OTUD4 regulates metastasis and chemoresistance in melanoma by stabilizing Snail1
Yuchen Gao and Jiaxin Tang are cofirst authors.
Abstract
Melanoma is the most aggressive form of skin cancer with rapidly increased incidence worldwide especially in the Caucasian population. Surgical excision represents the curative treatment choice in patients with early-stage disease. However, the therapeutic outcomes in patients with metastatic melanoma remains unsatisfactory. Thus, understanding molecular mechanisms contributing to metastasis and chemoresistance is critical for new improved therapies of melanoma. Snail1, an important epithelial-mesenchymal transition transcription factors (EMT-TFs), is critical to induce the EMT process, thereby contributing to cancer metastasis. However, the involvement of Snail1 in melanoma metastasis remains elusive and the underlying mechanism to regulate Snail1 in melanoma needs to be further investigated. Here, we identified OTUD4 as a novel deubiquitinase of Snail1 in melanoma. Moreover, the depletion of OTUD4 in melanoma cells markedly inhibited Snail1 stability and Snail1-driven malignant phenotypes both in vitro and in vivo. Overall, our study establishes OTUD4 as a novel therapeutic target in metastasis and chemoresistance of melanoma by stabilizing Snail1 and provides a rationale for potential therapeutic strategies of melanoma.
1 INTRODUCTION
Melanoma is the most lethal skin cancer with increased incidence worldwide. Although the curative rate is high in melanomas at early stage, the prognosis in patients with metastatic melanoma remains poor due to the limited response to current therapies and frequent resistance to target therapies such as BRAF inhibitor (Guo et al., 2021; Turner et al., 2018). Therefore, to elucidate key mechanisms underlying metastasis and therapeutic resistance in melanoma is urgently needed.
The epithelial-mesenchymal transition (EMT), a reprogramming of polarized epithelial cells into mesenchymal cells, endows cells with more invasive properties and contribute to the stemness, metastasis and chemoresistance. (Bakir et al., 2020; Chen et al., 2017; Nieto et al., 2016; Pastushenko & Blanpain, 2019). Snail1, a key EMT transcription factors (EMT-TFs), is pivotal to induce EMT and drive the EMT-associated malignancies (Kaufhold & Bonavida, 2014). The expression of Snail1 is tightly regulated at the transcriptional and post-translational levels. TGFβ, Wnt, Notch and growth factors can increase Snail1 transcription, thereby inducing EMT and promoting the progression of several malignant tumors (Kaufhold & Bonavida, 2014). In addition, multiple posttranslational regulatory mechanisms are critical to control the subcellular localization, degradation and its repressive function (Baulida et al., 2019). For instance, the phosphorylation events controlled by GSK3β, and the small C-terminal domain phosphatase could regulate Snail nuclear exit (Wu et al., 2009). The lactylation and oxidative deamination of Snail1 was demonstrated to affect its function (Fan et al., 2023; Herranz et al., 2016). As an unstable protein, Snail1 could undergo ubiquitin-proteasome dependent degradation, in which several E3 ligases such as β-TrCP1, Fbxl14, and Fbxl5 are essential to catalyze the ubiquitination and subsequent degradation of Snail1 (Baulida et al., 2019; Viñas-Castells et al., 2010, 2014). Recently, different deubiquitinase including DUB3, USP9X and USP27X were reported to cleave the ubiquitination of Snail1 and promote the Snail1 dependent malignant processes in triple negative breast cancer and pancreatic cancer respectively (Guan et al., 2022; Lambies et al., 2019; Liu et al., 2017).
The upregulation of Snail1 is observed in melanocytes undergoing malignant transformation (Poser et al., 2001), which enhances the mobility of cancer cells and drives melanoma metastasis (Hao et al., 2012; Maiques et al., 2018). Snail1-mediated inhibition of CYLD could activate RIP1 expression and contribute to the resistance to BRAF inhibitors in melanoma (Lei et al., 2018). The overexpression of Snail1 in murine and human melanoma cells promotes EMT and immunosuppression, which render Snail1-positive melanoma fail to respond to immunotherapy. Considering these important roles of Snail1 in the resistance to BRAF inhibitor and immunotherapy, to elucidate the underlying mechanism to stabilize Snail1 in melanoma could greatly benefit the clinical outcome of melanoma patients. Here, we identified OTUD4 as a novel DUB of Snail1 in melanoma, which directly binds, deubiquitinates and stabilizes Snail1 and promotes Snail1-driven tumor metastasis and chemoresistance in melanoma.
2 MATERIALS AND METHODS
2.1 Cell culture, plasmids, and antibodies
HEK293T, A375 and A2058 cells were purchased from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin (Sigma-Aldrich), and 100 mg/mL streptomycin (Sigma-Aldrich) at 37°C in a 5% CO2 air incubator. All cell lines were mycoplasma-free and authenticated by short tandem repeat DNA profiling analysis. Snail1 and OTUD4 were cloned into pLV.3-FLAG, pLV.5-S-HA, pCMV-HA, PET28A and pGEX4T-1 vectors. The OTUD4 shRNA sequences are 5′-GGAACTAGACACGTTGGAAGT-3′ and 5′-TTGAAGAATATTTAAAGCGTT-3′. Anti-Snail (3895, dilution: 1:500), anti-E-cadherin (3195, dilution: 1:1000) and anti-N-cadherin (13116, dilution: 1:1000) antibodies were purchased from Cell Signaling Technology. Anti-OTUD4 (HPA036623, dilution: 1:1000), anti-FLAG (M2, dilution: 1:1000), anti-β-actin (A1978, dilution: 1:5000), and anti-HA (H3663, dilution: 1:1000) antibodies were purchased from Sigma-Aldrich. Anti-Ub antibody (sc-8017, dilution: 1:1000) was purchased from Santa Cruz Biotechnology.
2.2 Transwell migration and invasion assays
Cells were resuspended in serum-free DMEM for inoculation onto the membranes of 24-well transwell inserts (pore size, 8 mm; Corning) in upper chamber. Inserts with non-coated membranes were used for migration assay, while inserts with Matrigel-coated membranes were used for invasion assay. The lower chambers were filled with DMEM containing 20% FBS served as chemoattractant. After 48 h, the cells that migrated or invaded the surface of lower chambers were fixed and stained with crystal violet and counted in three randomly selected microscopic fields.
2.3 Western blot analysis
The cells were harvested and were lysed with NETN buffer (pH 8.0, 300 mM NaCl, 20 mM Tris-HCl, 1 mM EDTA, 0.5% NP-40, and protease inhibitors (1x protease inhibitor cocktail [Roche], 1 mM sodium orthovanadate, 10 mM β-glycerophosphate, 1 mM phenylmetnylsulfonyl fluoride, and 10 mM sodium fluoride) for 30 min on ice. After centrifugation, the lysates were boiled in loading buffer for 5 min. Proteins were fractionated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes, then incubated with the indicated primary and secondary antibodies.
2.4 Immunoprecipitation
Cells were transfected with empty vector, FLAG-Snail1 or FLAG-OTUD4 as indicated. Cell pellets were lysed in NETN buffer and incubated with FLAG affinity gel at 4°C for 2 h. The precipitates were washed for three times and boiled in loading buffer for 5 min. The immunoprecipitates were subjected to Western blot.
2.5 Denaturing nickel resin chelated with intrilotriacetic acid (Ni-NTA) pulldown
Cells transfected as indicated were harvested and washed once with PBS. Cells were lysed in 8 M urea, 0.1 M NaH2PO4, 300 mM NaCl and 0.01 M Tris-HCL (pH 8.0). Lysates were briefly sonicated to shear DNA and incubated with Ni-NTA agarose (R90101, Invitrogen) for 2 h at 4°C. Ni-NTA agarose was washed four times with 8 M urea, 0.1 M NaH2PO4, 300 mM NaCl and 0.01 M Tris-HCL (pH 8.0). Input and Ni-NTA agarose was boiled in loading buffer and subjected to Western blot.
2.6 Denaturing immunoprecipitation for ubiquitination
Cells were lysed in 100 mL 62.5 mM Tris-HCl (PH 6.8), 10% glycerol, 2% SDS, 1 mM iodoacetamide and 20 mM NEM, boiled for 15 min, diluted 10 times with NETN buffer containing protease inhibitors, 20 mM NEM and 1 mM iodoacetamide, then centrifuged to remove cell debris. The cell extract was subjected to immunoprecipitation with the indicated antibodies and blotted as indicated antibody.
2.7 Cell Counting Kit (CCK)-8 assay
Cells were seeded at a density of 2000 cells/well in 96-well plates. The cell viability in each group was evaluated using a CCK8 assay kit (HY-K0301, MCE). The optical density values at a wavelength of 450 nm were measured using a microplate reader to determine cell viability.
2.8 Glutathione S-transferase (GST) pull-down assay
Indicated cDNA was cloned into a pGEX-4T-1 vector with GST-tag and purified from the Escherichia coli strain BL21 using Pierce Glutathione Agarose (Thermo Scientific). Bacterial-expressed GST and GST-OTUD4 protein bound to Pierce Glutathione Agarose was incubated with PET28A-Snail1, purified from the Escherichia coli strain BL21 for overnight at 4°C. Then the agarose was washed with NETN three times and PBS three times, subjected to Western blot.
2.9 Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cultured cells using TRIzol reagent (Thermo Fisher Scientific) and converted into cDNA using FastKing gDNA Dispelling RT SuperMix (Tiangen). qPCR was performed using FastFire qPCR PreMix (SYBR Green) using primers against Snail1. All experiments were performed in triplicate with GAPDH as an internal control. Primer sequences are listed. hSNAIL1 Forward: 5′-GGCCCTGGCTGCTACAAGGC-3′; Reverse: 5′-CTCGAGGGTCAGCGGGGACA-3′. GAPDH Forward: 5′-GATCGAATTAAACCTTATCGTCGT-3′; Reverse: 5′-GCAGCAGAACTTCCACTCGGT-3′.
2.10 Animal experiments
5 × 106 A375 cells were transfected as indicated and injected into the lateral tail vein of nude mouse (4 mice/group) to study the lung metastatic. The lungs were harvested 6 weeks after injection and visible lung metastatic nodules were examined macroscopically. All animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of the Jinan University (IACUC-20220526-05).
2.11 Statistical analysis
Statistical analysis were performed with GraphPad Prism 8.0 software. Each experiment was repeated three times and experimental data are presented as mean ± standard deviation (mean ± standard deviation). Differences between two groups of data were compared using Student's t-test. Differences among more than two groups were compared using one-way analysis of variance. analysis of variance and Tukey's test: compare all pairs of columns. *p < 0.05, **p < 0.01. ***p < 0.001 is considered statistically significant.
3 RESULTS
3.1 OTUD4 directly interact with Snail1
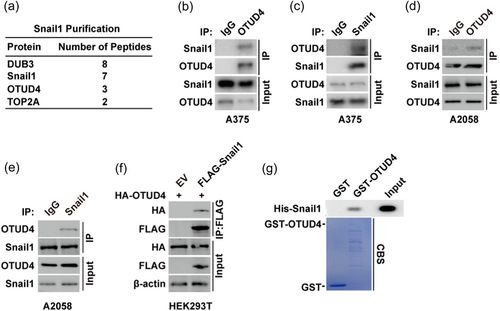
Considering the well-established role of Snail1 in EMT and cancer metastasis, we first assessed the cellular phenotype of Snail1 in melanoma cells. In A375 cells, knocking down of Snail1 could significantly increase the expression of E-cadherin while decrease the expression of N-cadherin (Supporting Information: Figure 1A), suggesting the regulatory role Snail1 in the EMT progression of melanoma cells. Consistently, depletion of Snail1 inhibit the migration and invasion capacity of melanoma cells (Supporting Information: Figure 1b−d). To identify novel regulator of Snail1 in melanoma, A375 cells stably expressing FLAG-Snail1 were constructed to perform tandem affinity purification and mass spectrometry analysis. In addition to previously reported Snail1 interacting proteins such as DUB3 (Liu et al., 2017), we identified OTUD4 as a potential Snail1 interacting protein (Figure 1a). We next confirmed the interaction of Snail1 and OTUD4 by endogenously reciprocal coimmunoprecipitation assay in A375 cells (Figure 1b−c) and A2058 cells (Figure 1d−e). Also, exogenous coimmunoprecipitation assay performed in HEK293T cells confirmed the in vivo interaction of Snail1 and OTUD4 (Figure 1f). Further, His-Snail1 could be pulled down by purified GST-OTUD4 but not GST in vitro (Figure 1c). Taken together, these results indicate the direct interaction between OTUD4 and Snail1 in melanoma cells.
3.2 OTUD4 regulates Snail1 protein stability in melanoma cells
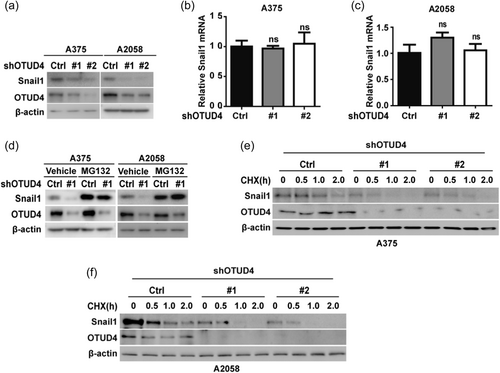
The direct interaction between OTUD4 and Snail1 prompted us to examine a potential role for OTUD4 in the regulation of Snail1 stability and the function. Firstly, we observed that depletion OTUD4 in A375 and A2058 cells significantly decreased the protein level of Snail1 (Figure 2a) without affecting snali1 mRNA levels (Figure 2b,c). In addition, the proteasome inhibitor, MG132 could rescue the decreased protein level of Snail1 in cells depleted of OTUD4 (Figure 2d). To further confirm the role of OTUD4 in Snail1 protein stability regulation, we treated A375 and A2058 cells with cycloheximide (CHX) and the results showed that Snail1 protein stability was significantly decreased in cells following the depletion of OTUD4 (Figure 2e,f). Taken together, these results suggested that OTUD4 could regulate Snail1 protein stability in melanoma cells.
3.3 OTUD4 deubiquitinates Snail1
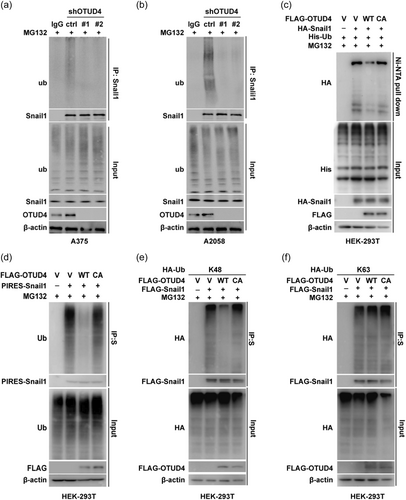
Next, we tested whether OTUD4 is a bona fide DUB of Snail1 in melanoma cells with the ubiquitination assay. In A375 and A2058 cells, downregulation of OTUD4 markedly reduced the ubiquitination level of endogenous Snail1 (Figure 3a,b). In addition, Ni-NTA pull down assay were perfomed in cells cotransfected with His-tagged ubiquitin, HA-Snail1 and FLAG-OTUD4 WT/C45A mutant (catalytically inactive mutant). The significantly decreased Snail1 ubiquitin level was observed in cells with stable expression of OTUD4 WT but not in cells with catalytically inactive C45A mutant (Figure 3c), indicating that OTUD4 could directly deubiquitinate Snail1. In line with this, similar phenotype were observed in A375 cells contrancefected with OTUD4 and Snail1 by pull-down assay with S-tag labeled Snail1 (Figure 3d). Further, we investigated which types of polyubiquitin chain on Snail1 was regulated by OTUD4. The ubquitination assay results showed that, in agreement with the role of OTUD4 in the Snail1 protein stability control, OTUD4 catalyze the cleavage of the K48 rather than the K63 polyubiquitin chain on Snail1. Conclusively, these results suggested that OTUD4 is a bona fide deubquitinase of Snail1 which stabilize Snail1 in melanoma cells through the catalytic cleavage of K48 polyubiquitin chain of Snail1.
3.4 OTUD4 regulates cell migration, invasion, and metastasis through Snail1
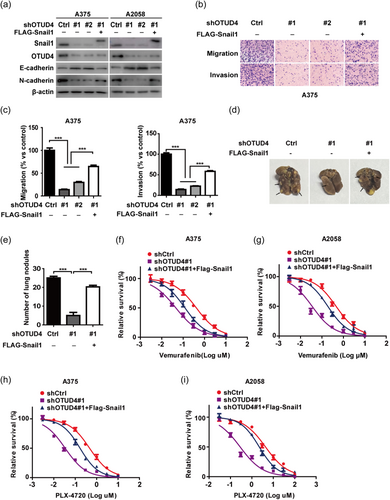
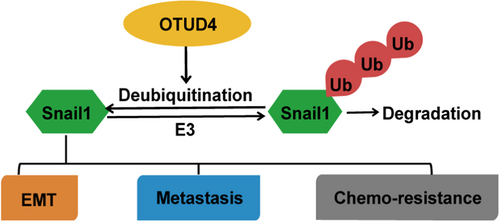
As a well known key factor of EMT, Snail1 plays a pivotal role in the migration and invasion progression of cancer cells. Therefore, we examined whether OTUD4 could regulate the migration, invasion, and metastasis through Snail1 in melanoma cells. Firstly, we observed that, in A375 and A2058 cells, depletion of OTUD4 result in the the significantly increased E-cadherin expression with the sharply decreased N-cadherin expression (Figure 4a). Considering both of E-cadherin and N-cadherin had been identified as the key marker of EMT, this result strongly suggested that OTUD4 could regulate the EMT progression in melanoma. Interestingly, reconstitution of Snail1 could largely rescue such an effect in A375 and A2058 cells (Figure 4a), suggesting that OTUD4 regulate melanoma EMT though stabilize Snail1. Moreover, through transwell assay, we observed that the depletion of OTUD4 significantly inhibited the migration and invasion of A375 and A2058 cells while the reconstitution of FLAG-Snail1 in OTUD4 depleted cells could largely reverse these cellular phenotype (Figure 4b–c; Supporting Information: Figure 2a−b). Next, we investigated the function of OTUD4 in xenograft metastasis model. We discovered that the depletion of OTUD4 markedly suppressed the number of metastatic lung nodules mediated by A375 cells administration, which could be abrogated by the reconstitution of FLAG-Snail1 (Figure 4d,e). Furthermore, concerning the important roles of Snail1 in the resistance to BRAF inhibitor in melanoma, we investigate the the cytotoxicity of melanoma cells with OTUD4 depletion in response to the B-Raf V600E-Specific Inhibitors. Vemurafenib and PLX-4720 were employed in the CCK assay and the results demonstrated that depletion of OTUD4 significantly sensitize both of the A375 and A2058 cells to vemurafenib (Figure 4f,g) and PLX-4720 (Figure 4h,i) which could be markedly reversed by reconstituting of FLAG-Snail1, implying the important pharmacological potential of targeting OTUD4-Snail1 axis to overcome the chemoresistance of melanoma therapy. Overall, our findings highlight an important role of OTUD4 in regulating EMT, metastasis and chemosensitivity through stabilizing Snail1 in melanoma (Figure 5).
4 DISCUSSION AND CONCLUSION
Here, we reveal several unexpected findings with important clinical implications. We demonstrate the previously uncharacterized tumor-promoting activity of the deubiquitinase OTUD4 through stabilizing Snail1 in melanoma harboring BRAF mutants. We further provide preclinical evidence demonstrating that targeting OTUD4 is an effective approach to suppress metastasis of melanoma (Figure 4).
Melanoma develops from transformed melanocytes as a result of several driver genetic alterations such as BRAF, RAS and NF1 mutations, which are caused by UV radiation and other risk factors. Despite the successful development of targeted therapies and immunotherapies for unresectable melanoma, the response rate of immunotherapies remains low and patients with advanced melanoma harboring BRAF-mutation will frequently experience a disease relapse and progression after several months of treatments targeting BRAF-mutations. Therefore, understanding the underlying mechanisms of metastasis and chemoresistance will benefit the poor outcomes of metastatic melanoma patients.
EMT has been demonstrated to enhance the migration and invasion ability of different types of tumor cells, thereby facilitating the invasion into extracellular matrix tissues and eventually metastasis to target organs (Huang et al., 2022). Loss of E-cadherin, one important cell adhesion molecule, is a functional event of EMT that contributes to the destabilization of adherent junctions. One of the early events in the development of melanoma is the loss of escargot dependent transcriptional regulation of e-cadherin. Loss of e-cadherin is accompanied by malignant transformation of melanoma cells (Kuphal et al., 2005). In addition, downregulating E-cadherin prevents CD103-mediated immune attack and may contribute to melanoma resistance (Shields et al., 2019). Among the critical EMT-TFs, the aberrant expression of Snail1 was implicated malignant transformation of melanocytes,melanoma metastasis,immunosuppression, as well as resistance to BRAF inhibitors (Hao et al., 2012; Kudo-Saito et al., 2009; Lei et al., 2018; Liu et al., 2011; Maiques et al., 2018; Shields et al., 2019). Our study and others evidence validates the therapeutic benefits of depleting Snail1 in melanoma (Supporting Information: Figure 1) Considering the well-established tumor-promoting function of Snail1 and the challenging to directly targeting Snail1, to target Snail1 stability might be an appealing strategy in the intervention of melanoma.
OTUD4 acts as a member of the OTU subfamily through its enzymatic deubiquitination activity. Human OTUD4 carries only serine, which means that OTUD4 is active through a catalytic triad (Ser-His-Asp) composed of serine rather than cysteine and histidine and aspartate (Louis et al., 2015). It has been associated with embryonic development in zebrafish and with human ataxia, dementia, and hypogonadotropin syndrome (Margolin et al., 2013; Tse et al., 2013). As a deubiquitination enzyme with a preference for K48-linked polyubiquitin chains, OTUD4 stabilizes the cellular antiviral response adapter protein MAVS and promotes innate antiviral signaling (Liuyu et al., 2019). In addition, OTUD4 regulates TGF beta signaling by deactivating SMURF2, a negative regulator of TGF beta, through a catalytic dependent and independent mechanism (Jaynes et al., 2020). In nasopharyngeal carcinoma (NPC), it deubiquitinated and stabilized gasdermin E (GSDME), promoting GSDME-dependent pyroptosis to enhance the radiosensitivity of NPC (Di et al., 2022). In addition, OTUD4 could regulate AlkB protein stability by serving as a scaffold for USP7 and USP9X and contribute to alkylation damage resistance in prostate cancer and other tumors (Zhao et al., 2015). Thus, OTUD4 is implicated in regulating cell proliferation, differentiation, migration and other cellular processes (Massagué, 2008; Seoane, 2006; Siegel & Massagué, 2003)\. However, the role of OTUD4 in melanoma remains elusive. Our study revealed the previously unrecognized function of OTUD4 in stabilizing Snail1 and demonstrate its clinical relevance as a therapeutic target in melanoma. The evidence we observed are as follows. First, we identify OTUD4 as a bona fide deubiquitinase of Snail1 through tandem affinity purification and mass spectrometry analysis. OTUD4 directly interacts with Snail1, decreases its ubiquitination level through the deubiquitinating enzyme activity, thereby leading to the stabilization of Snail1 (Figures 1-3, Supporting Information: Figure 2). Next, the depletion of OTUD4 markedly attenuate Snail1-driven EMT and metastasis both in vitro and in vivo animal models (Figure 4, Supporting Information: Figure 2). In addition, our results show that the reconstitution of Snail1 could significantly rescue such phenotypes caused by the depletion of OTUD4 (Figure 4, Supporting Information: Figure 2), implying that the tumor-promoting activity of OTUD4 in melanoma largely depends on its ability to stabilize Snail1. Although our study is mainly focused in BRAF-mutant melanoma, further investigation need warranted especially in melanoma with NRAS or NF mutants, which have even worse prognosis and no targeted therapies.
OTUD4 itself could be regulated at the transcriptional level. For instance, OTUD4 is downregulated by the ZIKV infection induced miR-103a-3p to stimulate replication (Liuyu et al., 2019). In addition, viral infection was reported to promote its transcriptional activation by IRF3/7-binding on the promoter of Otud4 (Ye et al., 2022). High level transcription of OTUD4 was significantly observed in tsc22d3-overexpressing embryos in Bmp-dependent dorsoventral patterning (Tse et al., 2013). Our finding highlighted the potential of OTUD4 as a more druggable target toward Snail1 stability in melanoma. Further efforts are warranted to investigate regulatory mechanisms to regulate the expression and activity of OTUD4 toward Snail1 in melanoma.
In conclusion, our study identifies the previously unknown oncogenic role of OTUD4 in stabilizing Snail1 in melanoma. Importantly, we provide preclinical evidence demonstrating that targeting OTUD4 might be an effective therapeutic strategy to destabilize Snail1 and suppress Snail1-dependent malignant activity in melanoma.
AUTHOR CONTRIBUTIONS
Yuchen Gao and Jiaxin Tang contributed equally to this manuscript. Haoxing Zhang and Tongzheng Liu conceived the study and designed the experiments. Yuchen Gao, Jiaxin Tang, Xiuqing Ma, Caishi Zhang, and Lei Huang performed most of the experiments. Yuchen Gao, Jiaxin Tang, Xiuqing Ma, Jingjing Che, and Yinci Zhang analyzed data and wrote the manuscript assisted by Caishi Zhang, Lei Huang, Yalei Wen, Yinjie Zhu, Tongzheng Liu, and Haoxing Zhang. All authors discussed the results and commented on the manuscript.
ACKNOWLEDGMENTS
This study was supported by National Natural Science Foundation of China (81773758, 81603133, and 82273127, 81772820), the project of State Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medicinal University (QJJ[2022] 420), Guangdong Basic and Applied Basic Research Foundation (2019A1515011934, 2021A1515011233, 2022A1515012371, 2020A1515010213, and 2022A1515011769), Guangzhou Basic and Applied Basic Rearch Foundation (2023A04J0645), the Fundamental Research Funds for the Central Universities (21622102), Medical Joint Fund of Jinan University (YXJC2022006), Shenzhen Science and Technology Innovation Commission Grants (JCYJ20200109114225087 and 20200812015744001).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.




