Proinflammatory macrophages promote degenerative phenotypes in rat nucleus pulpous cells partly through ERK and JNK signaling
Abstract
Intervertebral disc (IVD) degeneration is the major contributor to low back pain, a highly prevalent musculoskeletal problem that represents the leading cause of disability. Proinflammatory M1 macrophages were identified in degenerated IVDs. However, their role in the pathogenesis of IVD degeneration and the underlying mechanism was largely unknown. In this study, we explored the combined effects of molecules secreted by M1 macrophages on nucleus pulposus cells, by treating rat nucleus pulposus cells (rNP) with the conditioned medium collected from M1-polarized RAW264.7 cells (MФCM). We found that MФCM caused molecular changes associated with IVD degeneration, including increased expression of key matrix catabolic genes (Adamts4, Adamts5, Mmp3, and Mmp13), reduced the expression of major matrix-associated anabolic genes ( Sox9, Acan, and Col2a1), and upregulated transcription of inflammation-related genes ( IL-1b, IL-6, Ccl2, and Ccl3), in rNP cells. Moreover, we found that MФCM activated both ERK and JNK pathways in these cells, and that inhibition of JNK pathway attenuated MФCM-induced expression of both catabolic and inflammatory genes, whereas ERK inhibition only suppressed induction of catabolic, but not inflammatory genes. Together, our data demonstrated that proinflammatory macrophages promoted the degenerative phenotypes in rNP cells in part through ERK and JNK signaling, and suggested that inhibition of these pathways may serve as a potential therapeutic approach for the treatment of IVD degeneration.
1 INTRODUCTION
Low back pain, a highly prevalent musculoskeletal problem, represents one of the leading causes of musculoskeletal disability worldwide (Hartvigsen et al., 2018; Huang, Urban, & Luk, 2014; Risbud & Shapiro, 2014). Among its many contributors, degeneration of intervertebral disc (IVD) is widely accepted as the major one (Risbud & Shapiro, 2014). One early event of IVD degeneration is the upregulation of proinflammatory cytokines in the disc cells (Le Maitre, Freemont, & Hoyland, 2005; Phillips et al., 2015; Risbud & Shapiro, 2014; Wang et al., 2013), which in turn disrupts disc matrix homeostasis by suppressing expression of extracellular matrix (ECM)-related genes, and stimulating expression of MMPs (matrix metalloproteinases) and ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs), the key matrix-degrading proteases involved in IVD degeneration (Bachmeier et al., 2009; Pockert et al., 2009). Moreover, these cytokines upregulate chemokines in the disc cells (Le Maitre et al., 2005; Phillips et al., 2015; Risbud & Shapiro, 2014; Wang et al., 2013). These cellular and molecular changes are followed by the infiltration of inflammatory cells into the IVD (Nakazawa et al., 2017; Risbud & Shapiro, 2014). These inflammatory cells not only directly influence matrix catabolism and anabolism, but also indirectly affect matrix homeostasis by inducing expression of inflammation-related genes, such as chemokines and proinflammatory cytokines, in the disc cells (Risbud & Shapiro, 2014), therefore leading to exacerbation of the inflammatory and degenerative status of discs. Thus, blocking the action of inflammatory cells may be considered as a therapeutic approach to halt the progression of this degenerative disease. Clearly, a better understanding of the identities and functions of these inflammatory cells will facilitate the efforts to develop such therapies.
Macrophages have been recently shown as one type of inflammatory cells that could potentially play a pathological role in IVD degeneration (Kawaguchi et al., 2001; Koike, Uzuki, Kokubun, & Sawai, 2003; Nakazawa et al., 2017; Nerlich, Weiler, Zipperer, Narozny, & Boos, 2002; Peng et al., 2006; Shamji et al., 2010). In fact, inflammatory cells expressing the pan-macrophage marker CD68 were readily identified in the degenerated IVDs, particularly within the nucleus pulposus (NP), but not in the healthy IVDs (Nerlich et al., 2002; Shamji et al., 2010). Characterization of macrophage surface markers further revealed that three different macrophage phenotypes, including proinflammatory M1, remodeling M2c, and anti-inflammatory M2a phenotypes, appeared in the degenerative IVDs, but not in the normal IVDs (Nakazawa et al., 2017). But surprisingly, only M1 cells were positively correlated with degenerative grades in NP tissues, suggesting that proinflammatory M1 macrophage is the major type of macrophages involved in the degeneration of NP tissues.
M1 macrophages function as important inflammatory regulators in many tissues by secreting a variety of cytokines, many of which are implicated in the pathogenesis of IVD degeneration (Risbud & Shapiro, 2014). In recent years, progresses have been made in revealing the pathological roles of individual cytokines secreted by M1 macrophages, particularly TNF-α, IL-1β, and IL-6, in degeneration of IVDs (Le Maitre et al., 2005; Le Maitre, Hoyland, & Freemont, 2007; Phillips et al., 2015; Risbud & Shapiro, 2014; Seguin, Pilliar, Roughley, & Kandel, 2005). However, the combined effects of all cytokines secreted by M1 cells on IVDs, and the common downstream pathways that mediate their pathological effects are still poorly understood.
In this study, we explored the integrated effects of molecules secreted by M1 macrophages on NP cells, by using the conditioned medium collected from M1-polarized RAW264.7 cells (MФCM) and a rat nucleus pulposus cell line (rNP). Our data showed that MФCM inhibited expression of matrix anabolic genes, but increased expression of inflammation-related genes as well as matrix catabolic genes, in rNP cells.
2 MATERIALS AND METHODS
2.1 Cell culture and treatments
Immortalized rNP cells were established by Prof. Di Chen (Rush University; Oh et al., 2016), and grown in Dulbecco's modified Eagle medium (DMEM)/F-12(1:1) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Thermo Fisher Scientific, Rockford, IL). The murine macrophage cell line RAW264.7 was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin. All cells were maintained at 37°C in a humidified 5% CO2 incubator, and the culture medium was changed every 2–3 days unless otherwise indicated.
The conditioned medium of proinflammatory M1 macrophages was prepared as previously described (Zheng et al., 2018). Briefly, RAW264.7 cells (4 × 106) were seeded in 10 cm culture dishes and cultured overnight in normal growth medium (DMEM, 10% FBS, and 1% penicillin/streptomycin). RAW264.7 cells were then treated with 100 ng/ml lipopolysaccharide (LPS) for 24 hr. After that, LPS-containing medium was removed, and RAW264.7 cells were cultured in normal growth medium for an additional 24 hr. Subsequently, the culture medium was collected and centrifuged briefly to remove cell debris. The remaining supernatant was considered as 100% conditioned medium of proinflammatory macrophages (MФCM). The LPS-treated and untreated RAW264.7 cells were lysed with Trizol reagent and subjected to total RNA isolation and quantitative polymerase chain reaction (qPCR) analyses.
To test the effects of proinflammatory macrophages on NP cells, rNP cells were cultured in growth medium with or without 30% MФCM. At 24 hr after treatments, cells were harvested for RNA or protein analyses. For inhibitor experiments, rNP cells were cultured in growth medium with or without 30% MФCM in the presence of indicated inhibitors or their respective vehicles. At 24 hr after treatments, cells were harvested for RNA or protein analyses. MEK inhibitor U0126 and JNK inhibitor SP600125 were both from Sigma (Sigma, St. Louis, Missouri), and used at a final concentration of 10 μM.
2.2 Total RNA isolation and quantitative RT-PCR (qPCR)
qPCR analyses were performed as we previously reported (Gu et al., 2018). Briefly, total RNA was isolated using the Trizol reagent (Invitrogen, Carlsbad, California) following the standard protocol. Subsequently, 1 μg total RNA was reverse-transcribed to complementary DNA (cDNA) with the iScript cDNA synthesis kit (Bio-Rad, Hercules, California) according to the kit's instruction. The cDNA was then subjected to qPCR amplification using universal SYBR green supermix (Bio-Rad, Hercules, CA) in the presence of both sense and antisense primers specific to indicated genes. Relative expression of each gene was determined by the 2−ΔΔCt method. Expression of 18S rRNA was used as an internal loading control. Sequences for qPCR primers used in this study were listed in Table 1.
| Gene | Species | Forward primer 5′–3′ | Reverse primer 5′–3′ |
|---|---|---|---|
| 18s rRNA | Rat | GACTCTCGGCATGCTAACTAG | GGACATCTAAGGGCATCACAG |
| Acan | Rat | AGGATGGCTTCCACCAGTGC | TGCGTAAAAGACCTCACCCTCC |
| Col2a1 | Rat | CCTGGACCCCGTGGCAGAGA | CAGCCATCTGGGCTGCAAAG |
| Sox9 | Rat | TAAATTCCCAGTGTGCATCC | GCACCAGGGTCCAGTCATA |
| Adamts4 | Rat | CTTCGCTGAGTAGATTCGTGG | AGTTGACAGGGTTTCGGATG |
| Adamts5 | Rat | GGGTTATACTGACGTTGTGAGG | TCTAGTCTGGTCTTTGGCTTTG |
| Mmp3 | Rat | TCTTCCTCTGAAACTTGGCG | AGTGCTTCTGAATGTCCTTCG |
| Mmp13 | Rat | GCAGCTCCAAAGGCTACAA | CATCATCTGGGAGCATGAAA |
| IL-1b | Rat | TGCAGGCTTCGAGATGAAC | GGGATTTTGTCGTTGCTTGTC |
| IL-6 | Rat | AAGCCAGAGTCATTCAGAGC | GTCCTTAGCCACTCCTTCTG |
| Ccl2 | Rat | GGTCTCTGTCACGCTTCTG | TTCTCCAGCCGACTCATTG |
| Ccl3 | Rat | CACCCTCTGTTACCTGCTC | ATTTGCCGTCCATAGGAGAAG |
| Tnfa | Mouse | CAAGGAGGAGAAGTTCCCAA | CTCTGCTTGGTGGTTTGCTA |
| iNOS | Mouse | GTTCTCAGCCCAACAATACAAGA | GTGGACGGGTCGATGTCAC |
| IL-6 | Mouse | ATAGTCCTTCCTACCCCAATTTCC | GATGAATTGGATGGTCTTGGTCC |
| IL-1b | Mouse | GCAACTGTTCCTGAACTCAACT | ATCTTTTGGGGTCCGTCAACT |
| Gapdh | Mouse | GCACAGTCAAGGCCGAGAAT | GCCTTCTCCATGGTGGTGAA |
- Note. qPCR: quantitative polymerase chain reaction.
2.3 Total protein isolation and western blot
Western blot analyses were performed as previously described (Gu et al., 2018). Briefly, total proteins were extracted from cells with 1 × radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors (Roche, Mannheim, Germany). Concentrations of total proteins were determined with the Pierce Bicinchoninic Acid (BCA) Protein Assay kit (Thermo Fisher Scientific, Rockford, Illinois) following its instruction. 30 μg protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene fluoride (PVDF) membranes (Perkin Elmer, Boston, MA). After being blocked with 1 × Tris-buffered saline and Tween 20 (TBST) containing 5% blotting-grade blocker nonfat dry milk (Bio-Rad, Hercules, CA), the membranes were incubated overnight with appropriate primary antibodies at 4°C, followed by incubation with the horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling Technology, Beverly, MA, 1:2,000). Subsequently, protein bands on the blots were visualized using the Clarity Western ECL Kit (Bio-Rad, Hercules, CA), and imaged with the ChemiDoc Touch Imaging System (Bio-Rad).
Primary antibodies against phospho-JNK (4668), total-JNK (9252), phospho-ERK (4370), total-ERK (4695), Phospho-c-Jun (2361), and β-actin (4970) were purchased from Cell Signaling Technology (Beverly, Massachusetts), and were all used at 1:1,000 dilution. Primary antibodies against Collagen II (ab34712, 1:5,000), MMP3 (ab52915, 1:1,000), MMP13 (ab39012, 1:3,000), Sox9 (ab5535, 1:2,000), and phospho-p38 (ab4822, 1:1,000) were from Abcam (Cambridge, Massachusetts).
2.4 Statistical analysis
GraphPad Prism 5 software was used for statistical analysis. All quantitative results were shown as the mean ± standard deviation (S.D.). Student’s t test was used to determine the statistical difference between two groups. Any difference with a p-value < 0.05 was considered as statistically significant. All quantitative results were calculated from a minimum of three biological replicates.
3 RESULTS
3.1 Proinflammatory macrophages suppressed the expression of major matrix anabolic genes in rNP cells
To determine the combined effects of molecules secreted by proinflammatory macrophages on NP cells, we first treated RAW264.7 cells (a murine macrophage cell line) with 100 ng/ml LPS, a bacterial product that was shown to induce M1 polarization (Qin et al., 2012). The qPCR analysis showed that messenger RNA (mRNA) levels of M1-associated markers, including TNF-α, IL-1β, IL-6, and iNOS, were all remarkably upregulated in LPS-treated cells compared with those of untreated cells (Supporting Information Supplemental Figure 1), confirming that LPS treatment induced RAW264.7 into proinflammatory M1 cells.
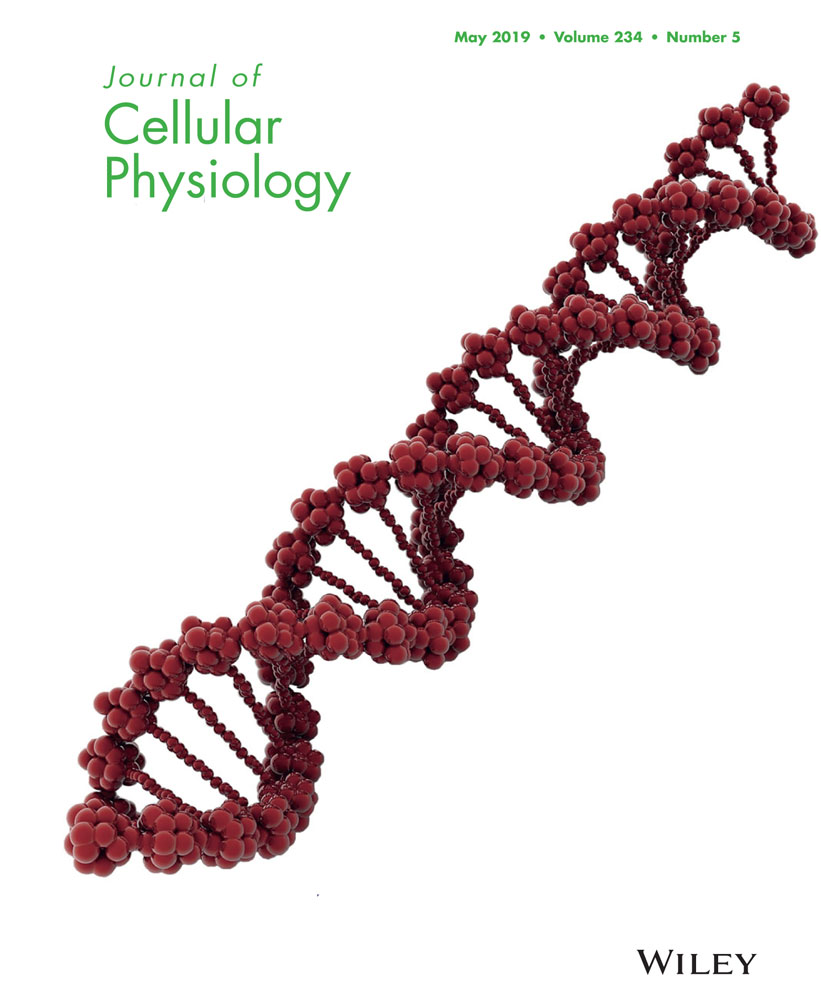
We then collected the conditioned medium from LPS-treated RAW264.7 cells (MФCM), and examined its effect on cell morphology and expression of matrix anabolic genes in rNP cells (a rat nucleus pulpous cell line). The rNP cells showed the small square-polygonal shape (Figure 1a), but became elongated and exhibited fibroblast-like morphology upon the MФCM treatment (Figure 1b). Collagen II and aggrecan are two major components of extracellular matrix synthesized by NP cells (Zheng et al., 2018). Interestingly, mRNA levels of Col2a1 (encoding collagen II) and Acan (encoding aggrecan) were significantly decreased (by 67% and 17%, respectively) in MФCM-treated rNP cells compared with those of untreated cells (Figure 1c,d). Consistently, the mRNA level of Sox9, an upstream regulator of Col2a1 and Acan genes, was also significantly reduced by 68% upon the MФCM treatment (Figure 1e). In keeping with these qPCR results, western blot analyses showed that MФCM treatment suppressed protein levels of Sox9 and Col2a1 by 77% and 78% (p < 0.05), respectively, in rNP cells (Figure 1f,g). Taken together, these data indicated that proinflammatory macrophages inhibited the expression of major matrix anabolic genes in rNP cells.
Proinflammatory macrophages suppressed the expression of major matrix anabolic genes in rNP cells. (a,b) Representative microscopy images of rNP cells treated without (a) or with 30% MФCM (b) for 24 hr. Scale bar: 100 μm. (c–e) qPCR analyses were performed to compare relative mRNA levels of Acan (c), Col2a1 (d), and Sox9 (e) genes, in rNP cells treated with or without 30% MФCM for 24 hr. Relative gene expression was determined by the 2−ΔΔCt method after normalizing first to 18 s rRNA, and then to the control group (untreated cells). Data were shown as means ± SD. *p < 0.05, n = 3. (f) Western blot analyses were performed to compare relative protein levels of Col2a1 and Sox9 in rNP cells treated with or without 30% MФCM for 24 hr. Blots shown were representative results of three independent experiments. (g) Quantification of (f). Data were shown as means ± SD. *p < 0.05, n = 3. mRNA: messenger RNA; qPCR: quantitative polymerase chain reaction; rNP: rat nucleus pulposus; rRNA: ribosomal RNA
3.2 Proinflammatory macrophages induced expression of key matrix-degrading enzymes in rNP cells
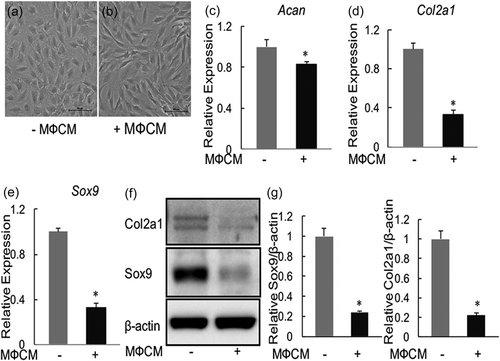
We next examined the effect of MФCM on the expression of Mmps (Mmp3 and Mmp13) and Adamts (Adamts4 and Adamts5). As shown in Figure 2, the MФCM treatment induced mRNA levels of Mmp3 and Mmp13 by 2,990-fold and 1,465-fold, respectively, in rNP cells. Similarly, MФCM significantly increased, though to a much lesser degree transcriptional levels of Adamts4 and Adamts5 genes (p < 0.05; Figure 2a,b). Consistent with these qPCR results, western blot analyses revealed that protein levels of Mmp3 and Mmp13 were significantly induced by the MФCM treatment (by 18-fold and 53-fold, respectively; Figure 2e,f). Collectively, our data demonstrated that proinflammatory macrophages promoted expression of key matrix-degrading enzymes, particularly Mmp3 and Mmp13, in rNP cells.
Proinflammatory macrophages induced the expression of key matrix-degrading enzymes in rNP cells. (a–d) qPCR analyses of Adamts4, Adamts5, Mmp3, and Mmp13, in rNP cells treated with or without 30% MФCM for 24 hr. Relative gene expression was calculated by the 2−ΔΔCt method after normalizing first to 18r rRNA, and then to the untreated control group. Data are shown as means ± SD. *p < 0.05, n = 3. (e) Western blot analyses of Mmp3 and Mmp13, in rNP cells treated with or without 30% MФCM for 24 hr. Blots shown were representative results of three independent experiments. (f) Quantification of (e). Data were shown as means ± SD. *p < 0.05, n = 3. qPCR: quantitative polymerase chain reaction; rNP: rat nucleus pulposus; rRNA: ribosomal RNA
3.3 Proinflammatory macrophages stimulated expression of inflammation-related genes in rNP cells
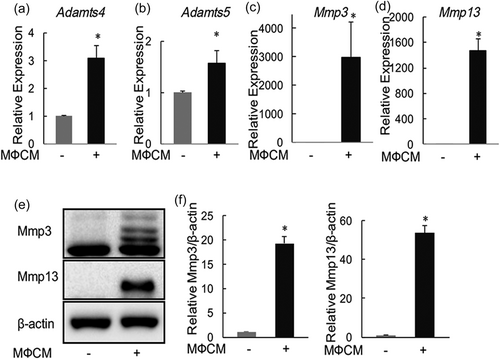
We next evaluated the effects of MФCM on the expression of proinflammatory cytokines and chemokines in rNP cells. Indeed, MФCM treatment induced mRNA levels of IL-1b and IL-6 (by 5-fold and 155-fold, respectively; Figure 3a,b) two important proinflammatory cytokines implicated in the pathogenesis of IVD degeneration, in rNP cells (Le Maitre et al., 2005, 2007; Risbud & Shapiro, 2014). Likewise, the MФCM treatment stimulated the transcription of Ccl2 and Ccl3 (by 1,790-fold and 18-fold, respectively; Figure 3c,d), two chemokines that played important roles in the recruitment of inflammatory cells into degenerating IVDs (Ohba et al., 2008; Wang et al., 2013). Thus, our data demonstrated that proinflammatory macrophages stimulated the expression of inflammation-related genes, including cytokines (IL-6 and IL-1b) and chemokines (Ccl2 and Ccl3), in rNP cells.
Proinflammatory macrophages stimulated mRNA expression of proinflammatory cytokines and chemokines in rNP cells. qPCR analyses were performed to compare relative mRNA levels of IL-1b (a), IL-6 (b), Ccl2 (c), and Ccl3 (d) in rNP cells treated with or without 30% MФCM for 24 hr. Relative gene expression was calculated by the 2−ΔΔCt method after normalizing first to 18 s rRNA, and then to the control group. Data were shown as means ± SD. *p < 0.05, n = 3. mRNA: messenger RNA; qPCR: quantitative polymerase chain reaction; rNP: rat nucleus pulposus; rRNA: ribosomal RNA
3.4 Proinflammatory macrophages activated both ERK and JNK MAPK pathways in rNP cells
Having established that proinflammatory macrophages promote degenerative phenotypes in rNP cells, we next sought to investigate the underlying mechanism. Mitogen-activated protein kinase (MAPK) pathways, including ERK, JNK, and p38 signaling cascades, are important mediators of cytokines, such as TNF-α and IL-1β, in promoting IVD degeneration (Risbud & Shapiro, 2014). To test whether proinflammatory macrophages activated these pathways in rNP cells, we examined the effects of MФCM on phosphorylation of p38, ERK, and JNK proteins, the commonly used readouts for activation of MAPK pathways, in rNP cells. Western blot analysis showed that the MФCM treatment did not affect the level of phosphorylated p38 (phospho-p38; p > 0.05; Figure 4), indicating that MФCM had no major effect on the activity of p38 pathway in rNP cells. By contrast, the levels of phosphorylated ERK (phospho-ERK) and phosphorylated JNK (phospho-JNK) were increased by 1.2-fold and 2.9-fold, respectively, after 24 hr of MФCM treatments (Figure 4), indicating that MФCM activated both ERK and JNK pathways in rNP cells. Taken together the results, our data demonstrated that proinflammatory macrophages activated both ERK and JNK MAPK pathways in rNP cells.
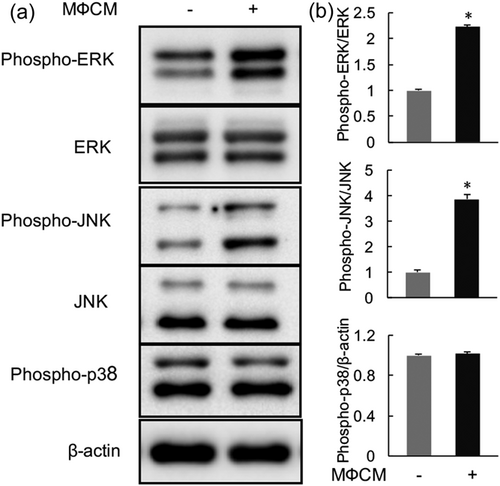
Proinflammatory macrophages activated both ERK and JNK pathways in rNP cells. (a) rNP cells were treated with or without 30% MФCM for 24 hr, and then subjected to western blot analysis analyses with specific antibodies against the indicated proteins. Blots shown were representative results of three independent experiments. (b) Quantification of (a). Data were shown as means ± SD. *p < 0.05, n = 3. rNP: rat nucleus pulposus
3.5 Pharmacological inhibition of ERK pathway alleviated the catabolic and antianabolic effects of proinflammatory macrophages on rNP cells
To determine whether increased activity of ERK pathway contributed to the degenerative phenotypes in rNP cells caused by the MФCM treatment, we first evaluated the effects of the MEK inhibitor U0126 on mRNA expression of anabolic (Sox9 and Col2a1,), catabolic (Mmp3 and Mmp13), and inflammatory (IL-6, IL-1b, Ccl2, and Ccl3) genes. qPCR analyses showed that U0126 relieved the inhibitory effect of MФCM on Col2a1 mRNA expression (p < 0.05; Figure 5a), but did not affect its ability to suppress mRNA expression of Sox9 gene (p > 0.05; Figure 5b). Moreover, U0126 significantly reduced MФCM-induced mRNA expression of catabolic Mmp3 and Mmp13 genes (Figure 5c,d). By contrast, U0126 had no effect on mRNA expression of all inflammatory genes examined (p > 0.05; Supporting Information Supplemental Figure 2). To further confirm the effects of U0126 on the expression of anabolic and catabolic genes at the protein level, we performed western blot analyses. As expected, U0126 treatment reduced both basal and MФCM-induced phosphorylation of ERK, indicating that U0126 potently inhibited the activation of ERK signaling (Figure 5e). Consistent with its effects on mRNA expression, U0126 partially restored the protein level of Col2a1 suppressed by the MФCM treatment, but did not rescue Sox9 protein expression (Figure 5e). Moreover, U0126 significantly reduced MФCM-induced expression of Mmp3 and Mmp13 proteins (Figure 5e). Collectively, these data indicated that ERK pathway at least partially contributed to catabolic ad antianabolic effects of proinflammatory macrophages on rNP cells.
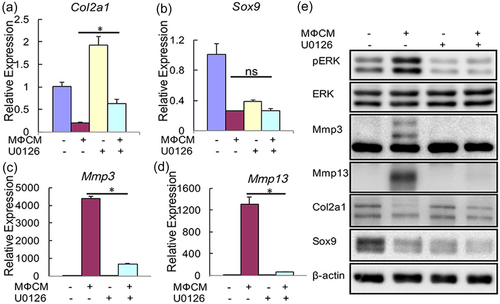
Pharmacological inhibition of ERK pathway alleviated the catabolic and antianabolic effects of proinflammatory macrophages on rNP cells. (a–d) qPCR analyses of Col2a1, Sox9, Mmp3, and Mmp13, in rNP cells that were treated with or without 30% MФCM in the presence or absence of 10 μM U0126 for 24 hr. Relative gene expression was calculated by the 2−ΔΔCt method after normalizing first to 18 s rRNA, and then to the control groups. Data were shown as means ± SD; ns: not significant, p > 0.05; *p < 0.05; n = 3. (e) Western blot analyses of rNP cells treated as described above with specific antibodies against the indicated proteins. Blots shown were representative results of three independent experiments. qPCR: quantitative polymerase chain reaction; rNP: rat nucleus pulposus; rRNA: ribosomal RNA [Color figure can be viewed at wileyonlinelibrary.com]
3.6 Pharmacological inhibition of JNK pathway attenuated MФCM-induced expression of catabolic and inflammatory genes in rNP cells
We next investigated the contribution of JNK pathway to MФCM-induced degenerative phenotypes in rNP cells. To this end, we first evaluated the effects of the JNK inhibitor SP600125 on mRNA expression of anabolic (Col2a1 and Sox9), catabolic (Mmp3 and Mmp13), and inflammatory (IL-1b, IL-6, Ccl2, and Ccl3) genes. qPCR analysis revealed that SP600125 did not notably affect MФCM-mediated suppression of Col2a1 and Sox9 mRNA expression (p > 0.05), but substantially reduced the MФCM-induced mRNA expression of Mmp3 and Mmp13 genes (p < 0.05; Figure 6a–d). Moreover, SP600125 significantly reduced the MФCM-mediated increase in mRNA expression of inflammation-related genes, including IL-1b, IL-6, Ccl2, and Ccl3 (p < 0.05; Figure 7). To further confirm the effects of SP600125 on the expression of anabolic and catabolic genes at the protein level, we then performed western blot analyses. As expected, SP600125 treatment blocked both basal and MФCM-induced phosphorylation of c-jun (a downstream target of JNK MAPK; Figure 6e), confirming that SP600125 potently inhibited the activity of JNK pathway. Consistent with qPCR results, SP600125 dramatically inhibited MФCM-induced expression of Mmp3 and Mmp13 proteins, but did not rescue MФCM-mediated suppression of Col2a1 and Sox9 proteins (Figure 6e). Taken together, our data indicated that JNK pathway at least partially contributed to catabolic and inflammatory effects of proinflammatory macrophages on rNP cells.
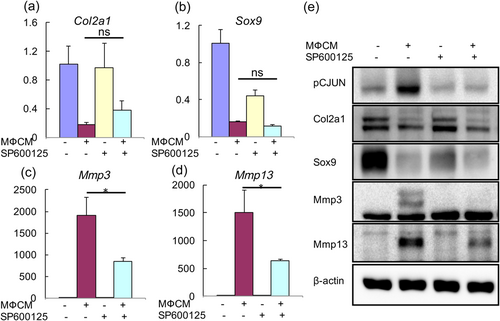
Pharmacological inhibition of JNK pathway attenuated MФCM-induced expression of catabolic genes in rNP cells. (a–d) qPCR analyses of Col2a1, Sox9, Mmp3, and Mmp13 in rNP cells that were treated with or without 30% MФCM in the presence or absence of 10 μM SP600125 for 24 hr. Relative gene expression was calculated by the 2−ΔΔCt method after normalizing first to 18 s rRNA, and then to the control group. Data were shown as means ± SD; ns: not significant, p > 0.05; *p < 0.05; n = 3. (e) Western blot analyses of rNP cells treated as described above with specific antibodies against the indicated proteins. Blots shown were representative results of three independent experiments. qPCR: quantitative polymerase chain reaction; rNP: rat nucleus pulposus; rRNA: ribosomal RNA [Color figure can be viewed at wileyonlinelibrary.com]
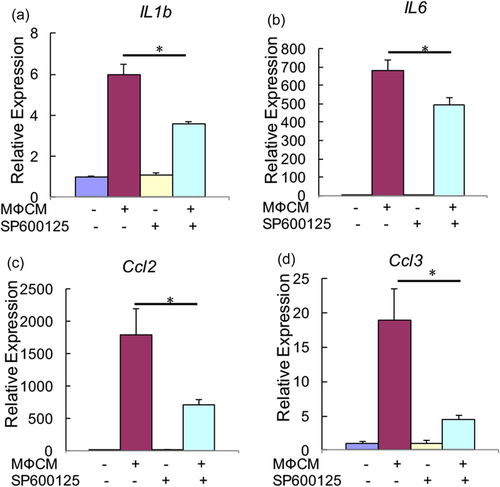
Pharmacological inhibition of JNK pathway attenuated MФCM-induced expression of inflammatory genes in rNP cells. qPCR analyses were performed to compare relative mRNA levels of IL-1b, IL-6, Ccl2, and Ccl3, in rNP cells treated with or without 30% MФCM in the presence or absence of 10 μM SP600125 for 24 hr. Relative gene expression was determined by the 2−ΔΔCt method after normalizing first to 18 s rRNA, and then to the control group. Data were shown as means ± SD. *p < 0.05, n = 3. mRNA: messenger RNA; qPCR: quantitative polymerase chain reaction; rNP: rat nucleus pulposus; rRNA: ribosomal RNA [Color figure can be viewed at wileyonlinelibrary.com]
4 DISCUSSION
Proinflammatory macrophages were recently identified as one type of inflammatory cells implicated in IVD degeneration. However, it is still unclear how they contribute to the pathogenesis of this degenerative disease. In this study, we demonstrated that proinflammatory macrophages promoted a full spectrum of molecular changes commonly associated with IVD degeneration, including increased expression of catabolic and inflammatory genes, coupled with the decreased expression of matrix anabolic genes. Moreover, we showed that these degenerative changes in rNP cells were partially attributed to increased activity of ERK and JNK signaling. Thus, our study identified ERK and JNK pathways as the potential therapeutic targets for the treatment of IVD degeneration.
Inflammatory cells, such as proinflammatory macrophages, secrete a variety of cytokines that act additively or synergistically to promote IVD degeneration (Risbud & Shapiro, 2014). It is conceivable that targeting a single cytokine will yield limited clinical effects, which was indeed shown by some human clinical studies (Risbud & Shapiro, 2014). A better therapeutic approach will be blocking actions of all or at least multiple cytokines on IVDs. Instead of focusing on a single cytokine, our study developed an in vitro cell culture model that mimicked the integrated actions of all molecules secreted by proinflammatory macrophages on NP cells. Thus, our cell culture model will likely serve as a useful tool for identifying additional novel targets for treating IVD degeneration.
Our data showed that MФCM did not affect the activity of p38 MAPK in rNP cells. By contrast, TNF-α or IL-1β was shown to activate p38 MAPK pathway (Studer et al., 2007). Moreover, blocking p38 MAPK activity modulated nucleus pulposus cell's response to TNF-α or IL-1β (Studer et al., 2007, 2008). The reason causing these discrepancies is still unclear. But it is possible that additional molecules secreted by proinflammatory macrophages neutralized the effects of TNF-α and IL-1β on p38 MAPK pathway. More studies are needed to test this possibility.
Our study has clearly shown both ERK or JNK pathways mediated the catabolic effect of proinflammatory macrophages on NP cells. It is still unclear whether they converged on a common downstream mechanism. If this is the case, inhibiting this common mechanism will be sufficient to block the catabolic effect of both ERK and JNK pathways. In addition to inducing catabolic enzymes, proinflammatory macrophages also suppressed the expression of anabolic genes. We found that inhibition of ERK pathway slightly reduced the inhibitory effect of MФCM on Col2a1, but not Sox9 gene expression, indicating that ERK only partially contributed to the antianabolic effect of MФCM, likely through a Sox9-independent mechanism. Thus, the major mechanism that underlies the antianabolic effect of proinflammatory macrophages is yet to be identified.
We found that inhibition of JNK pathway only modestly suppressed upregulation of inflammatory genes by M1 macrophages, suggesting that multiple mechanisms are involved in this regulation. In addition, activation of Notch signaling appeared to downregulate inflammatory genes in rNP cells (data not shown). It will be interesting to determine whether proinflammatory macrophages induced inflammatory gene expression by suppressing Notch signaling or its downstream effectors.
We should acknowledge that our current study has several limitations. First, we only examined the effects of MФCM on a rat NP cell line, but not on isolated human NP cells. Second, our findings were based solely on in vitro studies and were not confirmed in animal models or human samples. Third, we showed that either ERK or JNK inhibitor attenuated MФCM-induced degenerative phenotypes in rNP cells, but we did not test whether these two inhibitors had synergistic or additive effects. Clearly, future efforts are needed to address these limitations and test the potential of ERK and JNK inhibitors in treating IVD degeneration and low back pain. Moreover, more studies are warranted to elucidate the precise mechanism by which proinflammatory macrophages promote IVD degeneration, particularly the mechanism underlying their antianabolic and inflammatory effects.
ACKNOWLEDGEMENTS
This study was supported by grants from the National Key R&D Program of China (2016YFC1100203), and the National Natural Science Foundation of China (81772294 and 81572183).
CONFLICTS OF INTEREST
The authors confirm that there are no conflicts of interest.