA sequence database of clinically normal individuals reveals a number of rare/unique variants that alter the invariant nucleotides at the ±1 and 2 positions of acceptor and donor splice sites as well as a number of premature termination codons in a heterozygous state. These include c.1245+1G>A and c.1245+1G>T which is the same donor splice site that is altered in the patient described here. Also of note is the variant c.933+2T>C in the alternative transcript NM_080629.2. This variant alters the donor splice site of exon 7 (also called 6B) which is also alternatively spliced. It is present in 198 heterozygotes so, like the c.1245+2T>C variant described here, it too is likely to have a recessive effect. It is not clear if other COL11A1 constitutive exon splice site variants present in gnomAD such as c.3114+1G>A result in a frameshift and haploinsufficiency or the individuals with these changes have a mild form of Stickler syndrome, which can be difficult to diagnose.
Compared with the simple heterozygous dominant-negative COL11A1 variants that cause dominant type 2 Stickler syndrome (dSTL2) this patient appears to have a more severe ocular and auditory phenotype, which is consistent with the profound hearing loss found in other compound heterozygous COL11A1 variants involving exon 9 (Richards et al., 2013). Of patients with dSTL2, 50.6% have sensorineural hearing loss and another 26.6% have mixed hearing loss but this is usually mild to moderate (26–60 dB), rather than severe (60–80 dB) in these cases (Acke, Dhooge, Malfait, & De Leenheer, 2012). Her ocular phenotype includes widespread retinal atrophy, which is not typically associated with dSTL2. She also had early posterior vitreous detachment with retinal tears, significant myopia (−10D) and more visually significant cataract than might be expected at this age. Her global developmental delay is the feature that likely impacts most on her life, and this is not associated with dSTL2, but is possibly contributed to by her dual sensory impairment during childhood. In comparison her joints, which are likely spared the effect of the exon 9 variant are unaffected, although she is still young.
This case also highlights that de novo pathogenic variants may not be the only cause of the clinical presentation and that de novo changes along with an inherited change can occur even in rare disorders, which is important especially when both dominant and recessive modes of inheritance can result from changes to a gene. The patient described here had a de novo in frame deletion c.4109_4126del p.(Ala1370_Gly1375del) consistent with a dominant type XI collagenopathy. The second variant was also clearly pathogenic (recessive) affecting a donor splice site, albeit of an alternatively spliced exon. However pathogenic variants located deeper within COL11A1 introns have been reported (Akawi et al., 2012) and these types of change may not be routinely identified using exon capture sequencing protocols. Together these two pathogenic variants identified here highlight the need for functional and family analysis to confirm the mode of inheritance in COL11A1-related disorders, particularly for those variants that may alter normal pre-mRNA splicing.
ACKNOWLEDGMENTS
We thank the family for participating. We thank the staff at the NHS Medical Genetics laboratory at Addenbrooke's Hospital for technical and interpretative assistance. We thank Adrian Blackwell and Senjah Brown, clinic coordinators to the National Stickler syndrome diagnostic service, for administrative support. Thomas Nixon and the Vitreoretinal Research Group are supported by the Retinal Research Fund at the University of Cambridge.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
Thomas Nixon: investigation, writing—original draft presentation. Allan J. Richards: conceptualization, methodology, investigation, writing—original draft presentation. Adrian Lomas: investigation, data curation. Stephen Abbs: methodology. Pradeep Vasudevan: writing—review and editing. Annie McNinch: writing—review and editing. Philip Alexander: writing—review and editing. Martin P. Snead: supervision, writing—review and editing.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.