JAM2 is a prognostic biomarker and inhibits proliferation, metastasis and epithelial–mesenchymal transition in lung adenocarcinoma
Yanxin Dong and Jiale Zhang contributed equally to this work.
Abstract
Background
Junctional adhesion molecule 2 (JAM2) plays a pivotal role in various biological processes, including proliferation, metastasis and angiogenesis, contributing to tumor progression. While previous studies have highlighted the polarizing functions of JAM2 in different cancer types, its specific role in lung adenocarcinoma (LUAD) remains unclear.
Methods
In this study, we harnessed multiple public databases to analyze the expression and prognostic significance of JAM2 in LUAD. Using the Linkedomics database, Matescape database and R package, we explored the associated genes, the potential biological functions and the impact of JAM2 on the tumor microenvironment. Our findings from public databases were further validated using real-time quantitative PCR, western blot and immunohistochemistry. Additionally, in vitro experiments were conducted to assess the influence of JAM2 on LUAD cell proliferation, invasion, migration, apoptosis and epithelial–mesenchymal transition. Furthermore, we established a xenograft model to investigate the in vivo effects of JAM2 on tumorigenesis.
Results
Our results revealed a significant downregulation of JAM2 in LUAD, and patients with low JAM2 expression exhibited unfavorable overall survival outcomes. Functional enrichment analysis indicated that JAM2 may be associated with processes such as cell adhesion, extracellular matrix, cell junctions and regulation of proliferation. Notably, increased JAM2 expression correlated with higher tumor microenvironment scores and reduced immune cell abundance. Furthermore, overexpression of JAM2 induced apoptosis, suppressed tumor proliferation and exhibited potential inhibitory effects on tumor invasion and migration through the modulation of epithelial–mesenchymal transition. Additionally, in vivo experiments confirmed that JAM2 overexpression led to a reduction in tumor growth.
Conclusion
Overall, our study highlights the clinical significance of low JAM2 expression as a predictor of poor prognosis in LUAD patients. Moreover, JAM2 was found to exert inhibitory effects on various aspects of tumor progression. Consequently, JAM2 emerges as a promising prognostic biomarker and a potential therapeutic target for LUAD patients.
1 INTRODUCTION
Lung cancer stands as one of the most prevalent tumors and the leading cause of cancer-related mortality.1, 2 Lung adenocarcinoma (LUAD), comprising approximately half of all lung cancer cases, is characterized by an unfavorable prognosis, high metastatic potential and frequent recurrence. The current therapeutic approaches for LUAD encompass radiotherapy, chemotherapy, surgery, immunotherapy and molecular targeted therapy.3 However, the intricate interplay of tumor mutation profiles, the tumor microenvironment and immune dynamics underscores the pressing need for specific molecular markers and therapeutic targets. Molecularly targeted therapies, by selectively antagonizing tumor-specific molecules, offer the promise of curbing malignant growth and proliferation while sparing normal tissues—distinguishing them from conventional treatments such as radiotherapy and chemotherapy.4 Consequently, there is a critical need in tumor management for new biomarkers and treatment targets.
Tumor metastasis, the process by which cancer cells disseminate from their primary site to distant locations, plays a pivotal role in cancer progression. This intricate process involves the detachment of tumor cells from their primary site, migration through blood vessels or lymphatic vessels, and the colonization of distant organs or tissues.5, 6 Cell adhesion, mediated by cell–cell or cell–matrix interactions, profoundly influences tumor cell detachment and metastasis. Within this context, Junctional adhesion molecule 2 (JAM2) emerges as a key player.
Junctional adhesion molecule 2, located on the long arm of chromosome 21 at the 3q21 locus, is a versatile transmembrane protein belonging to the immunoglobulin superfamily and the JAM family. Its primary role lies in regulating the assembly of cellular junctions.7 Junctional adhesion molecule 2 is predominantly expressed on the cell membrane of eukaryotic cells, with a pronounced presence at cell–cell junctions. Its multifaceted functions include the modulation of tight junctions, regulation of cell permeability, control of cell proliferation, and participation in angiogenesis. Notably, hematopoietic stem cells establish interactions with the bone marrow through adherence to JAM2 expressed on bone marrow stromal cells, highlighting the pivotal role of JAM2 in sustaining the bone marrow mesenchymal microenvironment.8 In addition to its roles in cellular interactions, variations in the JAM2 gene have been associated with degenerative neurological disorders. Dysregulation of the neurovascular unit owing to JAM2 mutations can lead to Primary Familial Brain Calcification.9 Moreover, recent studies have demonstrated that Lactobacillus johnsonii MG can enhance the integrity of the intestinal barrier function in Caco-2 cells through GAPDH-JAM2 binding.10 In addition, dysregulation of JAM2 expression has been linked to tumor progression, exhibiting opposing effects in different tumor types.11 Despite the expanding understanding of the diverse functions of JAM2, its clinical significance and precise role in LUAD remain unclear. This study aimed to clarify the clinical significance and potential function of JAM2 in LUAD by integrating bioinformatics, examining clinical tissue specimens and performing in vitro experiments.
2 METHODS
2.1 Data sources and processing
The study's public data were sourced from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. The Cancer Genome Atlas database provided gene expression and detailed clinical information in 537 tumor samples and 59 normal samples. Meanwhile, gene expression comparisons between cancerous and normal tissue were derived from six GSE datasets—specifically, GSE10072, GSE19804, GSE18842, GSE27262, GSE63459 and GSE116959. For microarray data with detailed survival information from the GEO database, including GSE30219, GSE72094 and GSE68465, the following processes were performed that removed data older than 10 years and removed data with missing survival information. The study specifically incorporated tissue microarrays procured from individuals with LUAD who had not received radiotherapy or chemotherapy, spanning the period from 2017 to 2021.
2.2 Differential expression and prognostic analysis
Employing R software, Gene Expression Profiling Interactive Analysis (GEPIA) and the University of Alabama at Birmingham (UALCAN) database, this study identified the expression of JAM2 in LUAD. The overall survival of LUAD patients was depicted using Kaplan–Meier curves and Kaplan–Meier plotter database based on the expression of JAM2.
2.3 RNA extraction and real-time quantitative PCR
We performed the RNA extraction and real-time quantitative PCR according to the product instructions. Cellular and tissue RNA were extracted utilizing the Trizol reagent, followed by quantification of the extracted total RNA concentration via the NanoDrop spectrophotometer. This extracted RNA was then reverse transcribed into cDNA employing Vazyme's Reverse Transcription Reagents. Subsequent to this preparation, the cDNA, mix and primers were combined in an optimal ratio and allocated to a 96-well plate. Real-time quantitative PCR was then undertaken to quantify the expression levels of the targeted genes. To analyze the relative gene expression, we adopted the 2-ΔΔCt method. The primers utilized in this research were: GAPDH—F, GGAAGCTTGTCATCAATGGAAATC; R, TGATGACCCTTTTGGCTCCC; JAM2—F, GAAGCCCGCAATTCTGTTGG; R, AAGGCCACAACTACTACGGC.
2.4 Western blot
Fresh cells were washed twice with precooled phosphate-buffered saline before being lysed with an appropriate volume of radio immunoprecipitation assay lysis buffer (RIPA buffer) for 15–20 min on ice. Lysing was facilitated by conducting three to five sonications of 5 s each, utilizing an ultrasonic cell crusher. Then, the lysate was put into a low-temperature centrifuge and centrifuged at 12,000 rpm for 10 min, after which the supernatant from the centrifugation was transferred to a new centrifuge tube, which is the protein required for subsequent experiments. The proteins were separated using SDS–PAGE and then transferred to a PVDF membrane. Specific primary antibodies were applied to identify target proteins, which included JAM2 (ER65451, HuaBio, 1:500), vimentin (60330–1-Ig; Proteintech, 1:20000), N-cadherin (66219-1-Ig; Proteintech, 1:1000), b-actin (66009-1-Ig; Proteintech, 1:5000) and E-cadherin (60335-1-Ig; Proteintech, 1:1000). After incubation with the corresponding second antibodies, the relative expression of the proteins was detected using electrochemical luminescent solution according to the protocol.
2.5 CCK-8 and clone formation assay
Cell proliferation capacity was examined using cell counting kit-8 (CCK-8) assay. A negative control and overexpressing JAM2 cells were inoculated on 96-well plates. Subsequently, a 10% solution of CCK-8 was dispensed into each well and the plates were subsequently incubated for 1 h. The optical density of each well was measured at an absorbance wavelength of 450 nm using a microplate reader. Based on these optical density values, cell growth curves were constructed for each experimental condition.
To further evaluate cell growth, colony formation assays were conducted. Cells were inoculated in six-well plates with 800 cells per well. When the cells formed distinct colonies, the six-well plates were removed from the incubator, stained with crystal violet and photographed.
2.5.1 Apoptosis test
Cells from the experimental and control groups underwent apoptosis staining using Annexin V-IF647/PI, strictly following the instructions provided with the kit (G1514, Servicebio, China). The rate of apoptosis was subsequently quantified employing flow cytometry.
2.6 Wound healing experiment
The wound healing experiment was used to test cell migratory capacity. Cells were seeded uniformly into six-well plates and cultured until they reached over 90% confluence. At this point, a sterile 200 μL pipette tip was used to scratch the cell monolayer vertically with care, creating a wound. Subsequently, the detached cells were washed away, and the remaining cells were cultured in serum-free medium to prevent proliferation from influencing the results. Photographs were taken at 0 and 24 h to record the state of wound healing.
2.7 Cell migration and invasion assay
The transwell assay was utilized to evaluate the migratory and invasive properties of cells. Both experimental and control group cells were resuspended in serum-free medium and seeded at a density of 5 × 104 cells per transwell chamber. After adding 200 μL of serum-free medium to the upper chamber and 800 μL of medium containing 10% fetal bovine serum to the lower chamber, the 24-well plates were incubated in an incubator for 24 h. The migrated cells were subsequently visualized using microscopic observation.
The cell invasion assay requires the use of matrix adhesive to cover the polycarbonate membrane of the inner chamber in advance. Other steps were consistent with the cell migration assay.
2.8 Immunohistochemistry
The expression of JAM2 in tissue samples was investigated using immunohistochemistry. The primary JAM2 antibody was diluted to 1:150. The intensity of staining was categorized as negative, weak, moderate or strong. The samples were then evaluated and scored accordingly, based on these different levels of staining intensity.
2.9 Identification of co-expressed and differential genes
The LinkedOmics database is an online database that provides multidimensional analysis for information on TCGA data. In the LinkedOmics database, we evaluated the co-expressed genes of JAM2 using the Pearson correlation coefficient. In addition, we used the median expression value of JAM2 as a cutoff to identify differential genes in different subgroups by the DEseq2 package in R. The identification criteria we used were log2 foldchange ≥1 and p-value <0.05.
2.10 Functional enrichment
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed on the intersecting genes of co-expressed and differential genes using the matescape database. Gene Ontology analysis includes biological processes, cellular components and molecular function.
2.11 Tumor microenvironment scores and immune infiltration analysis
The tumor microenvironment (TME) was scored using the ESTIMATE algorithm, which includes three distinct scores: immune score, stromal score and ESTIMATE score. The relationship between the expression of JAM2 and the abundance of immune cells in the immune microenvironment was analyzed using the ssgsea algorithm. The association between JAM2 and immune-related molecules was assessed using the Pearson correlation coefficient.
2.12 Xenograft tumor-formation experiment
Female Balb/c nude mice, aged 5–6 weeks, received injections of either control or JAM2-OE H1299 cells (n = 5 per group). Upon the emergence of visible xenografts, the volumes were measured every 5 days. Subsequently, all mice were euthanized and weighed on the 21st day after the initial recording. The experiment has received approval from the animal ethics committee (MDKN-2023-022).
2.13 Statistical analysis
All statistical analyses were performed utilizing R, SPSS and GraphPad. Differences between the two groups were evaluated using t-tests. Furthermore, the Kaplan–Meier method was employed to illustrate the relationship between overall survival time and gene expression levels. The significance threshold was established at p < 0.05 for all tests.
3 RESULTS
3.1 Low expression of JAM2 in LUAD based on public databases
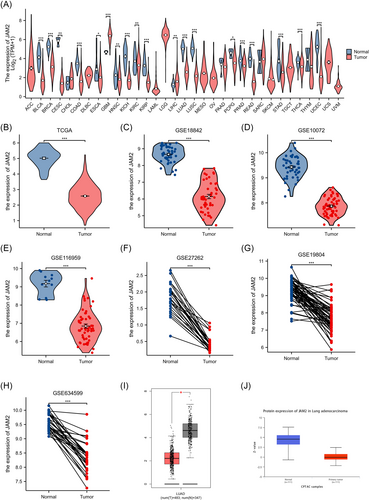
Based on a pan-cancer analysis of 33 different cancers sourced from TCGA, JAM2 expression exhibited a notable decrease in several tumor types (Figure 1A). Further examination of datasets from TCGA and GEO consistently indicated a significant downregulation of JAM2 messenger RNA (mRNA) in LUAD (Figure 1B–H). This finding was further validated using the GEPIA and UALCAN databases (Figure 1I,J).
3.2 Clinical characteristics and prognostic analysis based on public databases
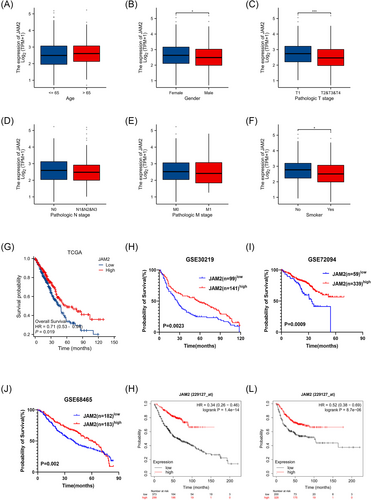
To assess the relationship between JAM2 expression and clinicopathological features, we analyzed JAM2 expression within various groups from TCGA database. The results revealed elevated levels of JAM2 expression in female patients, those with T1-stage tumors, and individuals with a history of smoking compared with their respective counterparts (Figure 2A–F). Additionally, within the LUAD patient cohort, stratification into low and high JAM2 expression groups demonstrated that lower JAM2 expression levels correlated with more adverse clinicopathological characteristics, including advanced T-stage and N-stage and overall TNM-stage (Table 1). Moreover, survival analyses across multiple datasets indicated that reduced JAM2 expression was associated with poorer prognosis (Figure 2G–J). The Kaplan–Meier plotter analysis reinforced these findings by revealing shorter overall survival and time to first progression in patients with low JAM2 expression (Figure 2K–L).
| Characteristics | Low expression of JAM2 | High expression of JAM2 | p-Value |
|---|---|---|---|
| n | 269 | 270 | |
| Pathological T-stage, n (%) | <0.001 | ||
| T1 | 70 (13.1%) | 106 (19.8%) | |
| T2, T3 and T4 | 198 (36.9%) | 162 (30.2%) | |
| Pathological N-stage, n (%) | 0.041 | ||
| N0 | 167 (31.9%) | 183 (35%) | |
| N1&N2&N3 | 99 (18.9%) | 74 (14.1%) | |
| Pathological M stage, n (%) | 0.607 | ||
| M0 | 185 (47.4%) | 180 (46.2%) | |
| M1 | 14 (3.6%) | 11 (2.8%) | |
| Pathological stage, n (%) | 0.032 | ||
| Stage I | 136 (25.6%) | 160 (30.1%) | |
| Stage II, Stage III and Stage IV | 130 (24.5%) | 105 (19.8%) | |
| Gender, n (%) | 0.052 | ||
| Female | 133 (24.7%) | 156 (28.9%) | |
| Male | 136 (25.2%) | 114 (21.2%) | |
| Age, n (%) | 0.380 | ||
| ≤65 | 134 (25.8%) | 123 (23.7%) | |
| >65 | 127 (24.4%) | 136 (26.2%) | |
| Smoker, n (%) | 0.105 | ||
| No | 32 (6.1%) | 45 (8.6%) | |
| Yes | 231 (44%) | 217 (41.3%) |
3.3 Function enrichment
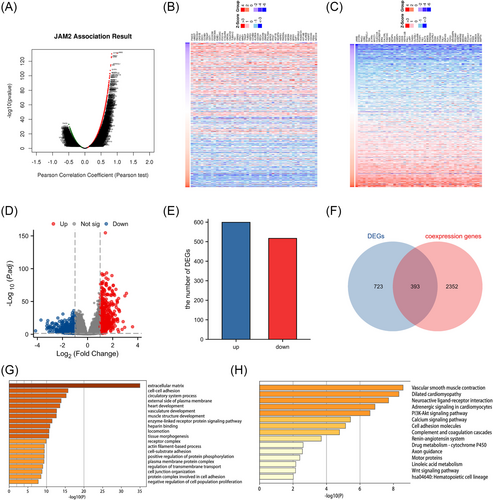
To explore the functions of JAM2, the co-expressed genes of JAM2 were analyzed by the co-expression analysis module in the LinkedOmics database. All co-expressed genes are visualized by volcano plots (Figure 3A). Employing rigorous criteria (|R-statistic| > 0.3 and false discovery rate < 0.05), we identified a total of 2745 co-expressed genes. Among these, 605 genes displayed a negative correlation, while 2140 genes exhibited a positive correlation with JAM2 expression. The top 50 genes, both positively and negatively associated with JAM2, were depicted in a heatmap (Figure 3B,C). Subsequently, LUAD patients were stratified into two groups based on JAM2 expression, leading to the identification of 1116 differentially expressed genes, including 599 upregulated and 517 downregulated genes (log2|fold change| > 1, p-value < 0.05) (Figure 3D,E). Among these, 393 genes intersected between the co-expressed and differentially expressed gene sets, signifying their significance for subsequent enrichment analyses (shown as Venn diagrams; Figure 3F). Functional enrichment analyses, including GO and KEGG, revealed that these genes were primarily involved in processes related to the extracellular matrix, cell adhesion, cell junctions, cell proliferation regulation, PI3K-Akt pathway and Wnt-related signaling pathways (Figure 3G,H).
3.4 TME scores and immune infiltration
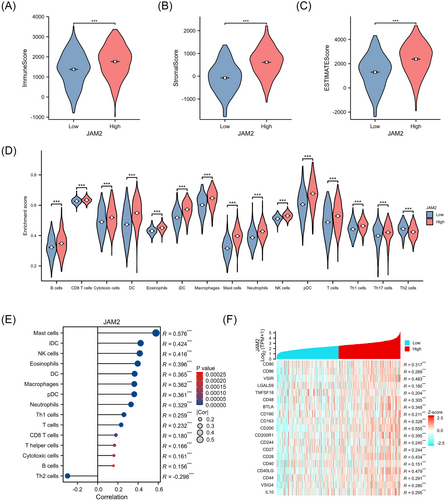
Within the JAM2 high-expression group, we observed elevated TME-related scores in LUAD, encompassing immune score, Estimate score and stroma score, compared with the other group (Figure 4A–C). JAM2 expression exhibited a negative correlation with Th2 cell abundance but a positive correlation with the infiltration of various immune cell types, including B cells, cytotoxic cells, DC cells, mast cells, macrophages, neutrophils, Th1 cells, NK cells and CD8+ T cells (Figure 4D,E). Additionally, JAM2 expression was associated with several immune-related molecules (Figure 4F).
3.5 The expression of JAM2 is low in LUAD tissues and correlates with an unfavorable prognosis
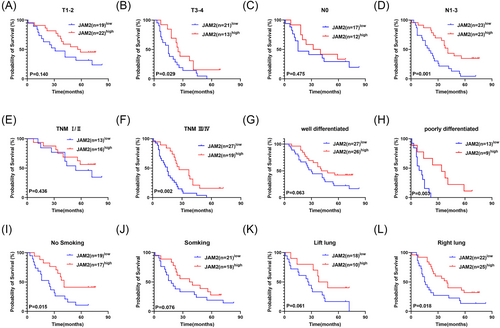
Immunohistochemistry analysis of 75 pairs of LUAD and adjacent paracancer tissues highlighted the low expression of JAM2 in LUAD specimens. Importantly, lower JAM2 expression was linked to a worse prognosis (Figure 5A–C). Furthermore, JAM2 emerged as an independent prognostic factor in LUAD patients (Figure 5D,E). Subgroup analyses demonstrated that low JAM2 expression was associated with unfavorable overall survival in LUAD with T3–4, N1–3, TNM III/IV stages, poor differentiation, non-smoking history or tumors located in the right lung (Figure 6A–L).

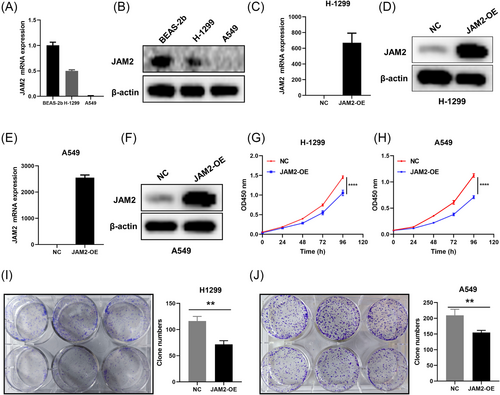
3.6 Overexpression of JAM2 inhibits cell malignant behavior in vitro and in vivo
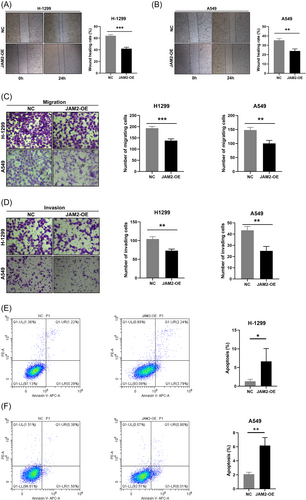
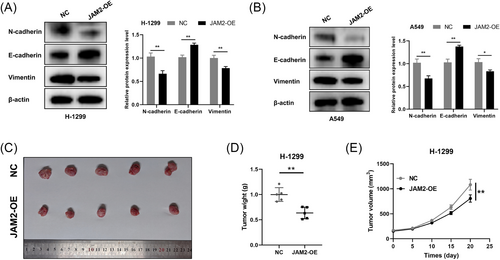
In vitro experiments utilizing LUAD cell lines H-1299 and A549, which displayed diminished JAM2 expression compared with normal bronchopulmonary epithelial cells BEAS-2b, revealed the functional impact of JAM2 (Figure 7A,B). We established LUAD cell lines that stably overexpressed JAM2 (Figure 7C–F). Stable overexpression of JAM2 resulted in inhibited cell proliferation, reduced cell viability and decreased colony-forming ability (Figure 7G–J). JAM2 also significantly suppressed wound-healing ability, cell migration and invasion while inducing apoptosis in LUAD cells (Figure 8A–F). Western blot analysis demonstrated that JAM2 promoted the expression of E-cadherin while inhibiting the expression of vimentin and N-cadherin in LUAD cells (Figure 9A,B). Notably, in a nude mouse xenograft tumor model, tumors in the JAM2 overexpressing group exhibited slower growth and reduced tumor weight, aligning with our in vitro findings (Figure 9C–E).
4 DISCUSSION
The JAM family plays a pivotal role in the formation of tight junctions and is essential for cell adhesion processes.12, 13 JAM2, a member of this family, is predominantly localized at the tight junctions of various small blood vessels.12 It exerts a significant influence on various facets of cancer biology, including cell migration, invasion, angiogenesis and proliferation. Notably, JAM2's role can vary considerably across different cancer types. For instance, in oral squamous cell carcinoma (OSCC), JAM2 levels were higher in the OSCC cell line with greater metastatic potential compared with the Tca8113 cell line, suggesting a potential role in OSCC metastasis.14 In melanoma, JAM2 promoted melanoma cell metastasis by binding to JAM-C on tumor cells, activating key molecules associated with metastasis.15 Similarly, JAM2 has been implicated in promoting tumor progression in cervical cancer, pancreatic cancer and glioma.16-18 In contrast, JAM2 plays a contrasting role in other solid tumors. In breast cancer, high JAM2 expression correlated with better patient prognosis and inhibited breast cancer cell invasion and migration by suppressing the epithelial–mesenchymal transition (EMT) process.19 Likewise, JAM2 exhibited a suppressive role in tumor progression in colorectal and esophageal squamous carcinomas.20, 21 Additionally, in a 21-trisomy Down syndrome mouse model, JAM2 overexpression inhibited the tumor effects of lung cancer by suppressing angiogenesis.22 Nevertheless, the clinical significance and in vitro role of JAM2 in LUAD remained elusive.
In this study, we explored the clinical significance and potential biological functions of JAM2 in LUAD using a multifaceted approach, combining bioinformatics analysis, in vitro experiments and in vivo studies. Our analysis of data from TCGA and GEO databases revealed the downregulation of JAM2 expression in LUAD, which was associated with worse clinicopathologic features and poorer prognosis. Importantly, immunohistochemistry results from our tissue microarrays validated these findings from public databases, further emphasizing the potential of JAM2 as an important biomarker for the diagnosis and prognosis of LUAD. Furthermore, our analysis suggested that JAM2 was linked to processes including cell adhesion, the extracellular matrix and the negative regulation of cell proliferation.
Tumor metastasis, a critical aspect of cancer progression, necessitates the ability of tumor cells to detach from their primary site, reduce cell-to-cell or cell-to-extracellular matrix adhesion and remodel the extracellular matrix to colonize new niches via blood vessels or lymphatic vessels.23 The enrichment of GO and KEGG pathways related to cell adhesion, extracellular matrix and cell proliferation indicated that JAM2 might play a role in tumor progression. Subsequent in vitro experiments corroborated this by demonstrating that JAM2 overexpression inhibited various biological functions in LUAD cells, including cell proliferation, migration and invasion. Importantly, JAM2 was found to induce apoptosis in LUAD cells. Analysis of EMT-related proteins suggested that JAM2 may suppress tumor cell metastasis by regulating the EMT process. The EMT is a critical process wherein epithelial cells undergo a transformation in cell polarity and morphology, ultimately gaining the ability to metastasize. This phenomenon is intricately linked to tumor metastasis, drug resistance and various other malignant behaviors in cancer.24 Thus, our findings strongly suggest that JAM2 may act as a suppressor of tumor cell metastasis by effectively inhibiting the EMT process.
Numerous previous studies have underscored the critical role of the tumor immune microenvironment in tumor progression.25-27 Tumor progression is not solely driven by genetic mutations within tumor cells themselves; it is also significantly influenced by the interactions among immune cells and mesenchymal stromal cells in the TME. In this context, JAM2 may actively participate in remodeling the tumor immune microenvironment. Our analysis revealed a strong positive correlation between JAM2 expression and immune infiltration, suggesting that JAM2 can induce an immune-activated microenvironment, thereby inhibiting tumor growth. Specifically, JAM2 exhibited positive correlations with various immune and inflammation-activating cell types, including mast cells, NK cells, iDS cells and T cells, while displaying negative correlations with immune-suppressive Th2 cells. Previous research has demonstrated that NK cells and T cells secrete a variety of pro-inflammatory factors that exert a tumor-killing effect.28, 29 Conversely, Th2 cells can produce pro-tumor progression factors, and their abundance has been linked to malignant progression in several cancer types, including LUAD, gastric cancer and colorectal cancer.30-32 Consequently, our findings suggest that JAM2 may function as a tumor suppressor and significantly modulate LUAD progression.
In summary, this study provides initial insights into the prognostic significance, immune characteristics and in vitro and in vivo functions of JAM2 in LUAD. However, it is important to acknowledge the limitations of this research. The relatively modest clinical sample may introduce some bias when assessing the prognostic value of JAM2. Furthermore, the conclusions regarding the effect of JAM2 on the immune microenvironment in LUAD should be subjected to further validation through subsequent experiments.
5 CONCLUSION
Our study presents reliable evidence for the role of JAM2 in LUAD. JAM2 is expressed at a low level and related to poor prognosis in LUAD. Meanwhile, JAM2 can affect the metastasis-related functions of LUAD and may exert a critical effect in the remodeling of the LUAD microenvironment.
AUTHOR CONTRIBUTIONS
Conception and design: Yanxin Dong and Jiale Zhang. Administrative support: Shouyin Di, Boshi Fan and Taiqian Gong. Provision of study materials or patients: Shouyin Di and Boshi Fan. Collection and assembly of data: Jiale Zhang, Yanxin Dong, Shun Xie. Data analysis, funding and interpretation: Shouyin Di and Boshi Fan. Manuscript writing: All authors; Final approval of manuscript: All authors.
ACKNOWLEDGEMENTS
We would like to thank all public databases, including TCGA, GEO, GEPIA, UALCAN, LinkedOmics and Matescape.
CONFLICT OF INTEREST STATEMENT
All authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
All relevant data in this study can be included in the published article.




