Circular RNA in cardiovascular disease
Abstract
Circular RNA (circRNA) are endogenous transcripts that display differential expression across species, developmental stages, and pathologies. Their lack of free ends confers increased stability when compared with linear transcripts, making them ideal candidates for future diagnostic biomarkers and therapeutic interventions. Increasing evidence has implicated circRNA in the pathogenesis of multiple cardiovascular diseases. In this paper, we summarize current understanding of circRNA biogenesis, properties, expression profiles, detection methods, functions, and their implication in cardiac pathologies including/ischemia reperfusion injury, myocardial infarction, cardiac senescence, cardiac fibrosis, cardiomyopathy, cardiac hypertrophy and heart failure, atherosclerosis, coronary artery disease, and aneurysm.
Abbreviations
-
- circRNA
-
- Circular RNA
-
- miR
-
- microRNA
-
- QK1
-
- RBP quaking 1
-
- ciRNA
-
- Circular intronic RNA
-
- SMCS
-
- mooth muscle cells
-
- PES1
-
- Pescadillo homolog protein
-
- Col1a2
-
- Collage type 1 alpha 2
-
- CTGF
-
- Connective tissue growth factor
-
- MICRA
-
- Myocardial infarction-associated circular RNA
-
- ARC
-
- regulated cytoskeleton associated protein
-
- HRC
-
- Heart-related circRNA
-
- siRNA
-
- Small interference RNA
1 INTRODUCTION
Circular RNAs (circRNA) are single stranded RNAs that form a covalently closed loop without free terminals. Many circRNAs are endogenous, stable, abundant, and exhibit cell type, tissue, and developmental stage-specific expression patterns in eukaryotic cells (Fischer & Leung, 2017). Rapid advancements in biochemical methods and the usage of high-throughput sequencing technologies in recent years have enabled more widespread circRNA isolation and identification (Hansen, Veno, Damgaard, & Kjems, 2016). Increasing studies have shown that circRNAs are linked to physiological and pathological development in many organisms. The circRNA is emerging as an important regulatory element at the transcriptional and posttranscriptional levels by serving as microRNA (miRNA) sponges, by holding RNA binding proteins (RBPs) and by controlling alternative splicing and parental gene expression (Abdelmohsen et al., 2017; Ashwal-Fluss et al., 2014).
Cardiovascular disease remains one of the leading causes of mortality worldwide (Onwuanyi, Clarke, & Vanderbush, 2003). Recent studies have shown that circRNAs are involved in a number of various cardiovascular diseases (Wang et al., 2016a; Devaux et al., 2017; Fan et al., 2017; Li et al., 2018). The impact of circRNA on the cardiovascular system remains poorly characterized. A more comprehensive understanding of circRNA will lay the foundation for the development of circRNA-based diagnostic and therapeutic interventions for cardiovascular diseases. In this paper, we will be discussing the biogenesis of circRNA, their detection methods, the different functions of circRNA, and the current understanding of circular RNA in cardiac pathologies specifically.
2 DISCOVERY OF CircRNA
The term circRNA was first documented in 1976 by Sanger and colleagues when they characterized the structure of viroids – infectious single-stranded covalently closed RNA molecules (Sanger, Klotz, Riesner, Gross, & Kleinschmidt, 1976). In 1980, circRNA was found in the mitochondrial genome of the yeast Saccharomyces cerevisiae (Arnberg, Van Ommen, Grivell, Van Bruggen, & Borst, 1980) and in the early 1990s circRNAs were sporadically discovered in mammals (Capel et al., 1993; Cocquerelle, Daubersies, Majerus, Kerckaert, & Bailleul, 1992; Nigro et al., 1991; Zaphiropoulos, 1996). It was not until 2012 that circRNA abundance and ubiquity in eukaryotes were recognized due to high throughout RNA sequencing technology and bioinformatics.
More than 32,000 circRNAs are currently known (Chen et al., 2016; Xu, Wu, Han, Zhao, & Song, 2017; Zeng, Lin, Guo, & Zou, 2017; Zou et al., 2017). CircRNA are now recognized as a special type of noncoding RNA, although current studies are showing that a certain subset of circRNA may be translated in vivo (Pamudurti et al., 2017).
3 BIOGENESIS OF CircRNA
Splicing is the mechanism by which nascent precursor messenger RNA (pre-mRNA) is edited into mature mRNA through the removal of introns. The 99% of splicing events involve a characteristic GU at the 5′ end of the intron, and an AG at the 3′ end of the intron; these two sites are termed the donor and acceptor sites, respectively (Burset, Seledtsov, & Solovyev, 2000). An additional necessary site, the branching point, involves a conserved adenosine residue 20–50 nucleotides upstream of the acceptor site in the intron. A protein–RNA complex known as the spliceosome mediates canonical splicing. Due to alternative splicing, a single pre-mRNA transcript can generate multiple distinct mature mRNA via the retention of different combinations of exons in the final transcript. Alternative splicing is present in about 95% of all multiexon pre-mRNA transcripts (Pan, Shai, Lee, Frey, & Blencowe, 2008). Other splicing mechanisms also exist including cryptic exon formation, exonic introns (exitrons), and recursive splicing (Sibley, Blazquez, & Ule, 2016).
In canonical splicing, the spliceosomal machinery catalyses the nucleophilic attack of the 2′ OH of the adenosine at the branching point onto the 5′ phosphate of the donor site. This forms a closed loop known as the lariat. The second step involves the nucleophilic attack of the free 3′ OH of the exon onto the 5′ phosphate of the acceptor site in the intron. The end result is the joining of the two exons in the mRNA and the release of the lariat. It is important to note that in canonical splicing the relative order of the exons in the mRNA matches the order of the exons in the genome. No shuffling of the exons is observed in the mRNA after the canonical splicing (Jeck et al., 2013).
CircRNA is generated through a unique mechanism of splicing known as backsplicing (Jeck et al., 2013). In contrast to canonical splicing, backsplicing involves the disruption of exon order in sequencing reads due to the circularity of the covalently closed structure of the circRNA. For instance, first-strand synthesis using random hexamer primers which begins in the middle of a linear mRNA transcript would only display the first half of the exon in sequencing reads. Alternatively, first-strand cDNA synthesis that begins in the middle of a circRNA will show the entire sequence of the circRNA in a disrupted order as the polymerase will loop back to where it began (Figure 1). Although exon shuffling in circRNA is heavily relied upon in bioinformatics to help distinguish circRNA from other linear transcripts, it is not an exclusive characteristic to circRNA (Jeck & Sharpless, 2014). Several processes occurring during transcription or splicing may result in pseudo circular transcripts with exons shuffled out of the genomic order.
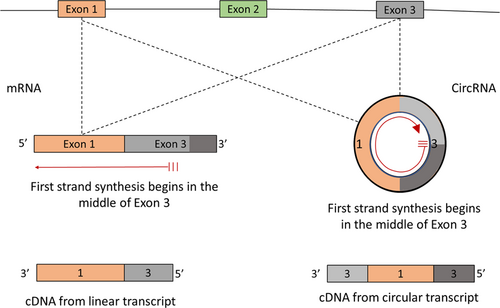
A hypothetical gene with two different products, one of which is linear and the other is circular. Using a specific primer to synthesize first strand cDNA from the middle of an exon, the circRNA will produce a different product from the linear transcript. Exon shuffling is observed in the cDNA from the circRNA, whereas the relative order of the exons is maintained in the cDNA from the linear transcript. cDNA, complementary DNA; circRNA, circular RNA [Color figure can be viewed at wileyonlinelibrary.com]
Two models of circRNA biogenesis through backsplicing have been proposed (Jeck et al., 2013): (a) The lariat precursor and (b) the direct backsplice models (or intron-pairing-derived circulation). In the lariat precursor or exon-skipping model, a minimum of two splicing events are required to generate a circRNA (Figure 2). The first transesterification reaction occurs between the donor site and the branching point of two distant introns separated by intronic and exonic sequences. A free 3′ OH is generated upstream of the donor site, which subsequently attacks the acceptor site, joining the two flanking exons together and releasing the exon-containing lariat with the 2′–5′ phosphodiester bond; this process is known as exon-skipping and is exhibited in the alternative splicing of many genes (Kelly, Greenman, Cook, & Papantonis, 2015). The second splicing event ligates two exons (or circularizes a single exon) in the released lariat intermediate in a mechanism analogous to that of the first splicing event. This process is driven by the restricted nature of the lariat as it aids in placing the acceptor and donor sites in close proximity to catalyze the splicing reaction. The result is the joining of any two exons to form a circRNA, with the release of an intronic double lariat by-product. Exon-skipping alone does not dictate circRNA formation as many other factors are involved, one of which is exon length which has been positively correlated with circularization (Barrett, Wang, & Salzman, 2015). Current evidence points to the involvement of the spliceosomal complex in the generation of circRNA, and this is consistent with the lariat-precursor model as it simply consists of two successive splicing events (Lasda & Parker, 2014, Starke et al., 2015).
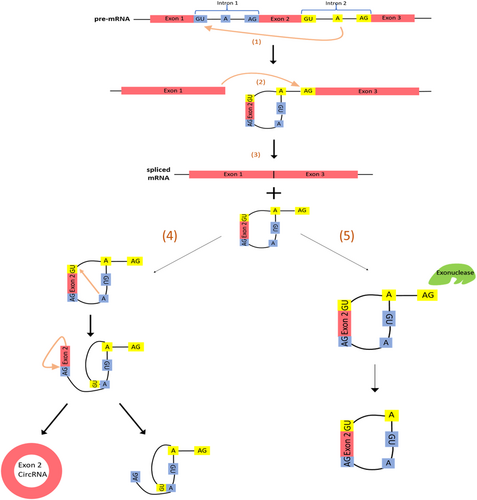
CircRNA and ElciRNA/ciRNA biogenesis. (a) Nucleophilic attack of the branching point onto the donor site of intron 1. (b) Attack of the free 3′OH of exon 1 onto the acceptor site of intron 2. (c) spliced mRNA product and a lariat intermediate with a 2′–5′ phosphodiester bond. (d) In the lariat-precursor model, a second splicing reaction occurs in the lariat intermediate analogous to that of the first splicing reaction. A double lariat and an exonic circRNA are the final products. (e) A riboexonuclease degrades the 3′ tail of the lariat, leaving behind an exonic/intronic circRNA termed ElciRNA. If exon-skipping was not exhibited (splicing reaction occurred between two adjacent exons) and the circRNA was completely intronic, it would be termed ciRNA. circRNA, circular RNA; mRNA, messenger RNA [Color figure can be viewed at wileyonlinelibrary.com]
In the direct backsplice model, flanking intronic sequence complementarity leads to the formation of secondary structures in the pre-mRNA transcript which primes the molecule for splicing via the appositional placing of two distant exons (Figure 3; Jeck et al., 2013). This effectively eliminates the need for a lariat precursor, and only one splicing event is required. A single transesterification reaction occurs between the branching point and the donor sites of the flanking introns to form a 2′–5′ phosphodiester bond. The free 3′OH of the exon then completes the circularization process, leaving a terminated fork product behind. It is important to note that the lariat-precursor model maintains a functional transcript in addition to the newly formed circular RNA, whereas the direct backsplice model ends with a terminated fork product in place of the pre-mRNA.
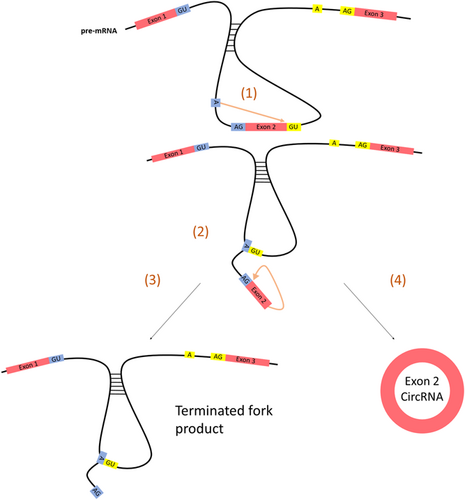
The direct backsplice model of circular RNA biogenesis. (a) Nucleophilic attack of the branching point onto the donor site of intron 2. (b) Attack of the free 3′OH of exon 2 onto the acceptor site of intron 1. (c) A terminated fork product and (d) an exonic circRNA are the final products [Color figure can be viewed at wileyonlinelibrary.com]
Under normal circumstances, the intronic-lariat released in canonical splicing undergoes debranching at the 2′–5′ phosphodiester bond and is subsequently degraded by exonucleases (Fica, Tuttle, Novak, Li, & Lu, 2013). However, lariats containing a 7 nt GU-rich motif at the 5′ splice site and an 11 nt C-rich motif at the branching point are protected from debranching and remain circular in nature (Shen, Han, Wei, & Ni, 2015). The 3′ tail of the lariat is degraded up to the branching point and the surviving circular molecule is termed circular intronic RNA (ciRNA) which is mainly present in the nucleus (Lasda & Parker, 2014).
Recently, multiple RNA binding proteins (RBPs) have been found to regulate the biogenesis of circRNA. The RBP quaking 1 (QK1), RBM20, and Muscleblind protein (MBL) have all been shown to increase circRNA expression and even induce their formation from otherwise linear transcripts via the binding to specific intronic motifs (Kelly et al., 2015, Khan et al., 2016). Knockdown of QK1 using siRNA or mutation of its binding sites reduced expression of circRNAs (Gupta et al., 2018). Muscleblind increases the expression of the circRNA from its own gene via the binding to conserved intronic motifs and certain exonic binding sites in its linear transcript (Ashwal-Fluss et al., 2014). Artificial constructs containing exons from other genes, which have been flanked with the muscleblind intronic motifs led to the circularization of the exons in the presence of muscleblind protein. The RBP RBM20 that is a splicing factor required in the canonical splicing of the titin gene (TTN) is also required for circRNA formation from the I-band of the same gene (Khan et al., 2016).
In addition, the RNA editing enzyme ADAR1 which mediates A-to-I substitutions has also been linked to circRNA biogenesis (Rybak-Wolf et al., 2015). Increased ADAR1 expression leads to a decrease in circRNA levels but the mechanism is not well understood. It is hypothesized that ADAR1 may be competing with other circularization-promoting RBPs to decrease the likelihood of circRNA formation. Another possible mechanism is ADAR1′s ability to reduce RNA complementarity through its A-to-I editing which reduces the chance of secondary structure formation and progression through the direct backsplice mode of biogenesis (Rybak-Wolf et al., 2015).
4 CLASSIFICATION OF CircRNA
CircRNA can be classified into three groups depending on their mode of biogenesis and their sequence: circRNA, EIciRNAs, and ciRNAs. CircRNAs are derived from exons in linear transcripts and lack any introns in their sequences. The majority of circRNA belong to this group and predominantly exist in the cytoplasm. In contrast, ciRNAs are not generated through backsplicing and lack exonic sequences. ciRNA exists in the nucleus and have little enrichment for miRNA target sites (Wu et al., 2014; Zhang et al., 2013). EIciRNA contain both introns and exons in their sequences (Jeck et al., 2013; Li et al., 2015). EIciRNA is predominately localize to the nucleus and interacts with U1 snRNP and Pol II, thus promoting the transcription of its parental gene (Li et al., 2015). The mechanism of EIciRNA formation remains unclear (Wu et al., 2014).
circRNA can also be grouped into intragenic and intergenic depending on the location from which they arise in the genome. circRNA, EIciRNA, and ciRNA are all intragenic circRNA arising from the sequences within the parental gene locus. Intergenic circular RNA is located in the genomic interval between two genes (Qu et al., 2017).
4.1 Properties of circRNA
- (1)
circRNA are abundantly expressed in many organisms ranging from the fly to the human (Jeck et al., 2013). Multiple circRNA isoforms could be processed from a single host gene via alternative splicing (Zhang et al., 2016a). TTN, the longest gene in the genome, alone generates up to 415 different exonic circRNA isoforms (Tan et al., 2017). The Ryanodine receptor 2 (RYR2) gene expresses over 100 circRNA isoforms in human hearts (Werfel et al., 2016). More than 400 host genes can generate more than one circRNA (Li et al., 2017). More than 32,000 circRNAs have been discovered in human cells to this day (Xu et al., 2017; Zeng et al., 2017).
- (2)
circRNAs are insusceptible to degradation by RNA exonucleases and are more stable with significantly longer half-lives than linear RNAs due to their covalently closed circular structure (Jeck et al., 2013, Suzuki & Tsukahara, 2014, Suzuki et al., 2006). These characteristics may place circRNA at the forefront of potential candidates for diagnostic and prognostic biomarkers for diseases, and especially since circRNA is present in the plasma (Salgado-Somoza, Zhang, Vausort, & Devaux, 2017).
- (3)
circRNAs often exhibit cell type, tissue, and developmental stage-specific expression in organisms (Jakobi, Czaja-Hasse, Reinhardt, & Dieterich, 2016; Li et al., 2017; Memczak et al., 2013; Xu et al., 2017). The expression profiles of circRNA were different at four heart differentiation stages: undifferentiated, mesoderm, cardiac progenitor, and definitive cardiomyocytes (Li et al., 2017). Werfel et al. observed significantly higher overall circRNA expression in rat neonatal hearts compared with adult rat hearts (Werfel et al., 2016); and remarkable changes in circRNA expression were also observed amongst different stages of induced pluripotent stem cell-derived cardiomyocytes (Siede et al., 2017).
- (4)
circRNAs seem to be evolutionarily conserved as evident by their homologous sequences across species (Jeck et al., 2013). ciRS-7 which originates from the vertebrate cerebellar degeneration-related 1 (CDR1) antisense transcript is highly expressed in human and mouse brain cells (Hansen et al., 2011). RNA-Seq data show that 1,288 circRNAs are commonly expressed in the hearts of humans, rats, and mice (Werfel et al., 2016), but many other circRNA expressed are specific to the species and do not show sequence homology.
- (5)
The majority of circRNAs are generated posttranscriptionally (Zhang et al., 2016b). Using metabolic tagging with 4sU of nascent RNAs, Zhang et al. showed that the majority of circRNAs are formed after transcription and splicing have terminated. Only a few circRNA are produced cotranscriptionally (Zhang et al., 2016b).
- (6)
Most circRNAs are not coexpressed with their linear transcripts. Only some circRNA are coexpressed with their parental genes (Li et al., 2017; Siede et al., 2017; Tan et al., 2017). The levels of the circular transcript in certain genes are even higher than the linear transcript of the same gene. circ-CDYL and circ-SMARRCA5 are examples of such circRNA (Siede et al., 2017).
- (7)
circRNAs expression profiles are significantly different between normal and pathological conditions in various disease models (Gupta et al., 2018; Siede et al., 2017; Werfel et al., 2016; Zou et al., 2017).
5 CircRNA DETECTION AND VALIDATION
Current methods to detect and quantify circRNAs include high throughout RNA-seq, circRNA microarray, and RT-PCR/qPCR using divergent primers and northern blot over the backsplice junction.
Circle-Seq uses next-generation sequencing (NGS), in combination with depletion of ribosomal RNA (rRNA) to reveal the presence and quantity of RNA. Sequencing data is then aligned to the genome of a species to identify circRNAs based on the presence of backsplice junction-spanning reads (Jeck & Sharpless, 2014). Eleven algorithms (e.g., PTESFinder, KNIFE, NCLscan, circRNA_finder, CIRCexplorer, DCC, find_circ, UROBORUS, CIRI, MapSplice, and segemehl) have been developed for detection of circRNA (Hansen et al., 2016). These different circRNA prediction algorithms show differences in term of their precision and sensitivity (Zeng & Lin, 2017). Due to the resistance of certain linear transcripts to RNase R digestion, a new method involving RNase R treatment followed by polyadenylation and poly(A) + RNA depletion (RPAD) was reported to isolate pure circRNA for RNA-Seq analysis (Panda et al., 2017). RPDA removes linear RNAs and enriches circular RNAs efficiently, which enhances the quantitative and qualitative analysis of circRNAs.
Microarray analysis uses known circular junction sequence-specific probes combined with linear RNA depletion by exonucleases to capture and quantify circRNA at high sensitivity and specificity (Jeck et al., 2013). The microarray can only detect known circRNA and results should be further confirmed by other methods. Northern blot using junction sequence-specific probes and qRT- PCR using outward primers are used to validate and quantify limited known circRNAs.
The use of exoribonucleases, such as RNase R, is a useful tool to help distinguish circRNA from other backsplice-containing linear RNA transcripts. The lack of a discrete 3′ ends in circRNA makes them immune to RNase R, whereas the linear transcripts remain prone to digestion. True circRNA show increased saturation following RNase R treatment as opposed to untreated samples. However, this phenomenon is not without exceptions. Certain circRNAs were found to be susceptible to RNase R, and some linear transcripts were found to be resistant to RNase R digestion (Jeck & Sharpless, 2014). Therefore, RNase R resistance alone cannot be used to confirm the circularity of the molecule.
In addition tools that aid in the verification of circRNA include 2D and gel trap electrophoresis (Hansen et al., 2011, Jeck & Sharpless, 2014). The slower migration of circular transcripts relative to linear transcripts of the same length aids in their separation in 2D gel electrophoresis. In gel trap electrophoresis, circRNA mixed with melted agarose becomes trapped by cross-links and is rendered immobile in a gel whereas linear transcripts are able to migrate freely (Jeck & Sharpless, 2014).
Validation of circularity requires not one, but multiple methods to concretely establish the structural nature of the molecule. The introduction of novel, improved methods with higher accuracy, and sensitivity are a prerequisite to overcome current and future challenges in circRNA identification and characterization studies.
6 CircRNA EXPRESSION
CircRNA are most abundant in the brain compared with other organs, with stress and aging playing significant roles in altering their expression profiles (Cortes-Lopez et al., 2018; Fischer & Leung, 2017). RNA-sequencing of human adult and fetal tissues (heart, kidney, liver, lung, colon, and stomach) show that up to 50% of circRNAs are tissue-specific and that both the number and the expression level of circRNA in fetal tissues are higher than adult tissues (Xu et al., 2017). In addition, the abundance of circRNA in gland tissues is higher than other adult tissues. However, circRNAs are expressed at very low levels in proliferating and human cancer cells (Bachmayr-Heyda et al., 2015; Rybak-Wolf et al., 2015). CircRNA's resistance to exonucleases grants them remarkably longer half-lives than linear RNA molecules, and they have been identified and detected in the plasma, which implies that circRNA may be a promising biomarker for disease (Memczak et al., 2013).
7 FUNCTION OF CircRNA
Recent research has revealed that circRNAs can function as competing endogenous RNAs or miRNA sponges, as target-RNA decoys by binding to RBPs, as regulators of splicing and transcription by binding srnRNA and enhancing Pol II activities, and as protein scaffolds and modifiers of parental gene expression.
7.1 CircRNA as miRNA sponges
miRNA negatively regulates gene expression by partial base pairing with the UTR of its complementary mRNA. Computational analysis has demonstrated the presence of miRNA binding sites in circRNAs. The interaction between complementary circRNA and miRNA sequences leads to the binding and holding of miRNA in what has been termed the “sponging effect.” miRNA captured by circRNA fails to bind to its target mRNA and loses its ability to repress gene expression, resulting in the increased expression of its target mRNA. For example, ciRS-7 or CRD1as, generated from the CDR1 gene and containing over 70 conserved binding sites for miRNA-7 (Hansen et al., 2013; Memczak et al., 2013). CRD1as RNA binds to the protein Argonaut 2 (AGO2) and miRNA, forming an RNA-induced silencing complex in the cell (Hansen et al., 2011). It is important to note that not all circRNAs are enriched for miRNA binding sites and repress miRNA (Khan et al., 2016).
7.2 CircRNA as protein (RBP) sponges
A number of circRNAs are produced from RBP genes and possess conserved binding sites for the host RBP. CircMbl and its flanking intronic sequences contain conserved binding sites for MBL, an RBP protein that regulates alternative mbl pre-mRNA splicing (Ashwal-Fluss et al., 2014). HuR is another RBP which binds to PABPN1 mRNA and enhances its translation. Massive binding of circPABPN1 to HuR blocks HuR binding to PABPN1 mRNA and lowers its translation (Abdelmohsen et al., 2017).
7.3 CircRNA as scaffolds for the assembly of other components
circRNAs also function as dynamic protein scaffolds facilitating contact and assembly of proteins. The first reported protein scaffolding circRNA is circFoxo3 (Du et al., 2017a). circFoxo3 from the forkhead box O gene shows high binding affinity to transcription factors including Id-1, E2F1, HIF-α, and FAK. Increased circ-Foxo3 levels inhibited nuclear translocation of Id-1, E2F1, HIF-α, and the mitochondrial translocation of FAK. circ-Amotl1 has also been reported to function as a protein scaffold by binding to AKT and PDK1 to form ternary complexes, thus facilitating the nuclear translocation of pAKT (Zeng et al., 2017).
7.4 Enhancing splicing and transcription
The majority of circRNAs are located in the cytoplasm where they may function as miRNA sponges, protein sponges, or protein scaffolds. However, ciRNA and EIciRNA (e.g., circEIF3J and circPAIP2) are retained in the nucleus, where they interact with U1 small nuclear ribonucleoproteins (snRNP) and enhance the RNA polymerase II (Pol II) transcription activity of their parental genes (Li et al., 2015). Still, the underlying mechanism of EIciRNA remains unclear.
7.5 Translation of circRNA
When circRNA molecules contain internal ribosomal entry site (IRES) elements or prokaryotic ribosome binding sites, they can be translated into proteins or peptides (Wilusz, 2018). Recent research has demonstrated that human circ-ZNF609, derived from the second exon of its host ZNF gene, is translated in a splicing-dependent and cap-independent manner (Legnini et al., 2017, Peng, Chen, Zhu, Shen, & Du, 2017). It has also been reported that N6-methyladenosine (m6A) can enhance or promote circRNA translation (Yang et al., 2017; Legnini et al., 2017).
8 CircRNA IN CARDIOVASCULAR DISEASE
Deep sequencing of cardiac tissue revealed the abundance, evolution, conservation, and developmental stage-specific differentiation of circRNA expressed in the heart (Jakobi et al., 2016, Werfel et al., 2016). A number of circRNAs are generated from genes associated with cardiovascular disease including Ryr2, Ttn, and Dmd (Jakobi et al., 2016). CircRNAs are differentially expressed in healthy and diseased human hearts (Zou et al., 2017), implying their roles in disease development.
8.1 Ischemia/reperfusion (I/R) injury and myocardial infarction (MI)
Ischemia results in anoxic injury to oxygen-dependent tissues, decreasing the mitochondrial ATP generation through oxidative phosphorylation. Reduced cellular ATP levels causes disturbances in ion homeostasis in the cell, leading to changes in cell membrane permeability, activation of hydrolases, proteases, and eventually apoptosis (de Groot & Rauen, 2007). Reperfusion, the restoration of blood supply is currently the mainstay treatment of ischemia caused by myocardial infarction, but it subjects the site of injury to further tissue (Kalogeris, Baines, Krenz, & Korthuis, 2012). Cardiac ischemia/reperfusion (I/R) injury and myocardial infarction (MI) are a leading cause of morbidity and mortality worldwide, and the literature supports the role of circRNAs in I/R injury through its interaction with miRNAs.
8.1.1 CDR1as
It has been reported that circRNA CDR1as expression is upregulated in hypoxic cardiomyocytes and in MI mice (Geng et al., 2016). Overexpression of CDR1as in vivo promoted an increase in cardiac infarct size and enhanced the expression of miR-7 target genes PARP and SP1. This is the first reported study on the functional role of circRNAs in MI and it suggests the potential of CDR1as as a new therapeutic target in the treatment of MI.
8.1.2 MICRA
Myocardial Infarction-Associated CircRNA (MICA) is downregulated in the peripheral blood samples of MI patients. Patients with low levels of MICRA developed left ventricular dysfunction, suggesting that MICRA levels may be used as a predictor of left ventricular dysfunction development in patients with MI (Vausort et al., 2016). This finding is supported by a more recent study where patients with low blood MICRA levels had a higher risk of left ventricular function loss at the time of reperfusion after percutaneous intervention (Salgado-Somoza et al., 2017). Although the function of MICRA is currently unknown, MICRA may act as a sponge of miR-150, which has been linked to the subsequent heart failure occurring after acute MI (Devaux et al., 2013, Salgado-Somoza et al., 2017).
8.1.3 CircRNA_081881
CircRNA_081881 levels were 12.5 folds lower in patients with MI (Deng et al., 2016). In silico analyses revealed that circRNA_081881 contains 7 different binding sites for miR-548. One of the potential targets of miR-548 is PPARγ, which was reduced in the plasma of acute patients with MI (Deng et al., 2016, Fliegner et al., 2008). The study also showed that siRNA silencing of circRNA_081881 reduced PPARγ expression, suggesting that circRNA_081881 positively regulates PPARγ through the sequestration of miR-548. This is yet to be experimentally confirmed.
8.1.4 MFCAR
Recently, Mitochondrial Fission and Apoptosis Circular RNA (MFCAR) was found to play a direct role in the expression of a mitochondrial membrane-associated protein, MTP18, which participates in apoptosis (Wang et al., 2017). Bioinformatics analysis and RNA pull-down assays demonstrated that MFCAR sequesters miR-652-3p to prevent it from binding to MTP18, thus increasing MTP18 expression.
8.1.5 CircSRY
Sex-determining region Y (SRY) circRNA confers protection to hypoxic cardiomyocytes by reducing apoptosis through repression of miR-138. miR-138 induces signaling through the MLK3/JNK/c-Jun pathway (He et al., 2013; Wang et al., 2016b). This signaling pathway is inhibited with circSRY overexpression.
Many other circRNA have been implicated in I/R injury. cZNF609, cZNF292, cAFF1, cDENND4C, and cTHSD1 (Boeckel et al., 2015) are differentially expressed in hypoxic human umbilical vein endothelial cells (HUVEC). Hypoxia and CoCl2 upregulated the expression of cZNF609, cZNF292, and cAFF, but decreased cTHSD1. Knockdown of cZNF292 using siRNA decreased spheroid angiogenic sprouting and cell proliferation, corroborating the findings that cZNF292 plays a role in I/R injury.
8.2 Cardiac senescence
Cellular senescence is the loss of mitotic activity of a cell induced by short telomere length or DNA damage secondary to stress, the latter of which is known as premature senescence.
8.2.1 CircFoxo3
circRNA screening of aged human and murine tissue, including lung, intestine, dermal, and heart tissue, revealed that aged tissues expressed significantly higher levels of circFoxo3 along with significantly higher levels of senescence associated β-galactosidase activity (SA-β-gal). SA-β-gal is used as the biomarker of cellular senescence, with younger tissues displaying virtually no positive SA-β-gal activity when compared with older tissues (Du et al., 2017b). Hydrogen peroxide treatment increased circFoxo3 expression as well as SA-β-gal activity in different cells including mouse cardiac fibroblasts and primary cardiomyocytes. siRNA specifically targeting the circular transcript of Foxo3 reduced SA-β-gal activity, indicating distinct effect of the circRNA from its linear counterpart.
circFoxo3 was also reportedly implicated in doxorubicin (Dox)-induced senescence (Du et al., 2017b). Dox treated mice displayed elevated left ventricular end diastolic and systolic diameters, whereas the left ventricular ejection fraction, left ventricular fractional shortening, left ventricular systolic pressure, and rate of rise of left ventricular pressure (dp/dt) were significantly decreased. Successive siRNA knockdown of circfoxo3 in Dox treated mice abrogated the damage.
The mechanism of action of circFoxo3 involves an interaction with a number of antisenescence proteins including ID1, E2F1, HIF1a, and FAK as confirmed by RNA pull-down assays (Du et al., 2017b). circFoxo3 affected the compartmental localization of these proteins. Overexpression of circfoxo3 increased antistress protein levels in cytoplasmic fractions, whereas decreasing their translocation into the nucleus, and consequently abolishing their transcriptional control over antisenescence genes.
8.2.2 Other circRNA
Recently, it has been reported that circTtn105-11, circFhod3, circArhgap32, and circStrn3 as well as the Qki host gene are downregulated in HL1 cells treated with Dox (Gupta et al., 2018). Qki plays a protective role in I/R induced apoptosis in cardiomyocyte (Darbelli & Richard, 2016) and Dox-mediated senescence (Gupta et al., 2018). Qki positively regulated circRNA expression from Ttn, Hhod3, and Strn3 leading to the inhibition of doxorubicin mediated injury. Over-expression of circTtn105-11 using lentiviruses reduced caspase 3/7 activity and increased cell survival. Finally, knockdown of circTtn105-11 increased cell susceptibility to Dox and attenuated the protective effect of Qki.
8.3 Cardiac fibrosis
8.3.1 CircRNA_00203
CircRNA_000203 is upregulated in the diabetic mouse myocardium, in mice with myocardial fibrosis and in Ang-II-induced mouse cardiac fibroblasts (Tang et al., 2017). In circRNA_000203 overexpressed tissues, cardiac fibrosis was increased and the expression of Col1a2 and CTGF was upregulated. It was demonstrated that circRNA_000203 is able to specifically sponge up miR-26b-5p, resulting in the reduction of free miR-26b-5p (Tang et al., 2017).
8.3.2 CircRNA_010567
circRNA-010567 is another miRNA sponge implicated in cardiac fibrosis and it plays a similar role to circRNA-000203. The miRNA target of circRNA_010567 is miR-141 which inhibits the translation of fibrosis-related proteins including TGF-β, Col I, Col III, and α-SMA (Zhou & Yu, 2017). By sequestering miR-141, circRNA-010567 increases the levels of profibrotic proteins.
Both circRNA_000203 and cirRNA_010567 may potentially act as new therapeutic targets for the prevention and treatment of cardiac fibrosis in diabetic cardiomyopathy.
8.4 Dilated cardiomyopathy and hypertrophic cardiomyopathy
Dilated cardiomyopathy and hypertrophic cardiomyopathy are the two most common types of cardiomyopathy. RNA-Seq data revealed that circRNA expression profiles were strikingly different in dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM) hearts when compared with normal control hearts (Zou et al., 2017). Particularly, the reduction of circRNAs produced from the genes CAMK2D (in DCM and HCM) and Titin (in DCM) was observed. RBM20 is an RBP that is responsible for the splicing of the I-band in the titin gene and promotes circRNA formation. Mutations in RBM20 lead to the formation of excessively compliant titin isoforms which resulted in DCM (Khan et al., 2016). Nonfunctional RBM20 caused the loss of multiple circRNAs from the titin gene. DCM due to RBM20 mutation was associated with circRNA loss, whereas HCM did not display critical losses in circRNA. Siede et al. showed that circSLC8A1, circ CHD7, and circATXN10 were increased in patients with DCM, whereas circDNAJ6C was decreased (Siede et al., 2017). The contradiction in results may be due to small sample sizes, and the use of different algorithms and detection methods which can lead to inconsistencies in findings. In the TAC-induced mouse pressure overload-induced cardiomyopathy model, circ-Foxo3 was found to be involved in the protective mechanism of Ganoderma spore oil against cardiomyopathy (Xie et al., 2016). The authors showed that Ganoderma spore oil improved heart function and decreased expression of circ-Foxo3 which is increased in doxorubicin (Dox) injured hearts (Du et al., 2017b).
More recently, Zeng et al. demonstrated the function of circ-Amotl1 derived from the angiomotin-like 1 gene (Amotl1), in Dox-induced animal cardiomyopathy (Zeng et al., 2017). circ-Amotl1 was observed to be highly expressed in neonatal hearts. Intraperitoneal injection of circ-Amotl1 relieved the aberrant effect of Dox on the heart, as shown by reduced apoptosis, hypertrophy, and fibrosis. circ-Amotl1 was found to interact with PDK1 and AKT1 and facilitate the nuclear translocation of AKP and PDK1, all of which are important for cell proliferation and survival. Their findings highlight the potential of circRNA as a therapeutic intervention in the treatment of cardiomyopathy.
8.5 Cardiac hypertrophy and heart failure
Cardiac hypertrophy, myocardial infarction, and myocardiomyopathy are all factors that contribute to heart failure, one of the main causes of morbidity and mortality worldwide. Previous research demonstrated that in the thoracic aortic constriction and isoproterenol-induced cardiac hypertrophy models, the expression of heart-related circRNA (HRCR, or mm9-circ-012559) was reduced, whereas miR-223 was increased, compared with sham or nonfailing control hearts (Werfel et al., 2016).
HRCR contains six binding sites for miR-223. Increased expression of HRCR sequesters free miR-223 levels leading to the attenuation of cardiac hypertrophy (Wang et al., 2016b). Over-expression of HRCR increased the expression of ACR (apoptosis repressor with CARD domain) and abrogated the inhibition of ACR by miR-223. This was the first study on the role of circRNA in hypertrophy and heart failure. Using circRNA microarray assays, Wu et al. later reported that 29 circRNAs were significantly upregulated, whereas 34 were downregulated in murine left ventricular tissues with myocardial infarction-induced heart failure (Du et al., 2017b). Ten over-expressed circRNAs and 10 downregulated circRNAs were randomly selected for qRT-PCR confirmation. The group successfully validated the upregulation of CDR1as, circRNA-004775, 007687, 013216, 010567, and 015506 and the downregulation of circRNA001135, 002007, 004768, 008398, 008640, and 014961. circRNA013216 and 010567 were the two most significantly upregulated circRNAs. Analysis using Arraystar's miRNA target predication program identified circRNA013216 as a sponging candidate for miR-181a-3p, miR486a-5p, and miR486b-5p, whereas circ01056 matched with miR124, miR141, and miR200a. The interaction between the two classes of molecules, however, was not confirmed.
RNA-Seq data also revealed overall circRNA expression was increased in human failing hearts and in murine hearts subjected to pressure overload hearts using transverse aortic constriction (TAC; Werfel et al., 2016). Total of 58 circRNAs, including three titin-derived circRNAs (m005501, m005492, and m005505) were significantly altered in TAC hearts. m005501 and m005492 were upregulated in in TAC hearts, whereas m005505 was decreased.
The expression profiles of circRNAs in the murine model of MI-induced heart failure (Du et al., 2017b) and TAC-induced heart failure (Werfel et al., 2016) were not consistent with one another. This discrepancy could be attributed to the differences in detection methods applied (microarray vs. RBA-Seq), the inherent differences in the models, sample collection time points (8 months after MI vs. 3 weeks after TAC), disease severity/progression, and the specific tissue used (left ventricle vs. a whole heart).
8.6 Atherosclerosis
8.6.1 ANRIL
Atherosclerosis is a narrowing of the arteries caused by buildup of plaque rich in cholesterol, fat, calcium, and other molecules. Genome-wide association studies identified a nonprotein coding region located on the gene desert in chromosome 9p21, which was strongly associated with cardiovascular disease, including coronary artery disease (Holdt & Teupser, 2012). One specific noncoding RNA in this locus, Antisense noncoding RNA in the INK4 locus (ANRIL), has been extensively studied for its strong involvement in atherosclerosis (Holdt & Teupser, 2012).
A circular form of ANRIL, circANRIL, is expressed in several isoforms, the most dominant of which is made up of exons 5, 6, and 7 of ANRIL. Individuals with more cardio protective SNPs in ANRIL have a higher expression of circANRIL and less severe coronary artery disease (CAD). Furthermore, the circANRIL to ANRIL ratio was highest in individuals with no signs of atherosclerotic vascular disease (Burd et al., 2010). circANRIL overexpression in smooth muscle cells (SMC) and macrophages differentiated from iPSC resulted in apoptosis and decreased cell proliferation. The same results were observed in SMC from patients with higher endogenous levels of circANRIL. circANRIL governs pre-rRNA maturation and nucleolar stress through interaction with multiple RBPs, thereby acting as a protective factor against atherosclerosis.
8.6.2 CircR-284
A recently published study showed that serum circR-284 levels were significantly elevated in patients with carotid plaque rupture, whereas miR-221 levels were downregulated (Bazan et al., 2017). miR-221 has been previously demonstrated to stimulate intimal thickening through downregulation of p27Kipi, a cyclin-dependent kinase inhibitor which reduces the proliferation of vascular SMCs (VSMCs). The ratio of circR-284: miR-221 was significantly and exclusively increased in the early stage of carotid plaque rupture, implying its potential as biomarker for rupture susceptibility.
8.6.3 CircWDR77 and has-circ_003575
Chen et al. have reported that CircWDR77 was upregulated in VSMCs treated with high glucose (Chen, Cui, Yuan, Zhang, & Sang, 2017). Silencing of circWDR77 inhibited VSMCs proliferation and migration by targeting miR-124/FGF2. Li et al. reported that oxLDL altered circRNA expression in HUVECs, specifically upregulating has-circ_003575. Silencing of has-circ_003575 enhanced HUVEC proliferation and angiogenesis. Whether CircWDR77 and has-circ_003575 are linked to atherosclerosis is still unknown.
8.7 Coronary artery disease
CAD is the major cause of cardiovascular disease related deaths. CircRNA microarray analysis on plasma from patients with CAD showed that 24 circRNAs are differentially expressed, 18 of which are upregulated, and 6 downregulated. The authors found that nine circRNAs could potentially bind to has-miR-130a-3p, resulting in an increase in TRMP3 expression (Pan et al., 2017).
Zhao et al. performed circRNA microarray assays using peripheral blood samples from 12 patients with CAD and 12 control individuals. They found that hsa_circ_0082081 and hsa_circ_0124644 were moderately correlated with the CAD, whereas hsa_circ_0113854 and hsa-circRNA5974 were weakly correlated. Moreover, circRNA-0124644 was identified as a biomarker for CAD in peripheral blood samples with a sensitivity and specificity of 0.861 and 0.626, respectively (Zhao et al., 2017). Further research on circRNA-0124644 is required to dissect its functional role and its relationship to the development and pathogenesis of CAD.
cZNF609 generated from the second exon of the Zinc finger protein was dysregulated in patients with CAD, diabetes, and hypertension (Boeckel et al., 2015; Liu et al., 2017). cZNF609 was abundantly expressed in normal HUVECs (Boeckel et al., 2015). High glucose and hypoxic stress significantly upregulated cZNF609 in ECs in vivo and in vitro. Knockdown of cZNF609 decreased retinal vessel loss and suppressed pathological angiogenesis, increased endothelial cell (EC) migration, and tube formation, and protected ECs against oxidative stress and hypoxic stress in vitro. cZNF609 acted as an endogenous miR-615-5p sponge to sequester and inhibit miR-615-5p activity, which led to increased MEF2A expression. cZNF609 has also been reported to function as a sponge of miR-150-5p, leading to the enhancement of AKT3 expression in other diseases (Peng et al., 2017).
8.8 Aortic aneurysm and thoracic aortic dissection
8.8.1 hsa-circ-000595
circRNA screening of aortic tissue from patients with aortic aneurysm revealed the differential expression of circRNAs, most notably hsa-circ-000595 which was 1.5 folds greater compared with healthy tissue (Zheng et al., 2015). CoCl2 treatment upregulated the levels of hsa-circ-000595 in VSMCs in a time and dose-dependant manner. Knockdown of hsa-circ-000595 with siRNA attenuated apoptosis in VSMCs and decreased the levels of proapoptotic proteins. In contrast, there were no changes in apoptosis in the absence of CoCl2, suggesting an active role of hsa-circ-000595 under hypoxia condition only. Knockdown of hsa-circ-000595 significantly increased the levels of miR-19a, an oncogenic miRNA serving as a biomarker for several types of cancer (Feng et al., 2014, Wu et al., 2014, Zhang et al., 2015) and an inhibitor of lipopolysaccharide-induced EC apoptosis (Jiang et al., 2015). The beneficial effects of has-circ-000595 knockdown may not only be limited to the reduction of VSMC apoptosis but could potentially be extended to the multitude of diseases associated with miR-19a including cancer and EC apoptosis. A sponging effect was not confirmed between has-circ-000595 and miR-19a, and further research must confirm the mechanism through which has-circ-000595 regulates miR-19a levels.
8.8.2 Other circRNA
Zou et al. performed a microarray assay to detect circRNA expression in in aortas with thoracic aortic dissection (TAD; Zou et al., 2017). Total of 156 circRNAs were upregulated whereas 106 were downregulated in TAD aortas compared with non-TAD aortas. Most altered circRNAs (~80%) were exonic circRNAs. qRT-PCR confirmed five upregulated circRNAs (hsa_circRNA_101238, hsa_circRNA_104634, hsa_circRNA_002271, hsa_circRNA_102771, and hsa_circRNA_104349) and three downregulated circRNAs (hsa_circRNA_102683, hsa_circRNA_005525, and hsa_circRNA_103458). GO analysis showed that the linear transcript isoforms of the upregulated circRNAs in TAD samples were associated with cell proliferation, cell adhesion, cell migration, and blood vessel development. In silico analysis showed that hsa_circRNA_101238 on chromosome 13 had five predicted miRNA targets: miR-320a, miR-320b, miR-138-5p, miR-593-5p, and miR-320c. miR-320a was downregulated in TAD tissues and is known to be an inhibitor of MMP9. It seems plausible that hsa_circRNA_101238 sequesters miR-320a, causing increased expression of MMP9 and leading to TAD. The function and mechanism of circ_101238 still requires further experimental evidence.
9 CONCLUDING REMARKS
CircRNA, once believed to be a by-product of aberrant splicing, has since become a major topic of research due to their wide range of biological functions. From gene expression regulation, to translation and mRNA competition, circRNA are proving to be useful molecules with the potential of being therapeutic targets and biomarkers for disease. There are several questions about circRNA that remain unanswered. Further studies are required to deduce the exact mechanisms through which circRNAs interact with proteins and other noncoding RNAs to mediate their effects.
This review provides scientists and clinicians with a summary of the ever-growing world of circRNAs and their role in cardiac disease.
ACKNOWLEDGEMENTS
This study is supported by funding from the Canadian Institutes of Health Research and The Natural Sciences and Engineering Research Council of Canada-Discovery Grant.
CONFLICTS OF INTEREST
The authors have declared that there are no conflicts of interest.