Contriving multiepitope subunit vaccine by exploiting structural and nonstructural viral proteins to prevent Epstein–Barr virus-associated malignancy
Abstract
Cancer is one of the common lifestyle diseases and is considered to be the leading cause of death worldwide. Epstein–Barr virus (EBV)-infected individuals remain asymptomatic; but under certain stress conditions, EBV may lead to the development of cancers such as Burkitt’s and Hodgkin’s lymphoma and nasopharyngeal carcinoma. EBV-associated cancers result in a large number of deaths in Asian and African population, and no effective cure has still been developed. We, therefore, tried to devise a subunit vaccine with the help of immunoinformatic approaches that can be used for the prevention of EBV-associated malignancies. The epitopes were predicted through B-cell, cytotoxic T lymphocytes (CTL), and helper T lymphocytes (HTL) from the different oncogenic proteins of EBV. A vaccine was designed by combining the B-cell and T-cell (HTL and CTL) epitopes through linkers, and for the enhancement of immunogenicity, an adjuvant was added at the N-terminal. Further, homology modeling was performed to generate the 3D structure of the designed vaccine. Moreover, molecular docking was performed between the designed vaccine and immune receptor (TLR-3) to determine the interaction between the final vaccine construct and the immune receptor complex. In addition, molecular dynamics was performed to analyze the stable interactions between the ligand final vaccine model and receptor TLR-3 molecule. Lastly, to check the expression of our vaccine construct, we performed in silico cloning. This study needed experimental validation to ensure its effectiveness and potency to control malignancy.
1 INTRODUCTION
Cancer is one of the most common lifestyle diseases that cause major global deaths. The various etiological agents of cancer are bacteria and virus. Epstein–Barr virus (EBV)-associated cancers account for approximately 1.5% of all cancer cases worldwide and are responsible for 1.8% of cancer-related deaths. According to the global burden of diseases report in 2010, approximately 142,979 deaths occurred because of the malignancies associated with EBV from the year 1990–2010 (http://www.healthdata.org/gbd). EBV attributed toward malignancies in 21 regions throughout the world. The highest mortality was reported in East Africa (47%) followed by the Southeast-Asia, South Asia, Western and Eastern Europe, North-Africa, North-America, Latin-America, and Sub-Saharan-Africa (Parkin, 2006). The virus can reactivate after encountering primary infection and has the capability to develop enduring latency within the infected population.
EBV is a member of the herpes virus family. Its genome consists of a linear dsDNA molecule. EBV has two distinct life cycles in the human host, a lytic form of infection that produces new infectious virion and a latent form of infection that allows the virus to persist in a dormant state for a lifetime in the host (Cohen, 2000). An EBV infection remains asymptomatic during childhood. EBV infection in adolescents and young adults often results in infectious mononucleosis, hepatomegaly, splenomegaly, and various malignancies, such as Burkitt’s lymphoma, Hodgkin’s disease, nasopharyngeal carcinoma (NPC), T-cell lymphoma, and gastric carcinoma. Symptoms of EBV infection include fever, sore throat, lack of appetite, rashes on the skin surface, swollen lymph node, weakness, and sore muscles. Infection with EBV usually occurs due to contact with oral and genital secretions. Recently, Pandey et al. reported the formulation of multiepitope subunits against parasitic and viral diseases, such as malaria, visceral leishmaniasis, and HIV (Kumar Pandey, Ojha, Mishra, & Prajapati, 2018; Pandey & Prajapati, 2018; Pandey, Ali, Ojha, Bhatt, & Prajapati, 2018; Pandey, Bhatt, & Prajapati, 2018). Here, in this research study we have selected eight proteins through a literature survey on the basis of their role in pathogenicity. Among these eight proteins, four proteins are structural and four are nonstructural. Structural proteins are part of the virion envelope, internal capsid proteins or the tegument and nonstructural protein participates in virus replication, transcription activation, or they cooperate in viral DNA synthesis. A special set of nonstructural proteins expressed in various forms of EBV latency, these proteins are involved in the maintenance of the viral DNA during latency and/or they participate in host-cell immortalization (Rajcani, Szenthe, Bánáti, & Szathmary, 2014). A structural protein, serine–threonine kinase (BGLF4) phosphorylates protein that leads to activation of condensin and topoisomerase, induces premature chromosome condensation. It is involved in efficient lytic DNA replication and modification in the structure of host nuclear lamina protein to facilitate the nuclear transport of EBV lytic proteins (Chang et al., 2012). Capsid vertex complex (CVC2) mediates the capsid docking to the nuclear pore, allowing entry of the viral genome into the host nucleus through binding to host nucleoporins NUP214. Nucleus egress complex 1 BFLF2 (NEC1), an early lytic protein plays an important role in virion release from the nucleus of the cell. It forms a complex with NEC2, promoting the encapsulation of the capsid and release of the capsid into the cytoplasm (Yadav et al., 2017). Triplex capsid complex (TRX1) is required to assemble the viral genome into the host cell and for efficient transport of TRX2 into the nucleus where the assembly of viral capsid occurs. Nonstructural protein includes early antigen protein D (BMRF1), involved in viral DNA replication, and acts as an accessory subunit to increase the viral DNA polymerase activity (Zhang et al., 1996). The RNA export factor ICP27 (SM) protein facilitates the nuclear export of early and late viral messenger RNAs from the host nucleus (Han et al., 2007). Latency-associated structural protein EBNA2 activates transcription by binding to the transcription factors TFIIB, TAF40, and TFIIH, and affects the activities of cell cycle regulators and retards the cell cycle progression at the G2/M phase (Huen, Henderson, Croom-Carter, & Rowe, 1995). To date, not even a single licensed vaccine is available to protect the vulnerable populations from this lifestyle disease. Although, some treatments are available, like the surgical removal of the affected part, immunotherapy, radiation therapy, chemotherapy, and so forth, these have many side effects. Several works on designing a vaccine based on the gp350 have been done, such as the use of the whole protein, use of random production epitopes, and so forth (Anyndita, Dluha, Rifa’i, Himmah, & Wahyuningsih, 2018). Gp350 was the first nonhuman primate EBV vaccine, but it was unable to produce neutralizing antibodies, and hence recurrence of the lymphoma was observed (Cohen, 2015). These glycoproteins based vaccines have not been used for vaccination due to their ineffectiveness toward a latent form of infection (Pandey, Sharma, Ojha, Bhatt, & Prajapati, 2018). Further, the therapeutics (nonadjuvanted) were developed by utilizing latent membrane protein 1 and the small portion of Epstein–Barr virus nuclear antigen, which was able to generate the T-cell responses in partial population, hence the outcome was inconclusive (Cohen, 2015). Thus, there is an urgent need of a vaccine that could elicit neutralizing antibody response through the activation of immune responses. So, in this study, we have designed a multiepitope based subunit vaccine by using immunoinformatic novel strategies. The vaccine developed by utilizing the immunoinformatic approaches have shown to be more dynamic and potent in comparison with conventional vaccines and hence, proceeding ahead into the clinical trials. Epitopes were predicted for B-cell, cytotoxic T-lymphocyte cells, and for Helper T-lymphocyte, and the antibodies secreted by the cells will specifically recognize their respective epitopes (Ojha, Khatoon, & Prajapati, 2018). The epitopes were selected on the basis of their scores, IC50 values, highest binding affinities, and population coverage. Consequently, these predicted epitopes were linked together with the help of linkers, and an adjuvant was added at the N-terminal of the construct to enhance the immunogenicity of the protein vaccine. Further, on the basis of allergenicity and antigenicity, we ensure the nonallergen and antigenic nature of the final vaccine construct. A molecular docking study was done to find out the interaction between the receptor (TLR-3) and the ligand (final vaccine construct; Kumar Pandey et al., 2018). Moreover, molecular dynamics (MD) simulation was performed to ensure the stability and flexibility of the receptor (TLR-3) and final vaccine construct (ligand) complex. Subsequently, by this approach, we confirmed the stability of the designed vaccine construct. Lastly, in silico cloning was performed to check the expression efficacy of vaccine construct after incorporating it in pET28a (+) vector. Hence, here we designed a multiepitope subunit model with the composition of immunogenic epitopes which may have the ability to activate the host humoral and cell-mediated immune systems.
2 METHODOLOGY
2.1 EBV protein exploration for vaccine construction
Initially, for designing the vaccine construct, a literature survey of the complete genome of the virus was done. A total of eight proteins of strain B95.8 were selected, including the structural and nonstructural proteins on the basis of their pathogenesis, which has been reported in the literature. The amino acid sequences of eight proteins were retrieved from the UniProt database (http://www.uniprot.org/) and the β-defensin (45 mer) amino acid sequence, which was used as adjuvant, retrieved from the National Centre for Biotechnology Information (https://www.ncbi.nlm.nih.gov/protein) in FASTA format.
2.2 Cytotoxic T lymphocyte epitope prediction
Cytotoxic T lymphocytes (CTLs) are a major component of the adaptive immune system and are capable of promoting the apoptotic death of cancerous cells with the help of a combination of the granule (perforin/granzyme) and receptor (Fas/tumor necrosis factor)-mediated (Narula, Pandey, Khatoon, Mishra, & Prajapati, 2018). The CTL epitope for eight proteins was predicted by NetCTL 1.2 server (http://www.cbs.dtu.dk/services/NetCTL/) at the sensitivity score of 0.8 (Larsen et al., 2007). MHC Class I binding epitope prediction was done by selecting three supertypes A2, A3, and B7 based on artificial neural networks (Larsen et al., 2007; Lund et al., 2004). The purpose behind the selection of these particular supertypes was that they cover approximately 90% population of the world (Sette & Sidney, 1999). For the prediction of CTL epitopes, the threshold value was set by default, that is, 0.75 (Larsen et al., 2007). Among these 8 proteins, only one epitope from each supertype was selected, based on the percentage conservancy of the peptide sequence.
2.3 Helper T lymphocytes (HTL) epitopes prediction
The HTL epitope for all the selected structural and nonstructural proteins was predicted by the Immune Epitope Database server (IEDB; http://tools.iedb.org/mhcii/). The human species/locus and allele reference set consist of 27 alleles; hence, all the alleles were selected for the prediction of HTL epitopes. For vaccine construction, we selected epitopes based on their IC50 value, percentile rank, and interferon gamma (IFN-γ) inducing nature. Epitopes with the least percentile rank, IC50 and IFN-γ positive, were sorted out for further analysis. Among these selected epitopes, two topmost epitopes were selected from each of the protein. The IC50 value for a potent antigenic peptide sequence should be <500 nM (Rana & Akhter, 2016).
2.4 IFN-γ inducing epitope prediction
IFN-γ plays an important role in viral and microbial infections. It activates both the adaptive and innate immune response by stimulating macrophages and natural killer cells and provides a heightened response to MHC antigens (Chatterjee, Ojha, Khatoon, & Prajapati, 2018). IFN-γ epitopes were predicted from the IFNepitope server (http://crdd.osdd.net/raghava/ifnepitope/scan.php). This server gives the output, which comprises both IFN-γ noninducing and inducing MHC Class II binder, which additional helps in activation of T-helper cells. (Dhanda, Vir, & Raghava, 2013). The prediction method is based on support vector machine (SVM) hybrid approach.
2.5 B-cell epitope prediction
BCPred (http://ailab.ist.psu.edu/bcpred/predict.html) and ABCpred (http://crdd.osdd.net/raghava/abcpred/) servers were used for the prediction of linear B-cell epitopes. The linear B-cell epitopes were predicted at the threshold of 0.5. BCPred server predicts epitope by exploiting three methods: (a) AAP method, (b) BCpred, and (c) FBCpred (Markstein, Xu, El-Manzalawy, Dobbs, & Honavar, 2008). AAP is the Amino Acid Pair antigenicity scale, which uses an SVM classifier to find the B-cell epitopes, which favor particular AAPs (Chen, Liu, Yang, & Chou, 2007). BCpred is based on five different kernel methods, having fivefold cross-validation by SVM. The performance of BCPred is higher (AUC 0.758) as compared with the AAP method (AUC 0.7). FBCpred utilizes subsequent kernel for the prediction of linear length B-cell epitopes (EL-Manzalawy, Dobbs & Honavar, 2008). The cutoff score of BCPred is >0.8 for the prediction of linear B-cell epitopes (Barh, Misra, Kumar, & Vasco, 2010). The ABCpred server uses the artificial neural network for the prediction of B-cell epitopes in an antigenic sequence. The epitopes that were conserved or partially common by both the prediction server in all strains were selected as the final epitopes for vaccine construction.
2.6 T-cell (HTL and CTL) epitope conservancy analysis
For the development of the subunit vaccine, conserved epitopes were selected; further, conservancy across antigen analysis was performed through an IEDB epitope conservancy analysis tool (http://tools.iedb.org/conservancy/). The predicted CTL and HTL epitopes were checked for their conservancy among other strains of EBV like strain CAO, strain RAJI, strain ag876, strain p3hr-1, type1, and type 2. Here, we selected the EBV-B95.8 strain because of their prevalence throughout the world and mostly reported in the literature. The reference protein set of other EBV strains were retrieved from the NCBI (http://tools.iedb.org/ncbi_seq_browser/).
2.7 Multiepitope subunit vaccine designing
To design a vaccine, the epitopes predicted by various immunoinformatic approaches were combined together with the help of their respective linkers. The B-cell, HTL, and CTL were linked together with the help of KK, GPGPG, and AAY linkers, respectively (Pandey, Ojha, Aathmanathan, Krishnan, & Prajapati, 2018c; Rana & Akhter, 2016). To make the vaccine more immunogenic, β-defensin (45 mer) amino acid sequence was added as an adjuvant at the N-terminal of the construct with the help of EAAAK linker (Barh et al., 2010). β-Defensin peptides activate the adaptive immunity that recruits the naive T cells and immature dendritic cells at the site of infection through the CCR6 receptor. It also provides innate host response in microbial infections (Mohan, Sharma, Bhat, & Rao, 2013). Linkers play a very important role in extended conformation (flexibility), protein folding, and separation of functional domains, and hence, making the protein structure stable (Nezafat, Ghasemi, Javadi, Khoshnoud, & Omidinia, 2014).
2.8 Antigenicity and allergenicity prediction
Multiepitope vaccine construct was evaluated for its nonallergic or allergic nature with the help of an online available server, AlgPred (http://www.imtech.res.in/raghava/algpred/). AlgPred analysis is based on the use of different algorithms (IgE epitope + MAST + SVM + ARPs BLAST) to predict the allergenicity of protein sequences. The position of the epitope in the given protein is predicted by IgE mapping. AlgPred uses MAST to search MEME/MAST allergen motifs and predict the allergen. It is an SVM module-based approach, which uses amino acid or dipeptide composition for the prediction of allergenicity. The BLAST of the query peptide sequence is done against the ARPs (allergen-representative peptide) database consisting of 2,890 allergen-representative peptides to see whether there is any BLAST hit in epitopes or not (Saha & Raghava, 2006). For evaluating the antigenicity of the vaccine construct, Vaxijen v2.0 server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used (Doytchinova & Flower, 2007). The antigenicity prediction method is simply based on the characterization of the physiochemical properties of the input protein sequence. The accuracy of this server varies from organism to organism and ranges from 70% to 89%.
2.9 Prediction of various physicochemical properties
To analyze the physiochemical properties of the final vaccine protein, ProtParam tool (http://web.expasy.org/protparam/; Gasteiger et al., 2005) was used. These physiochemical properties were computed on the basis of protein ionizable (pK) values of the amino acid present in the vaccine construct (Bjellqvist et al., 1993). The instability index of the protein indicates the stable nature of the protein. If the calculated instability index of peptide construct is below 40, it is considered as stable in nature while values above 40 are considered to be unstable in nature. In vivo half-life prediction for proteins was, by default, based on the principle of the “N-end rule.” According to this rule, degradation of protein is determined by the amino acids sequence present at the N-terminal end (Varshavsky, 1997). The aliphatic index of proteins is calculated by the percentage of aliphatic amino acids (valine, alanine, isoleucine, and leucine) present in the protein. The Grand average of hydropathicity (GRAVY) is a measure of the hydrophobic nature of the protein and is calculated by the sum of hydropathy index of all the hydrophobic amino acids on a total number of amino acids present in the protein.
2.10 Tertiary structure prediction of the designed vaccine construct
For the prediction of the 3D structure of subunit vaccine construct, a web-based server named RaptorX (http://raptorx.uchicago.edu/StructurePrediction/predict/) was used. The input query sequence was processed by the server, and the tertiary structure was predicted in the form of a three-domain structure. The best-suited template was used for the homology modeling. The single best template was used for the modeling purpose, which enhances the quality of alignment and tertiary structure (Peng & Xu, 2011). The quality of the predicted model was evaluated on the basis of confidence score, which consists of a p value and global distance test (GDT) for the relative global quality and absolute global quality, respectively. So, high-quality models were generated by the server with the help of multiple templates. The confidence score of the prediction gives an idea about the quality of predicted model (Källberg et al., 2012).
2.11 Tertiary structure refinement
The 3D structure of the final vaccine construct was further refined by the GalaxyRefine server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE). The refinement method used by the GalaxyRefine server has been successfully tested in CASP10 server and proved to be best performing for improving the structure quality. Refinement was done to enhance the template-based protein structure, by refining the whole protein structure for accuracy. Through the refinement process, the global and local structural quality of protein was improved. This relaxation method was used to refine the whole protein model and generate the refined models that were different from the initial structure. GalaxyRefine server provided five structure models, in which Model 1 was generated by the structural perturbation applied only to the clusters of side chains whereas, Models 2–5 was generated by more aggressive perturbations of loops and secondary structural elements. To avoid breakage in modeled structure perturbation, a triaxial loop closure method was used.
2.12 Tertiary structure affirmation
For the validation of the tertiary structure of the final vaccine protein, a Ramachandran plot was optimized by using RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) server. The Ramachandran plot explains about the probability of amino acid to form a secondary structure, based on the allowed and disallowed values of ф and Ψ angles; which are, in turn, calculated by the van der Waals radius of side chain atoms (Lovell et al., 2003).
2.13 Molecular docking of final vaccine protein with the immune receptor (TLR-3)
A molecular docking approach was used to check the binding affinity between the vaccine construct and the immune receptor. Protein–protein docking was performed with the help of ClusPro (https://cluspro.bu.edu/login.php?redir=/queue.php) server. Docking was performed to study the immune response between the TLR-3 (PDB ID: 2A0Z) present on the surface of immune cells and the multiepitope vaccine construct. Here, TLR-3 acts as an immunogenic receptor for antigenic response in case of EBV. The server basically selects those models which have good electrostatic interaction and desolvation energy. To reaffirm the stability of the designed vaccine construct, we again performed protein–protein docking between the vaccine construct and the TLR-3 receptor with the help of the Patch Dock server (https://bioinfo3d.cs.tau.ac.il/PatchDock/). The server predicted the potential complex on the basis of three major algorithms—molecular shape representation (detect the geometric patches on the surface of molecule), surface patch matching (form the hybrid geometric patches to match the surface with the previous one), and filtering, and scoring (filtering of the top complexes was done and further ranked on the basis of geometric shape complementarity score (Duhovny, Nussinov, & Wolfson, 2002). After obtaining the outcome from the patch dock server, the complexes were subjected to the FireDock (Fast Interaction Refinement in Molecular Docking) server for the refinement process. This server refined the complex on the basis of energy functions and hence, performed the large-scale flexible refinement (Andrusier, Nussinov, & Wolfson, 2007).
2.14 Molecular dynamics simulation of receptor-ligand complex
The MD simulation approach was used to calculate the motion of the atoms and molecules in a molecular assembly using Newtonian dynamics to determine the net force and acceleration experienced by each atom. It determines the stability of the protein-ligand structure (Karplus & McCammon, 2002). For carrying out the MD simulation of the multiepitope subunit vaccine, Groningen Machine for Chemical Simulations (GROMACS) 5.0 software package was used. During MD simulation, the intermolecular interactions between the ligand and receptor were analyzed by the using the GROMACS 96 43a1 forcefield. Energy minimization was performed to make the protein’s structure compact and hindrance free to avoid unphysical changes in potential energy during MD simulation. For this, bad contacts and very high energy configurations that create steric clash were removed. Initially, the system underwent constant number, volume and temperature (NVT) and constant number, pressure and temperature (NPT) ensemble simulation at a 300 K temperature and 1 bar pressure for a time period of 100 ps. Finally, MD simulation for the final confirmation of the vaccine–receptor was for 20 ns. The standard deviation and fluctuation of the protein backbone were measured by using RMSD root mean square deviation (RMSD) and root mean square fluctuation (RMSF) commands.
2.15 Codon adaptation and in silico cloning
For the purpose of cloning, codon adaptation of the vaccine construct was done for analyzing the codon usage by the prokaryotic organism (Escherichia coli). Codon optimization was done by using the Java Codon Adaptation tool (http://www.jcat.de/). For higher expression rate of the vaccine protein, E. Coli K12 strain was selected. For the efficient translation of the gene, we checked and avoided rho-independent transcription termination, prokaryote ribosome binding site, and cleavage site of restriction enzymes. The optimized codon sequence was reversed with the help of an online server, named reverse complement sequence, to ensure the complementation in the direction of the translation of the vector. Restriction sites XhoI and BamHI were added to N and C terminals, respectively, to the sequence. A snapgene restriction cloning module was used to insert the adapted sequence into pET28a (+) vector between the XhoI and BamHI. The flow chart of the designed work is shown in Figure 1.
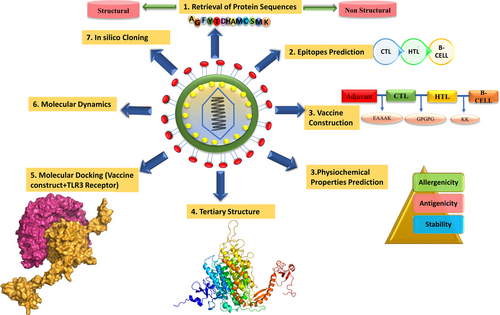
Flow chart of the designed work, representing the steps that were taken during the designing of the multiepitope subunit vaccine construction [Color figure can be viewed at wileyonlinelibrary.com]
3 RESULTS
3.1 Selection of structural and nonstructural viral proteins sequences for vaccine construction
From a total of 80 reported proteins of the virus, eight most pathogenic proteins were selected from the literature survey. Structural proteins like serine–threonine kinase (BGLF4; UniProt: P13288.2) and capsid vertex component (CVC2; UniProt: P03233.1) Nucleus egress complex1 BFLF2 (NEC1; UniProt: P0CK47.1), and triplex capsid protein (TRX1; UniProt: P03187.1) are involved in the invasion of virus into host and assembly of viral genome. Nonstructural proteins include early antigen protein D (BMRF1; UniProtID: P03191.1), RNA export factor ICP27 (SM) protein (UniProtID: Q04360.2), while latency-associated nonstructural proteins EBNA2 (UniProt: P12978.1) and LMP1 (UniProt: P03230.1) are involved in virus replication and pathogenesis. The sequences of the selected proteins were retrieved from the UniProt database. These selected proteins were further analyzed for epitope predictions.
3.2 Cytotoxic T lymphocyte (CTL) epitope prediction
The CTL epitope prediction was done with the help of the NetCTL server. The antigenic binders were predicted by considering three supertypes (A2, A3, and B7), which cover around 95% of the world population. We have selected eight proteins of the Epstein–Barr virus so, from each protein sequence, three epitopes were selected on the basis of highest score and supertypes. Hence, a total of 24 most conserved epitopes were selected from eight protein sequences (Supporting Information Table 1).
3.3 Helper T lymphocyte (HTL) epitopes prediction
HTL epitopes (15 mer) were predicted for HLA (HLA-DPA1*02:01/DPB1*01:01, HLA-DPA1*01/DPB1*04:01,HLA-DRB1*11:01, HLA-DQA1*04:01/DQB1*04:02,HLA-DPA1*03:01/DPB1*04:02, HLA-DRB1*09:01, HLA-DQA1*01:02/DQB1*06:02, HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*05:01/DQB1*03:01, HLA-DPA1*02:01/DPB1*01:01, HLA-DRB1*15:01, HLA-DRB1*03:01; Supporting Information Tables 2 and 3) and epitopes that had their peptide sequence highly conserved in all the existing strains of virus. Among the conserved epitopes, we selected 16 top-ranked epitopes having the lowest percentile score (low percentile rank = good binders with highest binding affinity), IC50 values less than 500 nM, and IFN-γ inducing positive epitopes. These highly conserved epitopes were selected as HTL epitopes for vaccine construction (Supporting Information Table 4). Interferon-γ epitopes were further analyzed because they are important activators of macrophages and inducers of HTL epitopes. A total of 16 IFN- γ positive HTL epitopes were selected for vaccine construction.
3.4 B-cell epitope prediction
First, we predicted B-cell epitopes by using two servers. Secondly, we compared all the predicted epitopes, and then, only the overlapped epitopes were selected for designing the vaccine. A total of 379 epitopes were predicted from ABCpred, and 134 epitopes were predicted from BCPred for all eight proteins. Among them, only 24 (10 mer) epitopes were found to be common or partially common in both the predictions. These common epitopes from ABCpred had the prediction score ranging from 0.52 to 0.85, and epitopes from BCPred had the prediction score ranging from 0.4 to 1. Among them, a total of 16 epitopes were predicted, and two epitopes from each protein with the best prediction score were selected for the vaccine construction. The epitopes with the prediction score of 0.8 or above were considered to be the best epitopes with highest binding affinity and were selected for vaccine construction (Supporting Information Table 5).
3.5 Multiepitope subunit vaccine construction
The epitopes with the highest prediction score were utilized for the development of the vaccine construct. A total of 16 B-cell epitopes, 24 CTL, and 16 HTL epitopes were selected based on their prediction score and were fused together with the help of their respective linkers. B-cell epitopes, CTL epitopes, and HTL epitopes were combined together with the aid of KK, GPGPG, and AAY linkers, respectively. To increase the immunogenicity of the vaccine, a 45 amino acid-long sequence of human β-defensin-3 (hBD-3) was added as an adjuvant at the N-terminal of the construct with the help of EAAAK linker (Weinberg, Quinones-Mateu, & Lederman, 2006). The final vaccine construct was found to be 879 amino acid long (Supporting Information Figure 1).
3.6 Evaluation of allergenicity, antigenicity, and a physiochemical parameter of the vaccine construct
The nonallergic nature of the vaccine construct was predicted through the Algpred server. The vaccine construct was predicted with a score of −1.434, which indicates its nonallergenic nature. Similarly, the antigenicity of the vaccine construct was predicted, and it confirmed that the protein was a good antigen with an overall prediction score of antigen 0.7432. The default set threshold value for antigenicity was 0.4 in the bacteria model. Further, the physiochemical parameters were evaluated, and the molecular weight of the vaccine construct was 92.7 kDa, which was considerable. The predicted instability index of the protein was 39.5, which is below 40 hence, denoting the protein as stable. The theoretical isoelectric point (pI) was predicted to be 9.94, indicative of the basic nature of the vaccine. The estimated half-life was found to be 30 hr in mammalian reticulocytes (in vitro). In vivo studies for estimated half-life in yeast and Escherichia coli were more than 20 and 10 hr, respectively. The half-life of the vaccine indicates the time taken for half of the protein to disappear after being synthesized in the cell, so the formulated vaccine has the capacity to recognize the antigen presenting cell within the period (Gasteiger et al., 2003). The aliphatic index of protein was 92.82, which indicates the thermal stability (higher the value of aliphatic index and higher the thermostability of the protein). The GRAVY of protein was 0.035; the positive value of GRAVY indicates the hydrophobic nature of the vaccine construct.
3.7 Modeling of the tertiary structure of vaccine construct
The input query amino acid sequence was partitioned into two domains, and the best among them was used as a template for 3D modeling. Here, the best template was “3rkoC,” which was further used for modeling. In addition, the relative quality of the model was evaluated by p value: a lower p value indicates a higher relative quality of the model. The calculated p value of the model was 9.23 × 10−5, which was pretty low, indicating its good quality. The secondary structure of the model had 35% helix, 10% beta-sheet, and 54% loop. A protein with uGDT unnormalized (GDT) and GDT score of greater than 50 is considered to be a good model. This vaccine model had the uGDT score of 121 and a GDT score of 35, indicating that the modeled protein was virtuous.
3.8 Tertiary structure refinement and its affirmation
The model generated from the 3D refine server was further analyzed in GalaxyRefine server, which generated five candidate models based on their RMSD and MolProbity score. From the refined models, one model was selected with the help of the RAMPAGE server (shown in Figure 2), and Model 2 was selected for the docking purpose. The selected model had 91% of residues in the favored region, 6.2% residues in allowed region, and only 2.6% residues in the disallowed region. The scores of several other parameters of this model were also good, that is, the GDT-HA score was 0.9265, the RMSD value was 0.463, the Mol probability was 3.033, the clash score was 49.6, and the poor rotamers was 2.5 (shown in Figure 3). Thus, the score of each parameter was found to be the best in case of this model, as compared with the other models (Figure 4).
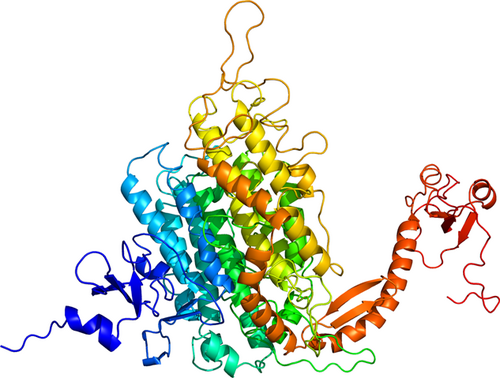
Refinement of multiepitope vaccine. 3D representation of tertiary structure of multiepitope vaccine after modeling and refinement [Color figure can be viewed at wileyonlinelibrary.com]
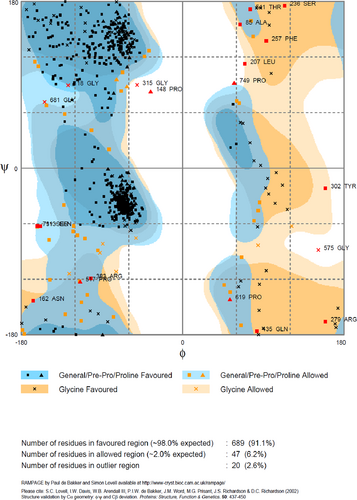
Affirmation of multiepitope vaccine tertiary structure by Ramachandran plot where 91% residues were found in favored region, 6.2% residues were found in allowed region and 2.6% residues were lies in outlier region [Color figure can be viewed at wileyonlinelibrary.com]
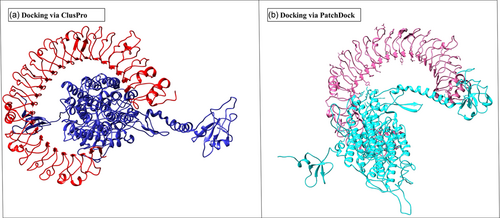
Representation of ligand-receptor docked complex (a) showing the molecular docking of the vaccine construct (Blue color) and TLR-3 receptor (red color; PDB ID: 2A0Z) via ClusPro (b) showing the molecular docking of the vaccine construct (cyan color) and TLR-3 receptor (pink color; PDB ID: 2A0Z) via PatchDock to check the stability of docked complex [Color figure can be viewed at wileyonlinelibrary.com]
3.9 Molecular docking of final vaccine construct (ligand) with the immunological receptor (TLR-3)
To optimize the interaction affinity between the final vaccine construct, and TLR-3 receptor molecular docking was performed. ClusPro server was used to predict the docked models by applying the Fourier correlation algorithm, which passes out the unwanted false positive structures by the combination of electrostatic and desolvation energies. The server then predicted a total of 29 candidate models with different binding scores and conformations, out of which one model complex with the lowest binding energy score of −1137.6 was chosen. The lowest binding energies denote the formation of a stronger complex. Further, the designed vaccine construct and TLR-3 receptor were subjected to the PatchDock server; it generates different models along with the solution or score table. The top 10 selected models were subjected to the FireDock server for the refinement process, and among the 10 models, the model with the lowest binding energy was selected. The predicted outcomes of the selected model were solution number 2, global energy (binding energy of the model) −8.96, attractive van der Waals energy (VdW) −6.68, repulsive VdW −0.15, and atomic contact energy −2.37.
3.10 Molecular dynamics simulation of receptor-ligand complex
MD simulation is the time-lapsed response of the physical movements of atoms and molecules (Shukla, Shukla, Sonkar, Pandey, & Tripathi, 2017). The energy components such as temperature, pressure, density, and volume of the simulation systems were evaluated. Further, RMSD and RMSF were also evaluated to confirm the stability of the docked complex. There was less fluctuation in temperature, up to 20 ps, but gradually it increased a bit in the time duration of 45 ps, and it again started to slow down as time lapsed and finally, became stable at the time duration of 100 ps. The fluctuation in pressure was found to be around 100 bars around the 35 ps in a time interval of 100 ps, and after that it became stable. The calculated RMSD value of the docked complex was 0.7 nm, which indicates the stability of the docked complex. Further, the plot of the RMSF suggested that initially, very mild fluctuations of 1.5 nm varied with the residue position of 100, after which there was no fluctuation at 390 amino acid residues. Then, at the 450 residue position, a slight fluctuation of 0.5 nm was observed. Finally, after that, there was no fluctuation observed. Hence, it can be concluded that the vaccine construct was stable in nature (shown in Figure 5).
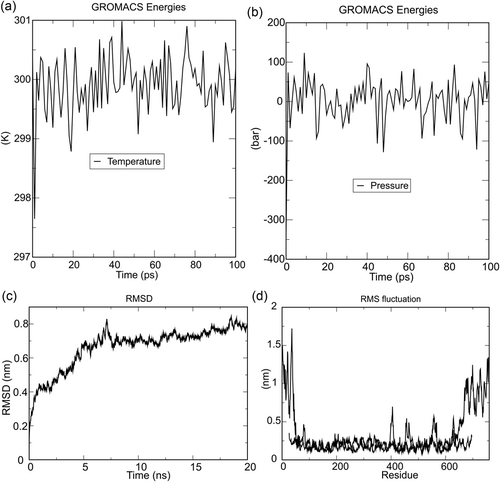
Molecular dynamics simulation of the ligand-receptor complex (vaccine and TLR-3). (a) Temperature progression plot of ligand-receptor complex shows that temperature of the system reaches 300 K and remains nearly constant around 300 K throughout the equilibration phase (100 ps). (b) Ligand-receptor complex pressure progression plot indicates fluctuation of pressure throughout the equilibration phase of 100 ps with an average pressure value 1 bar. (c) RMSD—root mean square deviation of docked complex shows very minute deviation, which reflects the stable microscopic interaction between ligand and receptor molecule. (d) RMSF—root mean square fluctuation plot of docked protein complex side chain fluctuation in plot generates peak, which reflects the flexibility of the side chain of docked protein complex
3.11 Codon optimization of the final vaccine construct
Codon adaptation was done for obtaining the gene sequence of the final vaccine construct. JCat (java codon optimization tool) was used to optimize the codon usage of the vaccine construct in E. Coli (strain K12). The optimized codon sequences were 2,637 nucleotides long, with a codon adaptation index value of 0.97. For good expression of the protein, the guanine-cytosine (GC) content should be in the range of 30%-70%; here, the vaccine protein had a GC content of 56%, indicative of its good expression possibility in the E. Coli host. While, optimizing the codon certain features related to transcription was avoided, like prokaryotic ribosome binding sites and cleavage sites of restriction enzymes to ensure uninterrupted protein expression of the gene sequence.
3.12 In silico cloning
To analyze the expression of multiepitope subunit vaccine into a vector, cloning was performed with the help of the Snapgene server. The codon sequence that was optimized by JCat was used here, and it was reversed with the help of an online server to ensure the complementation in direction of translation of the vector. Restriction site XhoI and Bam HI were added to N and C terminals of the adapted codon inserted into the pET28a (+) vector (Figure 6).
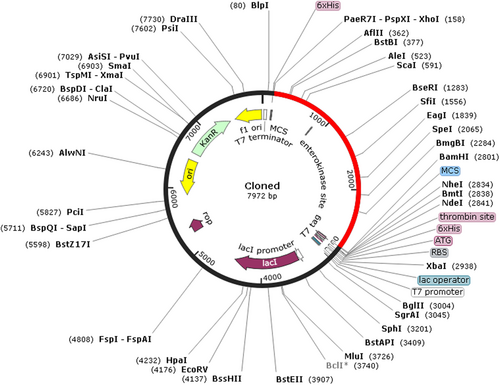
In silico cloning of the designed vaccine construct into the expression vector pET28a (+) was performed where red colored part represents the designed vaccine while the black part represents the expression vector pET28a (+) [Color figure can be viewed at wileyonlinelibrary.com]
4 DISCUSSION
EBV is an oncovirus, implicated in the pathogenesis of a number of human malignancies. Thus, a comprehensive and up-to-date analysis focused on the global burden of EBV-attributable malignancies because there is no significant preventive cure yet. The traditional vaccine development methods are generally very laborious, expensive, and demand a long period of time of their accomplishment. No effective vaccine has been developed yet to protect the wide global population from this disease. In our research study, we tried to design a multiepitope subunit prophylactic vaccine, with the help of computational tools to overcome the disadvantages of the traditional approach. The predicted B-cell, CTL, and HTL epitopes are immunogenic in behavior and potent for vaccine designing, which is ensured by using a different approach. The epitopes were joined together with the help of their respective linkers. To increase the immunogenicity of this subunit vaccine, an adjuvant β-defensin was added at the N-terminal with the help of an EAAAK linker. The final vaccine construct after adding the epitopes, adjuvants, and linkers constitutes of 879 amino acids. Moreover, antigenicity, allergenicity along with physicochemical properties were estimated to confirm the nonallergen, antigenic, and stable nature of the final vaccine construct. To make our vaccine competent for all the strains, we performed conservancy analysis during epitopes selection. The conserved epitopes in all strains were selected to avoid the problem of genetic diversification among the species. The tertiary structure of the final vaccine construct was optimized with the help of RaptorX, which can suitably interact with its specific human immune receptor. This vaccine model was further exposed to refinement by GalaxyRefine. The binding affinity of the vaccine construct with the receptor TLR-3 (present on the surface of immune cell) was confirmed by molecular docking through ClusPro server. This vaccine-TLR-3 complex was then tested for its stability by molecular dynamics simulation with the time duration of 20 ns using GROMACS. Finally, to check the translational efficiency of the final vaccine construct in an expression system, the vaccine was optimized for its codon usage with help of Java Codon Adaptation Tool. The optimized codon sequence was reversed to complement the direction of translation of the vector. Then, the sequence of XhoІ and BamHІ were added to the N and C terminals, respectively. For cloning, the gene sequence was then cloned in vector pET28a (+). In silico cloning ensures the expression of the final vaccine construct. Further, this study needs experimental validation to ensure the effectiveness and potency of the vaccine, in contrast to control the malignancies associated with EBV infection.
ACKNOWLEDGMENT
R. Ojha is thankful to the Central University of Rajasthan for providing research fellowship. V.K. Prajapati is thankful to the Central University of Rajasthan for providing computational facility.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
The protocol was designed by R. Ojha and V.K. Prajapati. The methodology was performed and the manuscript was written by R. Ojha, R. Nandani, and V.K. Prajapati.